Abstract
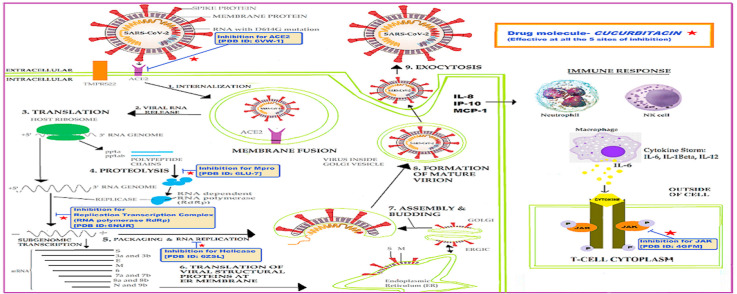
In the year 2019, the potent zoonotic virus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) began to rage globally, which resulted in the World Health Organization (WHO) declaring it as a pandemic on March 11th, 2020. Although extensive research is currently ongoing worldwide to understand the molecular mechanism and disease pathogenicity of SARS-CoV-2, there are still many nuances to elucidate. Therefore, developing an appropriate vaccine or therapeutic drug to combat coronavirus 2019 (COVID-19) is exceedingly challenging. Such scenarios require multifaceted approaches to identify suitable contenders for drugs against COVID-19. In this context, investigating natural compounds found in food, spices, and beverages can lead to the discovery of lead molecules that could be repurposed to treat COVID-19. Sixteen cucurbitacin analogues were investigated for activity against the SARS-CoV-2 main protease protein (Mpro), angiotensin-converting enzyme 2 (ACE2) binding receptor, nonstructural protein 12 (NSP12) RNA-dependent RNA polymerase (RdRp), NSP13 helicase, and Janus kinase 2 (JAK2)/signal transducer and activator of transcription 3 (STAT3) pathway using several relevant tools and simulated screening methods. All key proteins were found to bind efficiently only with cucurbitacin G 2-glucoside and cucurbitacin H with the lowest global energy. Further, the absorption, distribution, metabolism, and excretion (ADME) of all the cucurbitacins were analysed to explore their drug profiles. Cucurbitacin G 2-glucoside and H showed the best hits and all the analogues showed no adverse properties that would diminish their drug-likeness abilities. The encouraging results of the current study may lay the foundation for future research and development of effective measures and preventive medications against SARS-CoV-2.
Keywords: Coronavirus-2019, Cucurbitacin, Molecular docking, ADME/Toxicity analysis
Graphical abstract
1. Introduction
Over the past two decades, humans have grappled with the spread of zoonotic pathogenic strains of many viruses such as the severe acute respiratory syndrome coronavirus (SARS-CoV) in 2002, followed by the Middle East respiratory syndrome-related (MERS)-CoV in 2012 with ~10% and ~36% mortality rates, respectively [1]. Although the fatality rate was lower, the advent of SARS-CoV-2 in 2020 heralded the spread of a highly infectious and pathogenic strain of betacoronavirus causing acute respiratory distress syndrome (ARDS), often leading to lung dysfunction, arrhythmia, and death. To date, there are no marketable antiviral agents or vaccines available against SARS-CoV-2. Developing therapies for the SARS-CoV-2-induced coronavirus disease 2019 (COVID-19) requires the complete elucidation of the pathology and molecular mechanisms underlying the viral mode of infection.
Therefore, it is crucial to understand and identify potential targets for drug design against both SARS-CoV-2 and human proteins, wherever they interact. Searching for biologically promising compounds from natural sources could be an alternate strategy for developing remedies against SARS-CoV-2. Although a few candidate drugs are currently under consideration as effective antivirals for SARS-CoV-2, we believe that the evaluation of small molecule drug candidates from natural compounds could potentially provide durable, broad-spectrum host-directed therapies. Earlier studies have shown that cucurbitacins have remarkable inhibitory activity against the herpes simplex virus (HSV)-1 [2] and, thus, may have the potential to be developed as antiviral drugs against COVID-19.
Traditional belief as well as scientific evidences has proven that Cucurbita pepo, the oldest known cultivated species of the Curcurbiteae family, has immense medicinal and nutritional benefits [3]. Other species of the Curcurbiteae family, which are indigenous to the western hemisphere, include cucumbers, honeydew melons, cantaloupe, watermelons, and zucchini. To date, six cucurbitacins (B, D, E, E glucoside, Iso B, and Iso D) have been isolated from these species. Cucurbitacins are tetracyclic triterpenoids, and both the fruit and seeds of these plants are used as traditional medicines. Pulp with high carotenoid content and low fat content is known to have both beneficial physiological and immunomodulatory functions [4,5].
Pumpkin seeds have nutraceutical properties attributable to the rich content of various elements such as antioxidant vitamins, tocopherols, and carotenoids [6,7]. They are also known to contain proteins, polyunsaturated fats, and phytosterols [8,9]. Recently, cucurbitacins B, E, and D have been shown to exhibit potent antiviral activity against the bovine viral diarrhoea virus (BVDV), hepatitis C virus (HCV) [10], and HSV [11]. In the current unprecedented times of the COVID-19 pandemic, there is an urgent need to explore the effects of natural compounds such as cucurbitacins, and carefully bridging the gap in our understanding of all the interacting proteins of both SARS-CoV-2 and humans would facilitate this process.
Many strategic attempts have been made to interfere with the different steps in the coronavirus replication cycle. Previous outbreaks such as those of SARS-CoV and MERS have trained us to target key enzymes involved in viral replication such as proteases, polymerases, and helicases, as attractive therapeutic approaches [12]. A few antivirals with protease- and polymerase-related mechanisms of action are under investigation to counteract SARS-CoV-2 infection. One example is remdesivir, which is a nucleoside analogue and inhibitor of the RNA-dependent RNA polymerase (RdRp) of coronavirus [13].
Enzyme kinetics have shown that remdesivir triphosphate (RDV-TP) competes with natural ATP with four times more selectivity for position (i), thus, inhibiting RNA synthesis at position (i+5). This process delays chain termination and is a plausible mechanism for drug targeting in the SARS-CoV RdRp complex [[14], [15], [16]]. Thus, in the present study, we compared the cucurbitacin molecule with the RDV to assess its suitability as a target against the SARS-CoV RdRp complex. Cucurbitacin was also examined as a potential lead molecule to target the Mpro of SARS-CoV-2, which is the main enzyme mediating both viral replication and transcription [17]. Using combinatorial techniques of both structure-based virtual and high-throughput screening, innumerable pharmacologically active compounds, natural compound drug candidates, and repurposed drugs have been shown to inhibit Mpro proteases [18].
The various envelope proteins such as S, M, and E on coronaviruses exhibit extensive variability, but the NSPs are quite conserved, including the protein helicase (NSP13), which was first studied in SARS-CoV and is an NTP-dependent 5′ to 3′ direction enzyme that coverts duplex oligonucleotides (RNA or DNA) by catalysing the unwinding into single strands [19,20]. Thus, by targeting ATP-binding or direct NTPase activity, nucleic acids binding to the helicase, and blocking activities such as helicase translocation, inhibitors of NSP13 activity offer a potential therapeutic option against coronaviruses including SAR-CoV-2. Some natural compounds such as benzotriazole, imidazole, imidazodiazepine, phenothiazine, quinoline, anthracycline, triphenylmethane, tropolone, pyrrole, acridone, small peptide, and bananin derivatives are under consideration as potent inhibitors for NSP13 activity. Cucurbitacins have also been used as inhibitors of helicases.
Studies have proven that angiotensin-converting enzyme 2 (ACE2), a metallopeptidase, competently binds the S1 domain of the SARS-CoV “S” protein and mediates entry of their target cells. It has also been shown that the related enzyme ACE1 does not bind to the S1 domain and that the ACE2 receptor is the only functional receptor for SARS-CoV [21,22]. The entry of SARS-CoV-2 S protein is known to be blocked by the anti-ACE-2 antibody [23]. Recently, plasma therapy with infused convalescent antibodies has been used to treat severe COVID-19 patients [24]. We conducted in silico analysis to determine if cucurbitacin disrupts the interaction between the virus and ACE2 receptors and, thus, might be a potential effective therapy for COVID-19.
Finally, we also attempted to study the signalling mechanism mediating the release of pro-inflammatory cytokines in SARS-CoV-2 infection. The release of interleukin (IL)-6, IL-1β, and IL-12 is known to cause cytokine storm, inducing multiple organ failure in patients with acute conditions. These cytokines are released from various innate immune cells (monocytes, neutrophils, and NK cells), which in turn, activate T-lymphocytes via the JAK/STAT pathway [25]. Thus, antagonists of the JAK/STAT pathway may be correlated in reducing the cytokine storm and, thus, saving lives. In the present study, cucurbitacins were also explored as inhibitors of relevant signalling pathways.
2. Molecular modelling methods
2.1. Ligand and protein preparation
Cucurbitacin was selected for screening for activity against SARS-CoV-2, and its three-dimensional (3D) structure was retrieved from PubChem (https://pubchem.ncbi.nlm.nih.gov/) in the SDF format. The 3D structure of cucurbitacin was minimized with retained specified chirality using the default force field OPLS3 of ligprep/maestro and epik to generate the possible state at the default pH. The molecular enzymes of SARS-CoV-2 NSP12 (Protein Databank [PDB] Id 6NUR) [14] with bound cofactor NSP7 and NSP8, the main protease (PDB Id 6LU7), JAK2 (PDB Id 4GFM), ACE2 (PDB Id 6VW1), and NSP13 helicase (6ZSL) were targeted by the selected cucurbitacin to inhibit the viral infection of COVID-19.
These protein structures were retrieved from the PDB (http://www.rcsb.org/pdb) and prepared from protein preparation using the wizard function of Maestro 12.4 in Schrodinger 2020–2. The 3D structures of the proteins were pre-processed by choosing the default option and filling the missing side chains and loops with prime. Further, the structures were modified by removing hets/water within 5 Å, and finally, refined by assigning the H-bonds, removing water within 3 Å, and performing retrained minimization by choosing the OPLS3 force field.
2.2. Sitemap analysis
The protein-binding site was constructed using the standard default parameter setting of the sitemap maestro suite. The sitemap also facilitated the characterization of hydrophobic, hydrophilic, hydrogen donor, and hydrogen acceptor residues in the binding site. The top-ranked potential binding sites were identified, and the best predicted binding site was chosen based on a Dscore value > 1.
2.3. Receptor grid generation and ligand docking
The receptor grid was generated by using default parameter settings from maestro suite. Predicted sitemap binding sites were used for receptor grid generation, and further predicted receptor grids were used for ligand docking. Docking calculations were performed using the standard default parameter setting of the ligand-docking task of Maestro in which Cucurbitacin was docked into the predicted receptor grid with extra precision along with XP descriptor information. The ligand sampling was kept flexible while the proteins were considered as rigid structures and epik state penalties were applied. Finally, for the output file, a pose viewer file was chosen and post-docking minimization was performed. The best dock score was identified as a “hit”. Pymol 2.4.0 was also used for visualisation and figure generation.
2.4. Drug disposition analysis of top cucurbitacins as potent drug candidate
Drug performance and pharmacological efficacy are critically influence by four major parameters: absorption, distribution, metabolism, and excretion (ADME). Prior knowledge of the ADME and toxicological (Tox) parameters of drugs enables the control of their pharmacological activity and pharmacokinetics. Thus, pharmacology and effectiveness are mainly measured through the analysis of factors that influence the kinetics of drug doses and contact with the tissues in an organism. In this study, we used the qikprop function of maestro 12.4 to determine the ADME/Tox properties of all 16 of the cucurbitacin analogues. The server carefully predicts the toxicity endpoints by not only predicting the 2D similarity to compounds with known median lethal doses (LD50), but also by drawing parallels on fragment and molecular similarity and fragment tendency.
3. Results
3.1. Screening of cucurbitacins against various functional cellular machinery of SARS-CoV-2
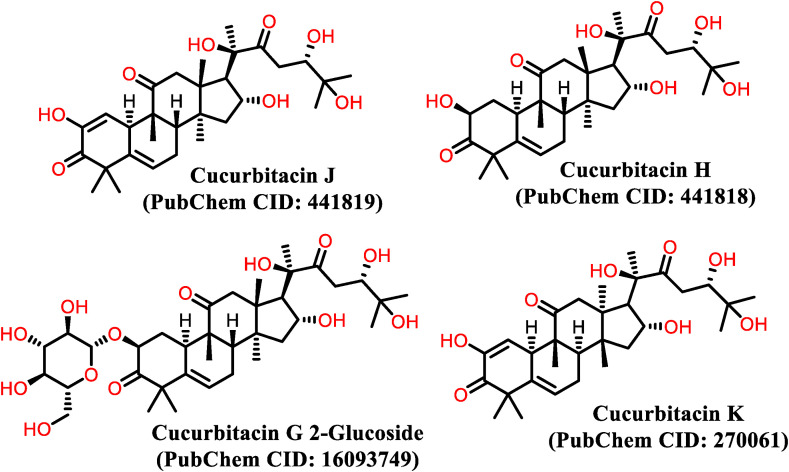
Considering the already known therapeutic activities of cucurbitacins and using XP docking methods, we identified all known cucurbitacins with structures that could be used to inhibit multiple targets and steps in the life cycle of coronaviruses (Table 1 ). In the present study, we selected 16 of the best cucurbitacins with known structural configurations designated as A, B, C, D, E, F, G, H, I, J, K, L, O, P, Q, R, and S (Fig. 1 ). Based on binding affinity and structure-based potential, we virtually screened using an effective procedure to identify ligand hits and conduct lead optimization for cucurbitacins as drug candidates. We docked 16 potential candidates and arranged a congeneric series for binding affinity prediction using methods such as macrocycle pose prediction by engaging polypeptides, predicting protein-ligand complex geometries, and free energy perturbation or MM-GBSA.
Table 1.
XP docking of cucurbitacins with Nsp12 RdRp, Nsp13 helicase, Mpro, ACE2, JAK2 of Covid-19.
| Entry | Molecules | RdRp |
Mpro |
ACE2 |
JAK2 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dock Score (kcal/mol) |
Emodel | H-Bond (kcal/mol) | Dock Score (kcal/mol) | Emodel | H-Bond (kcal/mol) | Dock Score (kcal/mol) | Emodel | H-Bond (kcal/mol) | Dock Score (kcal/mol) |
Emodel | H-Bond (kcal/mol) | ||
| 1. | Reference Ligands | Remdesivir (−8.63) | – | – | Ulinastain (−3.45) | – | – | HQC (−5.43) | – | – | Baricitinib (−6.49) | – | – |
| 2. | Cucurbitacin K | −9.57 | −66.33 | −6.21 | −7.95 | −60.21 | −4.31 | −9.61 | −85.69 | −5.53 | −6.87 | −56.70 | −3.84 |
| 3. | Cucurbitacin G 2 glucoside |
−8.17 | −64.76 | −4.05 | −9.43 | −63.79 | −4.86 | −9.92 | −83.71 | −6.52 | −8.06 | −65.59 | −5.26 |
| 4. | Cucurbitacin J | −8.06 | −75.16 | −4.38 | −6.93 | −60.64 | −3.95 | −9.01 | −82.11 | −4.7 | −4.33 | −44.14 | −2.38 |
| 5. | Cucurbitacin H | −7.5 | −75.65 | −3.73 | −7.72 | −64.51 | −3.48 | −9.87 | −85.60 | −5.49 | −5.7 | −29.93 | −3.2 |
| 6. | Cucurbitacin F | −6.83 | −62.16 | −3.84 | −5.46 | −49.69 | −2.61 | −8.12 | −69.69 | −3.83 | −4.56 | −38.63 | −2.76 |
| 7. | Cucurbitacin P | −6.66 | −66.24 | −2.8 | −7.04 | −46.87 | −3.16 | −7.62 | −60.79 | −4.28 | −4.8 | −37.44 | −2.3 |
| 8. | Cucurbitacin O | −6.36 | −70.83 | −2.78 | −5 | −54.70 | −1.57 | −7.65 | −73.40 | −3.64 | −3.55 | −48.13 | −2.1 |
| 9. | Cucurbitacin Q | −6.22 | −73.38 | −2.89 | −5.68 | −53.48 | −2.4 | −8.33 | −77.88 | −4.05 | −3.9 | −54.48 | −0.96 |
| 10 | Cucurbitacin D | −6.17 | −64.44 | −2.75 | −6.24 | −54.66 | −2.38 | −8.45 | −74.82 | −3.99 | −4.58 | −40.99 | −2.18 |
| 11. | Cucurbitacin E | −6.15 | −65.79 | −2.63 | −5.92 | −59.22 | −2.53 | −5.76 | −83.72 | −2.54 | −3.69 | −45.52 | −1.35 |
| 12. | Cucurbitacin B | −6.1 | −74.02 | −2.55 | −4.93 | −53.73 | −1.44 | −6.3 | −81.26 | −2.83 | −4.31 | −43.36 | −1.55 |
| 13. | Cucurbitacin A | −5.92 | −56.16 | −2.88 | −6.82 | −52.83 | −2.88 | −8.27 | −77.03 | −3.61 | −3.97 | −41.34 | −2.67 |
| 14 | Cucurbitacin L | −5.88 | −62.38 | −2.76 | −7.8 | −47.45 | −3.51 | −7.9 | −64.56 | −3.8 | −3.78 | −30.31 | −2.4 |
| 15. | Cucurbitacin C | −5.66 | −60.96 | −2.49 | −4.69 | −47.41 | −1.66 | −5.54 | −63.35 | −2.7 | −4.84 | −37.58 | −1.96 |
| 16. | Cucurbitacin S | −5.38 | −50.31 | −2.17 | −4.04 | −44.21 | −1.61 | −5.14 | −55.97 | −1.92 | −3.51 | −32.03 | −2.08 |
| 17. | Cucurbitacin I | −5.19 | −61.09 | −2.4 | −5.59 | −52.39 | −2.92 | −7.83 | −72.15 | −3.48 | −6.16 | −40.17 | −4.07 |
Fig. 1.
Structure of best hit cucurbitacins for SARS-CoV-2.
Consequently, we identified key molecules likely to bind well to the cucurbitacins. Thus, based on structure-based virtual screening and the induced fit docking protocols for the most effective ligand docking, we determined the most important functional cellular machinery for SARs-CoV-2 and host interaction. These were the SARS-CoV-2 polymerase, NSP12 (known for direct supply of positive sense RNA, which increases the rate of protein synthesis); the active viral protease, Mpro; COVID-19 protein, NSP13 helicase (known for repairing cellular components) and the host's ACE2 receptor (on the epithelial and Vero E6 cells that open door for entry of coronavirus); and the host signalling machinery, JAK2 (activates T-lymphocytes via JAK/STAT pathway.
3.2. Cucurbitacins inhibit JAK2 signalling and SARS-CoV-2 Mpro
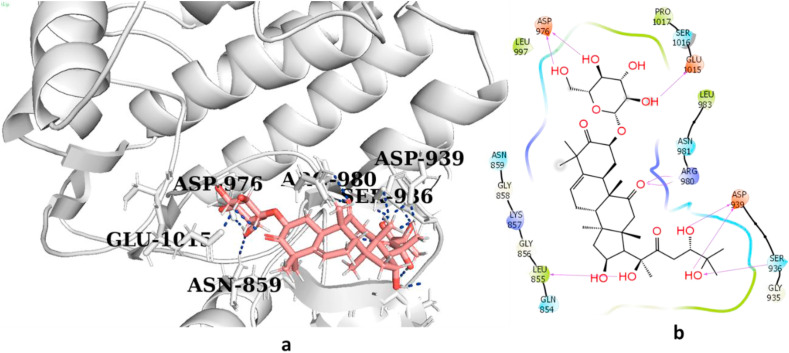
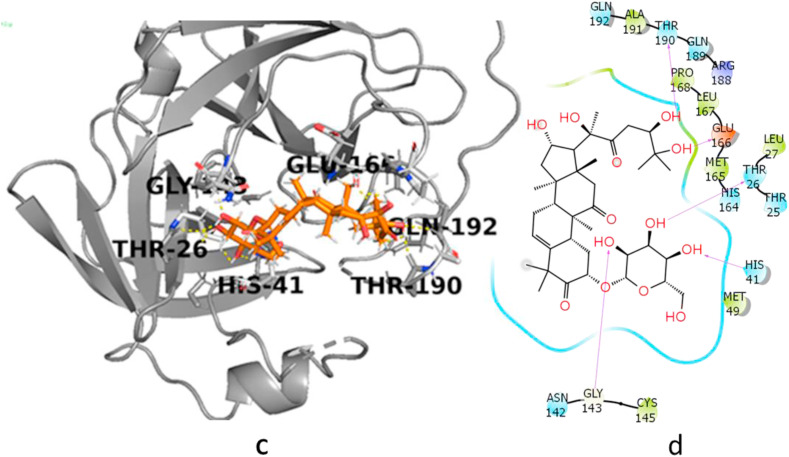
The sitemap showed that when all the selected cucurbitacins were bound to the kinase domain of JAK2, the binding mode and best pose (Emodel) of cucurbitacin G 2-glucoside were mostly similar to those of the other cucurbitacins (Fig. 2 a). The only difference was observed in the H-bond energy of G, which was slightly higher than that of the remaining cucurbitacins. Further, G formed hydrogen bonds with the Asp 976, Glu 1015, ARG 980, Asp 939, Ser 936, and Leu 855 residues with an excellent binding affinity (−8.06 kcal/mol). These results predicted that this molecule could be used as an inhibitor of SARS-CoV-2 infection. Similarly, among all the analogues, cucurbitacin G 2-glucoside with a Dscore of −9.43 kcal/mol (Fig. 2b) showed the best hit and appeared to be the most potent inhibitor of CoV- 2 Mpro protein. Thus, cucurbitacins are also active against the target Mpro of SARS-CoV-2.
Fig. 2a.
(a,b): 3D and 2D interaction diagram of cucurbitacin G 2-glucoside with high afinity for JAK2. (a, b: 4GFM_G); signalling forming hydrogen bonds with residues Ser 936, Arg980, Asp939, Leu855, Glu1015 and Asp976 and a hydrogen bond energy of (−9.43 kcal/mol).
Fig. 2b.
(c, d): 3D and 2D interaction diagram of cucurbitacin G 2-glucoside with main protease Mpro (c, d: 6LU7_G) with a Dscore of −9.43 kcal/mol.
3.3. Cucurbitacins inhibit ACE2 receptor
The docking of all cucurbitacins showed very good interaction with the RBD of ACE2 and spike (S1) glycoproteins of CoV-2 with a similar interaction profile. This was further corroborated by the sitemap, which determined that the RBD of ACE2 and spike glycoproteins had Dscore >1. In particular, cucurbitacin G 2-glucoside showed the best binding affinity for ACE2 and formed hydrogen bonds with Tyr510, Tyr516, Glu402, Asn51, and His378 residues (Fig. 2c) with hydrogen bond energy of −9.92 kcal/mol, which was much better than that the interaction of other reference molecules to date, including hydroxychloroquine.
Fig. 2c.
(e, f): 3D and 2D interaction diagram of cucurbitacin G 2-glucoside with ACE2 (e, f: 6VW1_G) with H-Bonding at Tyr510, Tyr516, Glu402, Asn 51 and His378 with hydrogen bond energy (−9.92 kcal/mol).
3.4. Cucurbitacins inhibit NSP12, SARS-CoV-2 RdRp
The binding mode and best pose (Emodel) of cucurbitacin G 2-glucoside was slightly higher than that of the remaining cucurbitacins. However, for RdRp, cucurbitacin-K formed hydrogen bonds with Phe396, Thr394, Hip256, and Ala252 residues with binding affinity −9.57 kcal/mol (Fig. 2d), whereas remdesivir was reported to form a hydrogen bond with binding affinity of −5.91 kcal/mol (Table 1). Cucurbitacins are triterpenes that may potentially bind to the allosteric site of RdRp and probably provide key acuities to NSP12 polymerase catalysis and reliability that may act as a template for the design of novel antiviral therapeutics.
Fig. 2d.
(g, h): 3D and 2D interaction diagram of cucurbitacin-K with RdRp (g, h: 6NUR_G) forming hydrogen bonds with residue Phe396, Thr394, Hip256 and Ala252 with binding affinity (−9.57 kcal/mol).
3.5. Cucurbitacins inhibit NSP13, the SARS-CoV helicase
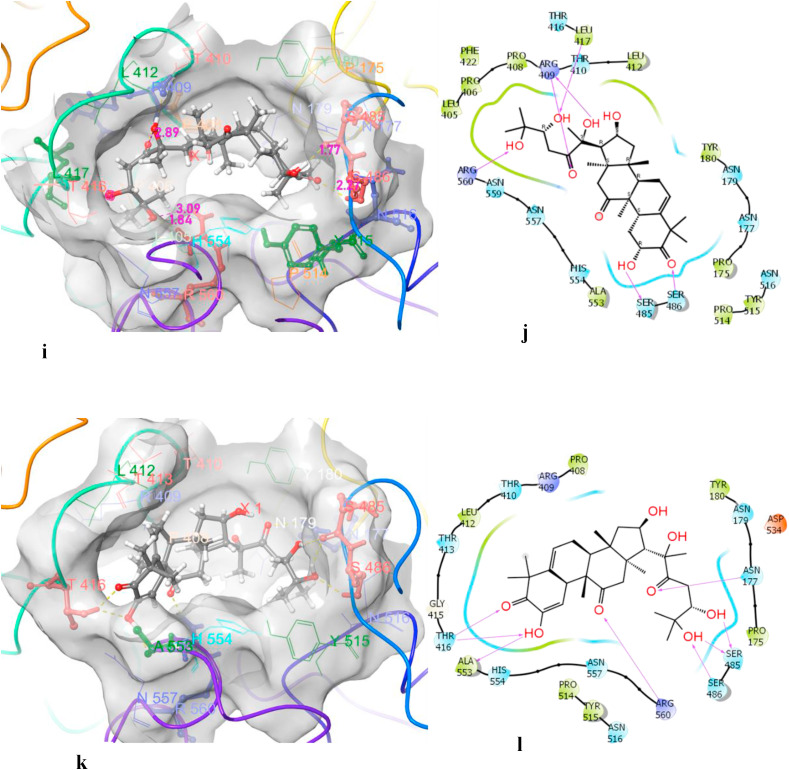
The sitemap was used to determine the possible nucleic acid binding site (Dscore = 1.07), which is sandwiched between RecA1 (241–443 amino acids [aa]) and RecA2 (444–596 aa) domain of the NSP13-helicase of COVID-19. In the nucleic acid binding site, Arg560 from motif IV was the only residue that might interact with the base and sugar moiety of the single strand DNA (ssDNA). Docking of the cucurbitacins led to the identification of the best possible hit for the NSP13 helicase of COVID-19 (Table 2 ). Our study clearly showed that one of the hydroxy groups of cucurbitacin H and cucurbitacin J interacted very strongly with Arg460 and revealed the best hit with a binding affinity of −10.069 kcal/mol (Fig. 3 ).
Table 2.
XP docking of 16 cucurbitacin in Nsp13 helicase.
| Entry | Compounds | docking score (kcal/mol) | Emodel | H-bond |
|---|---|---|---|---|
| 1. | MLS001181552 | −3.33 | −37.13 | −3.398 |
| 2. | Cucurbitacin J | −10.609 | −76.482 | −5.544 |
| 3. | Cucurbitacin H | −10.069 | −70.843 | −3.542 |
| 4. | Cucurbitacin O | −9.28 | −82.202 | −3.541 |
| 5. | Cucurbitacin P | −9.157 | −79.738 | −4.658 |
| 6. | Cucurbitacin F | −9.117 | −85.093 | −3.622 |
| 7. | Cucurbitacin C | −8.563 | −60.689 | −3.697 |
| 8 | Cucurbitacin A | −8.771 | −69.86 | −3.697 |
| 9. | Cucurbitacin E | −8.177 | −74.121 | −2.092 |
| 10. | Cucurbitacin I | −8.104 | −67.829 | −2.505 |
| 11. | Cucurbitacin Q | −7.877 | −83.515 | −3.185 |
| 12. | Cucurbitacin G 2-glucose |
−7.753 | −66.985 | −3.739 |
| 13. | Cucurbitacin K | −7.736 | −73.123 | −3.398 |
| 14. | Cucurbitacin B | −7.62 | −75.716 | −23.44 |
| 15. | Cucurbitacin L | −7.542 | −67.26 | −0.608 |
| 16. | Cucurbitacin D | −6.532 | −70.707 | −2.992 |
| 17. | Cucurbitacin S | −4.73 | −59.37 | 0 |
Fig. 3.
3D and 2D interaction diagram of cucurbitacin J (i, j) & H (k, l) with nsp13 helicase showing that one of hydroxy of cucurbitacin J and cucurbitacin H interacted very strongly with Arg 560 and revealed best hit with binding affinity of (10.609 kcal/mol) and (−10.069 kcal/mol) respectively.
3.6. Analysis of ADME data
The drug likeness of all 16 cucurbitacins was estimated using different ADME-related properties, namely physicochemical parameters, pharmacokinetics, lipophilicity, water solubility, and their medicinal chemistry. Analysis of the inhibitory effects of different cucurbitacins predicted that cucurbitacin G 2-glucoside and H are good candidates for antiviral drugs. The fast infection rate of pathogenic diseases has motivated drug discovery for these conditions, and recent technological advancements have increased the rate of drug discovery with prior knowledge of the drug candidate. We determined the anti-COVID-19 activity and drug-likeness of selected cucurbitacins by performing in-silico ADME/Tox calculations (Table 3 ).
Table 3.
Drug profile and ADME/toxicity analysis of all 16 analogues of cucurbitacins.
| Molecular Descriptor | Cucurbitacin G 2-glucside | Cucurbitacin H | Cucurbitacin J | Cucurbitacin K | Range | |
|---|---|---|---|---|---|---|
|
Physicochemical Parameters | ||||||
| FOSA (Å2) | 593.442 | 490.241 | 508.961 | 603.806 | 0.0–750.0 | |
| FISA (Å2) | 341.267 | 257.244 | 239.066 | 321.628 | 7.0–330.0 | |
| PISA (Å2) | 13.093 | 13.122 | 29.498 | 15.896 | 0.0–450.0 | |
| SASA (Å2) | 947.802 | 760.607 | 777.525 | 941.33 | 300.0–1000.0 | |
| #rotor | 16 | 10 | 10 | 16 | 0–15 | |
| QPpolrz | 62.601 | 49.792 | 50.491 | 61.691 | ||
| #rtvFG | 1 | 0 | 0 | 1 | 1–2 | |
| mol_MW (g/mol) | 696.831 | 534.689 | 532.673 | 534.689 | 130.0–750.0 | |
| dipole | 5.395 | 5.658 | 7.999 | 8.697 | 1.0–12.5 | |
| Volume (Å3) | 1971.601 | 1555.272 | 1568.832 | 1948.185 | 500.0–2000.0 | |
| Donor HB | 7 | 4 | 4 | 7 | 0.0–6.0 | |
| acceptor HB | 20.1 | 11.6 | 10.65 | 19.15 | 2.0–20.0 | |
| PSA (Å2) | 241.738 | 164.728 | 163.083 | 247.699 | 7.0–200.0 | |
| Rule Of Five | 3 | 1 | 1 | 3 | Maximum 4 | |
| Rule Of Three | 2 | 1 | 1 | 2 | Maximum 3 | |
| EA (eV) | −0.036 | −0.139 | 0.381 | 0.395 | −0.9–1.7 | |
| IP (eV | 9.694 | 10.018 | 9.537 | 9.884 | 7.9–10.5 | |
| Lipophilicity | ||||||
| QPlogPw | 30.932 | 18.467 | 17.56 | 29.866 | 4.0–45.0 | |
| QPlogPo/w | 0.412 | 2.181 | 2.739 | 0.76 | −2.0–6.5 | |
| QPlogS | −3.846 | −4.372 | −4.879 | −3.993 | −6.5–0.5 | |
| CIQPlogS | −5.191 | −5.465 | −5.909 | −5.635 | −6.5–0.5 | |
| QPlogPoct | 42.865 | 29.325 | 29.215 | 42.252 | 8.0–35.0 | |
| QPlogPC16 | 20.259 | 15.258 | 15.4 | 19.958 | 4.0–18.0 | |
| ACxDN∧.5/SA | 0.056108 | 0.030502 | 0.027395 | 0.053824 | 0.0–0.005 | |
| Pharmacokinetics | ||||||
| QPlogHERG | −4.794 | −3.965 | −4.265 | −4.822 | Concern below −8 | |
| QPPCaco (nm/sec) | 5.75 | 36.013 | 53.559 | 8.829 | <25 poor, >500 great | |
| QPlogBB | −3.936 | −2.393 | −2.299 | −3.74 | −3.0–1.2 | |
| QPlogKp | −6.227 | −5.254 | −4.861 | −5.855 | −8.0–−1.0 | |
| Human Oral Absorption | 1 | 2 | 2 | 1 | 1 = low, 2 = medium | |
| %Human Oral Absorption | 4.08 | 54.614 | 60.968 | 9.45 | <25% is poor >80% is high |
|
| CNS | −2 | −2 | −2 | −2 | −2 - +2 | |
| QPPMDCK | 1.874 | 13.616 | 20.911 | 2.979 | <25 poor, >500 great | |
| QPlogKhsa | −0.574 | 0.189 | 0.348 | −0.487 | −1.5–1.5 | |
| #metab | 13 | 9 | 9 | 12 | 1–8 | |
SASA: Total solvent accessible surface area; FOSA: Hydrophobic component of the SASA; FISA: Hydrophilic component of the SASA; PISA: π (carbon and attached hydrogen) component of the SASA; #rotor: Number of non-trivial (not CX3), non-hindered (not alkene, amide, small ring) rotatable bonds; #rtvFG: Number of reactive functional groups; PSA: Van der Waals surface area of polar nitrogen and oxygen atoms; IP(ev): PM3 calculated ionization potential; EA(eV): PM3 calculated electron affinity; QPlogPw: Predicted water/gas partition coefficient; QPlogPo/w: Predicted octanol/water partition coefficient; QPlogS: Predicted aqueous solubility; CIQPlogS: Conformation-independent predicted aqueous solubility; QPlogPoct: Predicted octanol/gas partition coefficient; ACxDN∧.5/SA: Index of cohesive interaction in solids; QPlogHERG: Predicted IC50 value for blockage of HERG K+ channels; QPPCaco: Predicted apparent Caco-2 cell permeability; QPlogBB: Predicted brain/blood partition coefficient; QPlogKp: Predicted skin permeability; CNS: Predicted central nervous system activity; QPPMDCK: Predicted apparent MDCK cell permeability; QPlogKhsa: Prediction of binding to human serum albumin; #metab: Number of likely metabolic reactions.
For cucurbitacin H, the solvent accessible surface area (SASA), predicted water–gas partition coefficient (QPlogPw), octanol/water partition coefficient (QPlogPo/w), aqueous solubility (QPlogS), half-maximal inhibitory concentration (IC50) value for blockade of the human ether-a-go-go-related gene (HERG) K+ channels (QPlogHERG), apparent Caco-2 cell permeability (QPPCaco), brain/blood partition coefficient (QPlogBB), skin permeability (QPlogKp), percentage human oral absorption, and Lipinski rule of five (max. 4) values are 760.60, 18.46, 2.18, −4.37, −3.96, 36.01, −2.39, −5.25, 54.61%, and 1 respectively.
For cucurbitacin H, which was identified as a hit for Mpro, the ACE2, JAK2, and RdRp, the SASA, QPlogPw, QPlogPo/w, QPlogS, IC50 of QPlogHERG, QPPCaco, QPlogBB, QPlogKp, percentage human oral absorption, and Lipinski's rule of five (max. 4) values are 947.80, 30.93, 0.41, −3.84, −4.79, 5.75, −3.93, −6.22, 4.08%, and 3, respectively. These results suggest that both cucurbitacin G 2-glucoside and H are promising candidates for future drug development, and further in vivo assays may prove their utility.
4. Discussion
Reports by experts had already predicted in 2018 that a flu pandemic was only a matter of time and there could be numerous undiscovered viruses that could kill millions worldwide. In the past, we witnessed MERS and SARS and there is no shortage of articles regarding their mechanism of action. Extensive research are now been followed to understand the molecular mechanism and disease pathogenicity of SARS-CoV-2, but still developing an appropriate vaccine or therapeutic drug to combat coronavirus 2019 (COVID-19) is a challenging task. A multifaceted approach is required to rein this virus and we believed that the natural compounds found in food, spices, and beverages can lead to the discovery of lead molecules that could be repurposed to treat COVID-19. In our present study, we investigated sixteen cucurbitacin analogues for activity against the SARS-CoV-2 main protease protein (Mpro), angiotensin-converting enzyme 2 (ACE2) binding receptor, nonstructural protein 12 (NSP12) RNA-dependent RNA polymerase (RdRp), NSP13 helicase, and Janus kinase 2 (JAK2)/signal transducer and activator of transcription 3 (STAT3) pathway using several relevant tools and simulated screening methods. We thus, could identify the host ACE2 receptor, CoV-2 RdRp polymerase, CoV-2 Mpro protease, CoV-2-helicase protein, and JAK2 signalling machinery as the best hits for specific cucurbitacins. All key proteins were found to bind efficiently only with cucurbitacin G 2-glucoside and cucurbitacin H with the lowest global energy. Also, the absorption, distribution, metabolism, and excretion (ADME) of all the cucurbitacins showed that Cucurbitacin G 2-glucoside and H showed the best hits and all the analogues showed no adverse properties that would diminish their drug-likeness abilities.
Plant-derived natural compounds have played an essential role in disease therapy. Cucurbitacins are triterpenes with various chemical structures found in the members of the Cucurbitaceae and numerous other plant families, such as Trichosanthes, Cucurbita, Cucumis, and Citrullus, which possess numerous medicinal properties [26]. Cucurbitacin B has been shown to inhibit HSV-1 and, thus, holds promise in the development of antiviral drugs [2]. The current COVID-19 pandemic has precipitated an urgent need for scientific strategies to repurpose already known lead compounds to develop possible therapies with activity against the SARS-CoV-2 virus.
Although cucurbitacins are known toxic compounds, they have been shown to possess various bioactivities and pharmacological efficiency against cancer, inflammation, diabetes, and atherosclerosis. There have been instances of severe poisoning in cattle that consumed bitter fruits of the Cucumis and Cucurbita species. The dose range of the toxicity of cucurbitacins based on the few in vivo toxicity reports has been reported to be between 2 and 12.5 mg/kg. Double bonds at C-23 and the acetyl group at C-25 have been found to modulate the toxicity of cucurbitacins, and their bioactive and toxic dose have been found to be close [3]. Among the various bioactivities shown by cucurbitacins, plant species such as Momordica are held in high esteem in many countries as traditional medicine for certain metabolic diseases such as diabetes [27].
The antidiabetic property of cucurbitacins is mediated by activation of the AMP-activated protein kinase (AMPK) pathway (a major regulatory pathway for GLUT4 translocation) [28,29]. These compounds also show anti-atherosclerosis by inhibiting the generation of lipid oxidation products such as malonaldehyde (MAD) and 4-hydoxynonenal (4-HNE) [3]. Cucurbitacin D improves blood circulation through inhibition of Na+/K + -ATPase [30]. The concentration of cucurbitacin C in the leaves of plants has also been reported to be an important parameter in the spider mite resistance of C. sativus, perhaps by acting as an antagonist of the spider mite ecdysteroid receptor [31]. However, the most promising therapeutic properties of cucurbitacins are their anticancer [32,33] and anti-inflammatory [34, 35] activities.
The diverse pharmacological activity of cucurbitacins is due to the variety of their side chain derivatives [36,37], which can be used to group these compounds and their derivatives into 12 main categories [33]. There are 16 main molecules designated cucurbitacin A to T, and hundreds of derivatives of the parent molecule. Chemically, cucurbitacins are derived from the basic cucurbitane ring skeleton, a triterpene hydrocarbon (International Union of Pure and Applied Chemistry [IUPAC] name: 19 [10-9β]-abeo-5α-lanostane), which is modified by groups containing oxygen and double bonds to produce numerous cucurbitacins with distinctive characteristics [38]. Pure cucurbitacins and their glycosidic forms have been isolated from plant tissues using non-polar solvents to remove undesired components [39]. Most cucurbitacins are soluble in non-polar solvents, but are insoluble in ether, whereas they are only slightly soluble in water. The majority of cucurbitacins are usually crystalline or exist as needle-shaped particles at 68–77 °F (20–25 °C) except for cucurbitacin H, which is an amorphous solid. Most cucurbitacins are derived as 2-O-glucoside, except for cucurbitacin D, which lacks the acetyl group at 25-OH, but is the most ubiquitous cucurbitacin isolated. Cucurbitacin G 2-glucoside and H have the same structures but differ in the configuration of their hydroxyl groups at position C-24, which has not been established [33]. Cucurbitacin R was identified to be 23,24-dihydrocucurbitacin D (DHCD), hence, has been moved to the cucurbitacin D group based on this description [40]. Similarly, Cucurbitacin J and K differ only in the configuration of their hydroxyl groups at position 24 and their chemical validation is still pending [41].
The earliest studies during the onset of the pandemic were conducted to understand whether the new SARS-CoV-2 binds to the host cells in a similar manner to that of the known SARS-CoV. Stefan Pohlmann and his team were among the first to discover that both viruses used ACE2 surface receptors to penetrate cells and the same protease to become infectious [42]. Binding of the CoV-2 homotrimeric spike glycoprotein (comprising a S1 subunit and S2 subunit in each spike monomer) to the host ACE2 receptor is the first critical step in the entry of SARS-CoV into target cells [[43], [44], [45]]. In addition to the lungs, ACE2 receptors are expressed in many extrapulmonary tissues, including the heart, kidney, endothelium, and intestine [[46], [47], [48], [49], [50]]. The receptor-binding domain (RBD) of the S1 subunit is responsible for the interaction between the CoV spike and ACE2 receptor [49]. Structural analysis of various spike residues has identified highly conserved or identical side chain residues in the SARS-CoV-2 RBD to those of the SARS-CoV RBD, suggesting convergent evolution and improved binding to the ACE2 receptor [51]. Notably, ACE2 is widely expressed on the luminal surface of intestinal epithelial cells, functioning as a co-receptor for nutrient uptake, particularly for amino acid resorption from food [52]. Therefore, the intestine could be a potent entry point for CoV-2, which gave us the idea of using oral formulations of natural products such as cucurbitacins as antagonist of the ACE2 receptor. Docking results showed that most of the selected cucurbitacins interacted well with the RBD of ACE2 and S1 glycoprotein of CoV2, thus, providing evidence supporting its promising role as a target site.
In the last decade, the world witnessed the emergence of numerous coronavirus infections, such as SARS and MERS, which showed that SARS-CoVs possess the largest RNA genomes and require fidelity for the RNA synthesis complex to replicate. The new emerging viruses of diverse CoV families require innovative research approaches to generate antiviral strategies that target conserved components of the viral life cycle. This includes strategies mainly focusing on the viral machinery responsible for the replication and transcription of the positive-strand viral RNA genome. NSP12 is a complex RNA synthesis machinery of CoV formed as a product of the ORF1a and ORF1ab viral polyproteins [53]. Sequence conservation and homology modelling studies have revealed mostly highly conserved surfaces on the NSP12 CoV family of viruses at the template entry, template-primer exit, NTP tunnels and the polymerase-active site [14]. Interestingly, SARS-CoV NSP12 contains two metal-binding sites with two zinc atoms, which are distal to known active sites as well as protein–protein and protein–RNA interactions. Coronavirus NSPs usually have bound zinc atoms in NSP3, NSP10, NSP13, and NSP14 that point to the extensive utilisation of zinc ions for folding proteins of the viral replication complex [[54], [55], [56]]. To date, the nucleotide-based prodrug “remdesivir” is the only known inhibitor of COVID-19, which inhibits the NSP12 CoV-RdRp by creating a covalent bond between the 5′hydroxy of remdesivir and 3′hydroxy of the RNA ribose in the RNA-binding site [57]. Cucurbitacin K forms a high-affinity hydrogen bond with NSP12 polymerase, thus, targeting these NSPs may hold the key to blocking viral replication.
In continuation of experiments targeting viral NSPs, subsequent studies in SARS/MERS did not clearly specify the function of NSPs such as NSP13 or helicases. Based on in silico modelling and virtual screening, Mirza and Froeyen [58] predicted a similarity in the amino acid/protein profile of SARS-CoV-2 NSP13 to those of SARS- and MERS-NSP13. NSP13 was thought to probably interact with other NSPs such as NSP7, NSP8, and NSP12 to contribute to viral replication [59,60]. Adedeji et al. [61] demonstrated that the helicase activity of NSP13 is enriched in the presence of RdRp NSP12, suggesting that these proteins interact in the functional replication complex and that the protein–protein interaction contributes to the efficiency of viral replication. SARS-CoV NSP13 catalyses the NTP-dependent 5′–3′ unwinding reaction and converts double-stranded oligonucleotides into single strands [19,20]. It has been proposed that molecules with the potency to inhibit putative ATP binding sites associated with NTPase activity could be used for future therapeutics. NTPase active site residues of SARS-CoV-2, including Lys288, Ser289, Asp374, Glu375, Gln404, and Arg567 are conserved throughout the coronavirus family. Thus, many natural compounds that inhibit ATP hydrolysis could be considered potential inhibitors of SARS-CoV2 replication. The present study also showed that one of the hydroxy groups of cucurbitacins H and J interacted very strongly with Arg460 and, thus, these compounds have the potential to inhibit SARS-CoV2.
SARS-CoV-2 replication and transcription requires encoding of two overlapping polyproteins (pp1a and pp1ab), which undergo extensive autolytic cleavage and proteolytic processing at 11 conserved sites through the action of 33.8 kDa Mpro (also known as 3C-like protease) to release from the polyproteins. The 3CL protease of coronaviruses facilitates viral assembly by cleaving polyproteins and this functional importance of Mpro in the viral life cycle makes it an attractive target for antiviral drug design [18,62]. The crystal structure of the Mpro of SARS-CoV-2 is highly similar to that of SARS-CoV Mpro, with only a 0.53 Å r.m.s difference between the two free enzymes [63]. In our study, cucurbitacin G showed potent inhibitory activity against the CoV2 Mpro protein.
Viral entry into the host normally activated inflammatory pathways, creating an anti-viral environment to stop the virus infection pathogenesis; however, an aberrant or uncontrolled host immune response may cause host cell and tissue damage [64]. The severity of COVID-19 infection is due to an exaggerated inflammatory response with the release of a large amount of pro-inflammatory cytokines. Most studies of SARS-CoV-2 have related to this cytokine storm to lung injury and multiple organ failure, which is activated via the JAK/STAT pathway [[65], [66], [67], [68], [69]]. Phosphorylation of JAK signalling molecules activates STAT proteins, which induces a cascade of cytokines [70,71]. Cucurbitacin B was reported to be an anti-proliferative agent in pancreatic cancer by inhibiting the activation of JAK2, STAT3, and STAT5 components [72]. Suppression of the JAK/STAT pathway increased the expression of p21WAF1 independently of p53 activity, thus, inducing cell cycle arrest. This mechanism could be exploited to stop the proliferation and recruitment of T-lymphocytes, thereby inhibiting cytokine production. The mechanisms of the anticancer activity of cucurbitacins are possibly mediated by inhibition of the JAK/STAT3 signalling pathway [73,74]. Interestingly, the same pathway is used by coronaviruses to activate T-lymphocytes at the later stage of infection, which leads to cytokine storm and subsequent mortality of the infected person. Moreover, cucurbitacins are also used as immunosuppressants, and they have been shown to inhibit the expression of the surface markers CD69 and CD25 required for the activation of T lymphocytes [75]. In the present study, we attempted to show that cucurbitacins can inhibit the JAK/STAT3 pathway and, thus, could inhibit SARS-CoV2 infection. Intriguingly, cucurbitacins have been recognised to possess anti-inflammatory activity and inhibit the expression of TNF and proinflammatory mediators such as nitric-oxide (NO) synthase-2 and cyclooxygenase-2-mediated inhibition of NO generation [34,35]. A similar strategy could be employed to curb the surging production of cytokines in SARS-CoV-2 infection, where cucurbitacins could be a critical source of future anti-inflammatory drugs for severe viral infections.
5. Conclusion
The past two decades have seen a surge in the emergence of novel viral diseases such as the global incidence of viral infections caused by SARS-CoV and MERS, which belong to the coronavirus family. In addition, repeated outbreaks of other coronaviruses have affected the human population such as the 229E, HKU1, NL63, and OC43. Incidentally, the current SARS-CoV-2 has an 80% nucleotide identity with the previous infective viruses, and there is a fairly good understanding of effective therapies against this family of viruses. The information acquired from investigations of these coronaviruses suggest that a promising therapeutic strategy would be to target the fairly conserved key enzymes involved in the replication cycle of the virus. This suggests that known therapies that target these enzymes, even in closely related coronavirus species, could be used in SARS-CoV-2 infection. Proteases and polymerases of SARS-CoV2 as targets for antiviral drugs, have gained attention in recent weeks in clinical trial of drugs such as indinavir, saquinavir, lopinavir/ritonavir, and remdesivir for COVID-19 with. In contrast, helicases have not been explored as much as the proteases and polymerases for this purpose. Moreover, the structural proteins of coronaviruses such as spike (S) proteins are known to show considerable variability among coronavirus species, and targeting the spike proteins has been an extremely laborious task. Key NSPs such as the helicase NSP13 have shown similar homology to helicases of other coronavirus species. Our present perspective is to present the potential application of helicase as one of the target enzymes that could be inhibited using natural compounds. Moreover, currently, there are very limited known studies of the use of natural compounds as repurposed drugs with antiviral activity administered as a single native molecule or in combination therapy. There are multiple steps in the life cycle of a virus and in the present study, cucurbitacin analogues were targeted against the virus at every possible process, such as attachment, entry, replication, morphogenesis, and exit from the host cell. The results suggest that the natural molecule cucurbitacin has potential as a therapeutic drug against COVID-19, especially considering the very encouraging outcome observed with cucurbitacin G 2-glucoside, cucurbitacin-H and cucurbitacin-J. However, all investigational agents for SARS-CoV-2 are under strict regulation by the World Health Organization (WHO) and this study urges the scientific community to continue conducting further in vivo trials for the experimental validation of our findings.
Declaration of competing interest
The authors declare that they have no conflict of interest.
Acknowledgement
The authors would like to acknowledge the Principal, Hindu College, University of Delhi, and the Schrödinger for the technical support provided for this research work.
References
- 1.Dyall J. Middle East respiratory syndrome and severe acute respiratory syndrome: current therapeutic options and potential targets for novel therapies. Drugs. 2017;77(18):1935–1966. doi: 10.1007/s40265-017-0830-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hassan S. Cucurbitacin B interacts synergistically with antibiotics against Staphylococcus aureus clinical isolates and exhibits antiviral activity against HSV-1. South Afr J Bot. 2017;108:90–94. [Google Scholar]
- 3.Kaushik U., Aeri V., Mir S.R. Cucurbitacins–an insight into medicinal leads from nature. Phcog Rev. 2015;9(17):12. doi: 10.4103/0973-7847.156314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaturvedi P. Antidiabetic potentials of Momordica charantia: multiple mechanisms behind the effects. J Med Food. 2012;15(2):101–107. doi: 10.1089/jmf.2010.0258. [DOI] [PubMed] [Google Scholar]
- 5.Dhiman K. A review on the medicinally important plants of the family Cucurbitaceae. Asian Journal of Clinical Nutrition. 2012;4(1):16–26. [Google Scholar]
- 6.Stevenson D.G. Oil and tocopherol content and composition of pumpkin seed oil in 12 cultivars. J Agric Food Chem. 2007;55(10):4005–4013. doi: 10.1021/jf0706979. [DOI] [PubMed] [Google Scholar]
- 7.Glew R. Amino acid, mineral and fatty acid content of pumpkin seeds (Cucurbita spp) and Cyperus esculentus nuts in the Republic of Niger. Plant Foods Hum Nutr. 2006;61(2):49–54. doi: 10.1007/s11130-006-0010-z. [DOI] [PubMed] [Google Scholar]
- 8.Phillips K.M., Ruggio D.M., Ashraf-Khorassani M. Phytosterol composition of nuts and seeds commonly consumed in the United States. J Agric Food Chem. 2005;53(24):9436–9445. doi: 10.1021/jf051505h. [DOI] [PubMed] [Google Scholar]
- 9.Sabudak T. Fatty acid composition of seed and leaf oils of pumpkin, walnut, almond, maize, sunflower and melon. Chem Nat Compd. 2007;43(4):465–467. [Google Scholar]
- 10.Alsayari A.S. 2014. Anticancer and antiviral activities of cucurbitacins isolated from Cucumis prophetarum var. Prophetarum growing in the southwestern region of Saudi Arabia. [Google Scholar]
- 11.Mathibe L., Botha J., Naidoo S. Effects of Z-venusol, isolated from Gunnera perpensa L., on IL-6 and cAMP activity in human breast cancer cells in vitro. South Afr J Bot. 2017;108:96–99. [Google Scholar]
- 12.Katsiki N., Banach M., Mikhailidis D.P. Lipid-lowering therapy and renin-angiotensin-aldosterone system inhibitors in the era of the COVID-19 pandemic. Arch Med Sci: AMS. 2020;16(3):485. doi: 10.5114/aoms.2020.94503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gordon C.J. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J Biol Chem. 2020;295(15):4773–4779. doi: 10.1074/jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat Commun. 2019;10(1):1–9. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferron F. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc Natl Acad Sci Unit States Am. 2018;115(2):E162–E171. doi: 10.1073/pnas.1718806115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Subissi L. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc Natl Acad Sci Unit States Am. 2014;111(37):E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xue X. Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design. J Virol. 2008;82(5):2515–2527. doi: 10.1128/JVI.02114-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin Z. Structure of M pro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020:1–5. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 19.Singleton M.R., Dillingham M.S., Wigley D.B. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 20.Jia Z. Delicate structural coordination of the severe acute respiratory syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Res. 2019;47(12):6538–6550. doi: 10.1093/nar/gkz409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riordan J.F. Angiotensin-I-converting enzyme and its relatives. Genome Biol. 2003;4(8):225. doi: 10.1186/gb-2003-4-8-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li W. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khatri I., Staal F.J., Van Dongen J.J. Blocking of the high-affinity interaction-synapse between SARS-CoV-2 Spike and human ACE2 proteins likely requires multiple high-affinity antibodies: an Immune perspective. Front Immunol. 2020:11. doi: 10.3389/fimmu.2020.570018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wooding D., J., Bach H. Treatment of COVID-19 with convalescent plasma: lessons from past coronavirus outbreaks. Clin Microbiol Infect. 2020;26:1436–1446. doi: 10.1016/j.cmi.2020.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ragab D. The COVID-19 cytokine storm; what we know so far. Front Immunol. 2020;11:1446. doi: 10.3389/fimmu.2020.01446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kupchan S.M., Meshulam H., Sneden A.T. New cucurbitacins from Phormium tenax and Marah oreganus. Phytochemistry. 1978;17(4):767–769. [Google Scholar]
- 27.Raman A., Lau C. Anti-diabetic properties and phytochemistry of Momordica charantia L.(Cucurbitaceae) Phytomedicine. 1996;2(4):349–362. doi: 10.1016/S0944-7113(96)80080-8. [DOI] [PubMed] [Google Scholar]
- 28.Ye J.M., Ruderman N.B., Kraegen E.W. AMP-activated protein kinase and malonyl-CoA: targets for treating insulin resistance? Drug Discov Today Ther Strat. 2005;2(2):157–163. [Google Scholar]
- 29.Iglesias M.A. AICAR administration causes an apparent enhancement of muscle and liver insulin action in insulin-resistant high-fat-fed rats. Diabetes. 2002;51(10):2886–2894. doi: 10.2337/diabetes.51.10.2886. [DOI] [PubMed] [Google Scholar]
- 30.Chen R.J. Active ingredients in Chinese medicines promoting blood circulation as Na+/K+-ATPase inhibitors. Acta Pharmacol Sin. 2011;32(2):141–151. doi: 10.1038/aps.2010.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balkema-Boomstra A. Role of cucurbitacin C in resistance to spider mite (Tetranychus urticae) in cucumber (Cucumis sativus L.) J Chem Ecol. 2003;29(1):225–235. doi: 10.1023/a:1021945101308. [DOI] [PubMed] [Google Scholar]
- 32.Soto-Hernández M. Lead compounds from Cucurbitaceae for the treatment of cáncer. In: Rao A.V., Rao L.G., editors. Phytochemicals—isolation, characterization and role in human Health. 2015. pp. 289–303. [Google Scholar]
- 33.Chen J.C. Cucurbitacins and cucurbitane glycosides: structures and biological activities. Nat Prod Rep. 2005;22(3):386–399. doi: 10.1039/b418841c. [DOI] [PubMed] [Google Scholar]
- 34.Bernard S.A., Olayinka O.A. Search for a novel antioxidant, anti-inflammatory/analgesic or anti-proliferative drug: cucurbitacins hold the ace. J Med Plants Res. 2010;4(25):2821–2826. [Google Scholar]
- 35.Qiao J. Cucurbitacin E exhibits anti-inflammatory effect in RAW 264.7 cells via suppression of NF-κB nuclear translocation. Inflamm Res. 2013;62(5):461–469. doi: 10.1007/s00011-013-0598-z. [DOI] [PubMed] [Google Scholar]
- 36.Stuppner H., Moller E. Cucurbitacins with unusual side chains from Picrorhiza kurroa. Phytochemistry. 1993;33(5):1139–1145. [Google Scholar]
- 37.Dinan L. Cucurbitacins are insect steroid hormone antagonists acting at the ecdysteroid receptor. Biochem J. 1997;327(3):643–650. doi: 10.1042/bj3270643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teuscher E. In: Biogene gifte-biologie, chemie, pharmakologie. und Lindequist U., editor. Gustav Fischer Verlag; 1994. [Google Scholar]
- 39.Subbiah V. Google Patents; 1999. Method of isolating cucurbitacin. [Google Scholar]
- 40.Roa M., Meshulam H., Lavie D. Constituents of ecballium elaterium XXIII: cucurbitacins and hexanorcucurbitacin. J Chem Soc. 1974;22:2552. [Google Scholar]
- 41.Stecher P.G. Merck and Co., Inc; The Merc index-An encyclopedia of Chemicals and drugs; 2009. Rahway; pp. 296–297. [Google Scholar]
- 42.Hoffmann M. Cell; 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walls A.C. Cell; 2020. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nature microbiology. 2020;5(4):562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou G., Chen S., Chen Z. Springer; 2020. Back to the spring of Wuhan: facts and hope of COVID-19 outbreak. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hamming I. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol: A Journal of the Pathological Society of Great Britain and Ireland. 2004;203(2):631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ding Y. Organ distribution of severe acute respiratory syndrome (SARS) associated coronavirus (SARS‐CoV) in SARS patients: implications for pathogenesis and virus transmission pathways. J Pathol: A Journal of the Pathological Society of Great Britain and Ireland. 2004;203(2):622–630. doi: 10.1002/path.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gu J. Multiple organ infection and the pathogenesis of SARS. J Exp Med. 2005;202(3):415–424. doi: 10.1084/jem.20050828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Danilczyk U. Essential role for collectrin in renal amino acid transport. Nature. 2006;444(7122):1088–1091. doi: 10.1038/nature05475. [DOI] [PubMed] [Google Scholar]
- 50.Crackower M.A. Angiotensin-converting enzyme 2 is an essential regulator of heart function. Nature. 2002;417(6891):822–828. doi: 10.1038/nature00786. [DOI] [PubMed] [Google Scholar]
- 51.Wan Y. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020;94(7) doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hashimoto T. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012;487(7408):477–481. doi: 10.1038/nature11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ziebuhr J. Coronavirus replication and reverse genetics. Springer; 2005. The coronavirus replicase; pp. 57–94. [Google Scholar]
- 54.Hao W. Crystal structure of Middle East respiratory syndrome coronavirus helicase. PLoS Pathog. 2017;13(6) doi: 10.1371/journal.ppat.1006474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ma Y. Structural basis and functional analysis of the SARS coronavirus nsp14–nsp10 complex. Proc Natl Acad Sci Unit States Am. 2015;112(30):9436–9441. doi: 10.1073/pnas.1508686112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Joseph J.S. Crystal structure of nonstructural protein 10 from the severe acute respiratory syndrome coronavirus reveals a novel fold with two zinc-binding motifs. J Virol. 2006;80(16):7894–7901. doi: 10.1128/JVI.00467-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gordon C.J. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J Biol Chem. 2020;295(20):6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mirza M.U., Froeyen M. Structural elucidation of SARS-CoV-2 vital proteins: computational methods reveal potential drug candidates against main protease, Nsp12 polymerase and Nsp13 helicase. Journal of Pharmaceutical Analysis. 2020;10(4):320–328. doi: 10.1016/j.jpha.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peng X. Genome-wide analysis of protein-protein interactions and involvement of viral proteins in SARS-CoV replication. PloS One. 2008;3(10):e3299. doi: 10.1371/journal.pone.0003299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.von Brunn A. Analysis of intraviral protein-protein interactions of the SARS coronavirus ORFeome. PloS One. 2007;2(5):e459. doi: 10.1371/journal.pone.0000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Adedeji A.O. Mechanism of nucleic acid unwinding by SARS-CoV helicase. PloS One. 2012;7(5) doi: 10.1371/journal.pone.0036521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mengist H.M., Fan X., Jin T. Designing of improved drugs for COVID-19: crystal structure of SARS-CoV-2 main protease M pro. Signal Transduction and Targeted Therapy. 2020;5(1):1–2. doi: 10.1038/s41392-020-0178-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hsu M.-F. Mechanism of the maturation process of SARS-CoV 3CL protease. J Biol Chem. 2005;280(35):31257–31266. doi: 10.1074/jbc.M502577200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Braciale T.J., Hahn Y.S. Immunity to viruses. Immunol Rev. 2013;255(1) doi: 10.1111/imr.12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang C. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ruan Q. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020;46(5):846–848. doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen G. Clinical and immunologic features in severe and moderate forms of Coronavirus Disease. J Clin Invest. 2019:137244. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gao, Y., et al., Diagnostic utility of clinical laboratory data determinations for patients with the severe COVID-19 [published online March 17, 2020]. J Med Virol. doi. 10. [DOI] [PMC free article] [PubMed]
- 69.Chen L. Analysis of clinical features of 29 patients with 2019 novel coronavirus pneumonia. Zhonghua jie he he hu xi za zhi= Zhonghua jiehe he huxi zazhi= Chinese journal of tuberculosis and respiratory diseases. 2020;43 doi: 10.3760/cma.j.issn.1001-0939.2020.0005. E005-E005. [DOI] [PubMed] [Google Scholar]
- 70.Sun J. Cucurbitacin Q: a selective STAT3 activation inhibitor with potent antitumor activity. Oncogene. 2005;24(20):3236–3245. doi: 10.1038/sj.onc.1208470. [DOI] [PubMed] [Google Scholar]
- 71.Toyonaga T. Blockade of constitutively activated Janus kinase/signal transducer and activator of transcription-3 pathway inhibits growth of human pancreatic cancer. Canc Lett. 2003;201(1):107–116. doi: 10.1016/s0304-3835(03)00482-8. [DOI] [PubMed] [Google Scholar]
- 72.Thoennissen N.H. Cucurbitacin B induces apoptosis by inhibition of the JAK/STAT pathway and potentiates antiproliferative effects of gemcitabine on pancreatic cancer cells. Canc Res. 2009;69(14):5876–5884. doi: 10.1158/0008-5472.CAN-09-0536. [DOI] [PubMed] [Google Scholar]
- 73.Chan K.T. Cucurbitacin B inhibits STAT3 and the Raf/MEK/ERK pathway in leukemia cell line K562. Canc Lett. 2010;289(1):46–52. doi: 10.1016/j.canlet.2009.07.015. [DOI] [PubMed] [Google Scholar]
- 74.Liu Y. Cucurbitacin E inhibits Huh7 hepatoma carcinoma cell proliferation and metastasis via suppressing MAPKs and JAK/STAT3 pathways. Molecules. 2020;25(3):560. doi: 10.3390/molecules25030560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang Y. Cucurbitacin IIb exhibits anti-inflammatory activity through modulating multiple cellular behaviors of mouse lymphocytes. PloS One. 2014;9(2) doi: 10.1371/journal.pone.0089751. [DOI] [PMC free article] [PubMed] [Google Scholar]