Abstract
Cells utilize transcriptional and post-transcriptional mechanisms to alter gene expression in response to environmental cues. Gene-specific controls, including changing the translation of specific mRNAs, provides a rapid means to respond precisely to different conditions. Upstream open reading frames (uORFs) are known to control the translation of mRNAs. Recent studies in bacteria and eukaryotes have revealed the functions of evolutionarily conserved uORF-encoded peptides. Some of these uORF-encoded nascent peptides enable responses to specific metabolites to modulate the translation of their mRNAs by stalling ribosomes, and through ribosome stalling may also modulate the level of their mRNAs. In this review, we highlight several examples of conserved uORF nascent peptides that stall ribosomes to regulate gene expression in response to specific metabolites in bacteria, fungi, mammals, and plants.
Keywords: uORF, nascent peptide, translation stalling, uCC, protein synthesis, ribosome
Introduction
Regulation of gene expression plays crucial roles in cellular stress responses, homeostatic control of biosynthetic processes, and in development. In addition to regulation by transcriptional control, gene expression can be translationally controlled. One key advantage of translational versus transcriptional control of gene expression is speed. Whereas transcriptional control relies on the synthesis of new mRNAs that must then be engaged by the ribosome to synthesize proteins, translational control bypasses this first step, as well as the second step in eukaryotic cells of transporting the mRNAs from the nucleus to the cytoplasm. Another advantage is that translational control directly regulates the production of proteins. In addition to global mechanisms of translational control involving altered functions of ribosomes or the factors that assist their function, gene-specific translational control can also be mediated by elements that function in cis in the regulated mRNAs. In this review, we will focus on cis-acting translational control mechanisms in bacteria and eukaryotes that use upstream open reading frames (uORFs) in mRNA leaders that encode regulatory peptides that control gene expression by acting on the ribosomes that translate them.
The mechanisms of translation initiation, and the structures of mRNAs, differ greatly between bacteria and eukaryotes. In bacteria, many mRNAs are polycistronic and encode multiple proteins from different ORFs. Ribosomes are recruited directly to ORF start codons. Base-pairing interactions between the ribosomal rRNA and nucleotides in a region slightly upstream of the start codon in the mRNA enhance the efficiency of initiation (102). In some cases, translation of sequential ORFs in bacterial mRNAs is coupled – for example, by transfer of a terminating ribosome from the 5’ ORF to the start codon of the subsequent ORF, or by translation of the preceding ORF melting an mRNA secondary structure that otherwise would restrict ribosome access to the downstream ORF. In addition to these translational coupling mechanisms operating between adjacent long protein-coding ORFs in an mRNA, translation of short uORFs in the 5’-leaders of mRNAs can be coupled to the translation of the main protein-coding ORFs (mORFs). Importantly, as discussed below, ribosome stalling during the translation of bacterial uORFs can control access to a downstream translation initiation site by controlling the structure of the mRNA (Fig. 1).
Figure 1. uORF-mediated control of bacterial gene expression.
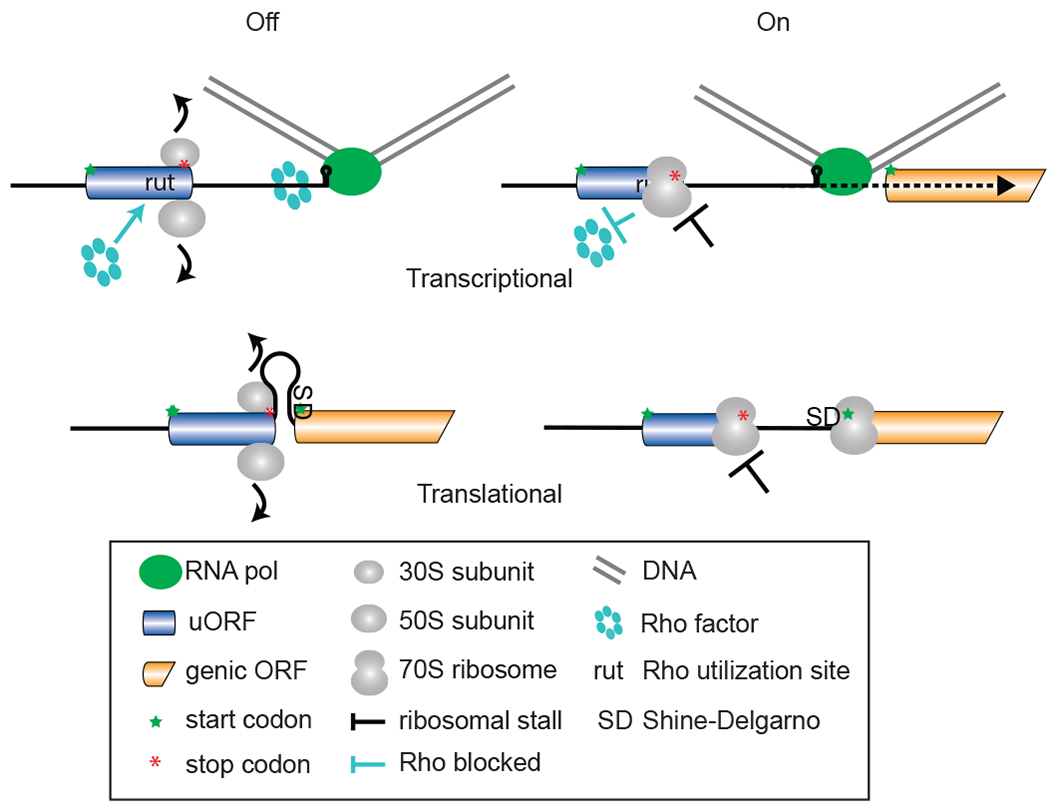
Transcriptional Off: Rho-dependent termination of transcription in the mRNA 5’-leader occurs when Rho has access to its binding site in the RNA (rut site) as occurs when there is no uORF-mediated ribosome stalling. RNA polymerase pauses at a GC-rich stem structure; if Rho binds to the RNA and by moving 5’ to 3’ on the mRNA catches up to the paused RNA polymerase, transcription will terminate in the 5’-leader. On: Ribosome stalling on the uORF blocks access of Rho to the rut site. RNA polymerase is not impacted by Rho and, therefore, can resume transcription of the downstream protein-coding regions after pausing. Translational Off: Elements (SD, Shine-Dalgarno region) important for downstream translation initiation are masked in RNA secondary structure as occurs when there is no uORF-mediated ribosome stalling. On: Ribosome stalling on the uORF changes RNA secondary structure to unmask the SD and enable downstream translation initiation.
In bacteria, the processes of transcription and translation are coupled. Ribosomes engage and translate an mRNA while the mRNA is still being transcribed. Because of this coupling, the translation of bacterial uORFs (also known as bacterial leader peptides) can affect mRNA production. As exemplified by the paradigmatic Escherichia coli Trp attenuation mechanism (reviewed in 139), stalling of translating ribosomes on Trp codons in the trpL leader peptide (uORF) of the trp operon mRNA under conditions of limiting tryptophan – when the levels of Trp-tRNA are low – favors formation of an RNA secondary structure termed the anti-terminator. Antiterminator formation enables the synthesis of the full-length trp operon mRNA and thus the production of Trp biosynthetic enzymes. In contrast, when Trp levels are high, efficient translation of the uORF leader peptide favors the formation of a Rho-independent transcription terminator structure in the mRNA 5’-leader and premature termination of trp operon transcription. Thus, in bacteria, translation of uORFs can both regulate ribosome access to the mRNA and control synthesis of the full-length mRNA (Fig. 1).
In eukaryotes, transcription occurs in the nucleus and translation in the cytoplasm, so these processes are not directly coupled. However, the translation of uORFs can regulate both translation of the mORF and stability of the mRNA. As opposed to the direct binding of ribosomes to the start codon of ORFs as in bacteria, eukaryotic mRNAs are translated via the scanning mechanism (Fig. 2, reviewed in 39; 40; 41). The small ribosomal subunit, with bound initiator Met-tRNAiMet and associated translation initiation factors, binds near the cap of an mRNA and scans in a 3’direction. Base-pairing interactions between the anticodon loop of tRNAiMet and a start codon (typically AUG) triggers translation start site selection. Typically, the scanning ribosome selects the first start codon it encounters during scanning (Fig. 2, scheme 3). However, this is not always the case. The efficiency of start codon selection is influenced by the identity of the start codon and its immediate sequence context (19; 51; 62; 63; 70; 84; 130). An A or G at the −3 position relative to the A+1 of the AUG codon is the most important determinant of a favorable initiation context, and a G at +4 also favors start codon selection. Scanning ribosomes will scan past AUG start codons in poor context without initiating (leaky scanning) and then initiate at downstream start codons (Fig. 2, scheme 2). Scanning ribosomes can also initiate at near cognate codons that differ from AUG by a single nucleotide change; however, even the most efficient near cognate codon (CUG in mammalian cells) is used at <20% the efficiency of a good-context AUG codon (51).
Figure 2. uORF-mediated translational control in eukaryotes.
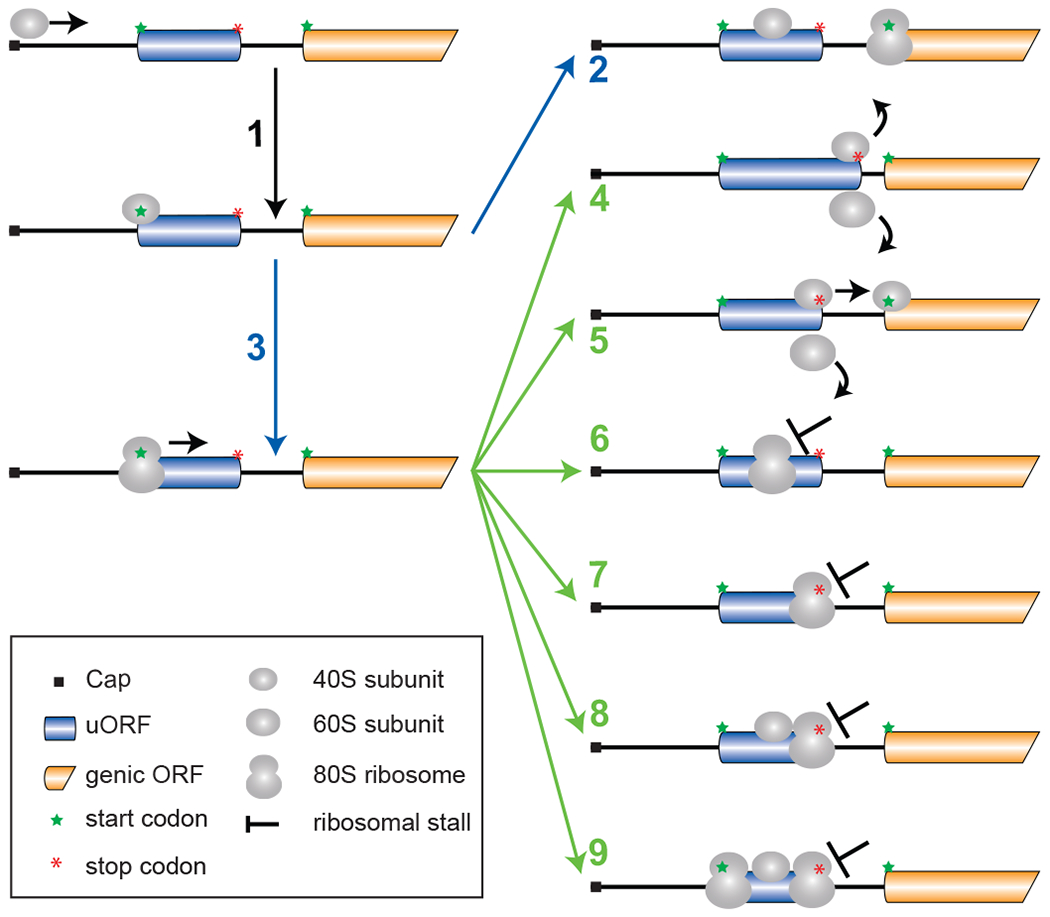
Translation initiation in eukaryotes begins with the small (40S) ribosomal subunit binding near the cap of the mRNA. The ribosome then scans the mRNA (1) inspecting for a start codon. The scanning ribosome either (2) skips over the start codon of the uORF without initiating (leaky scanning) and then translates the mORF or (3) translates the uORF. Multiple possible consequences of uORF translation include: (4) disengagement of the ribosome from the mRNA following translation of a long uORF (mORF translation is repressed) or (5) resumption of scanning by the small ribosomal subunit following translation of a short uORF enables reinitiation of translation at the mORF. If the peptide sequence encoded by the uORF causes ribosomes to pause (or stall) during elongation (6) or termination (7), a short uORF can inhibit mORF translation like a long uORF. Both the elongation and termination pausing events can prevent subsequent ribosomes from leaky scanning (8) to the mORF, and in some cases via ribosome queuing promote initiation at the uORF start codon (9) to create a positive feedback loop that enhances uORF translation and represses mORF translation.
How does eukaryotic uORF translation impact mORF translation? If the uORF terminates before the start codon of the mORF, then several features of the uORF and the mRNA leader determine the impact of the uORF on mORF translation. There are at least six ways (Fig. 2) that translation of a uORF can affect translation from a downstream start codon. The length of a uORF or the time spent translating it is inversely correlated with the efficiency of reinitiation following translation of the uORF (64; 73; 95). Accordingly, the translation of long uORFs (Fig. 2, scheme 4) is associated with much poorer reinitiation potential than the translation of short uORFs (Fig. 2, scheme 5). In a similar manner, stalling of a translating ribosome during elongation (Fig. 2, scheme 6) or termination (Fig. 2, scheme 7) can enhance the inhibitory activity of a short uORF. For example, rare codons or secondary structures that impede translation elongation on a uORF (64; 69) or sequences that interfere with peptide release at termination (9; 17) lower the probability of translation reinitiation. Stalling of translation elongation or termination on a uORF, in addition to lowering the probability of reinitiation, can block subsequent scanning ribosomes from leaky scanning the uORF and reaching the mORF start codon (Fig. 2, scheme 8). Interestingly, impeding the progress of the leaky scanning ribosome can trigger the formation of a ribosome queue. When the queue reaches the uORF start codon, the longer time spent by a scanning ribosome in the vicinity of the start codon will enhance initiation on the uORF (Fig. 2, scheme 9). The increased uORF translation will trigger a positive feedback loop that reinforces the stall, which in turn triggers queuing by subsequent ribosomes and enhanced translation of the uORF. As will described below, regulated elongation pausing and ribosome queuing underlies the regulation of antizyme inhibitor (AZIN1) mRNA translation (52). Notably, both the elongation-stalling and termination-stalling mechanisms can be consequences of the actions of nascent peptides specified by uORFs.
In addition to controlling ribosome access to the mORF, translation of uORFs in eukaryotes can impact mRNA levels. Cells employ a variety of quality control pathways to turnover defective mRNAs and ribosomes (104). The nonsense-mediated decay (NMD) pathway is responsible for degrading mRNAs in which a translating ribosome encounters a premature stop codon (a stop codon that is not the mORF stop codon). While this pathway’s major function is considered to be a quality-control surveillance mechanism to trigger degradation of aberrant mRNAs in which a nonsense (stop) codon occurs within the mORF, the key trigger for NMD is the presence of a stop codon upstream of the normal mORF stop codon. Thus uORFs, which by definition terminate with premature stop codons, can trigger NMD (36). Although not all uORFs trigger NMD, the parallel abilities of eukaryotic uORFs to cause NMD and, by doing so, to regulate transcript levels, and the abilities of bacterial uORFs to regulate transcript levels by controlling transcription termination, highlight the universal properties of uORFs to control both translation and mRNA levels.
In this review, we will focus on a special class of uORF-mediated translational control mechanisms that involve uORFs which encode conserved polypeptides. Interestingly, conserved peptide uORFs (CPuORFs), as well as upstream conserved coding regions (uCCs, conserved uORFs that initiate with near cognate start codons), are found in mRNAs from bacteria, fungi, viruses, mammals and plants. While the conserved peptide sequences could have reflected functional roles of the encoded proteins after their synthesis, the evidence indicates that amino acid sequence conservation of the uORFs exists to enable translational control via specialized requirements for translation factors or through nascent peptide interactions with the translational machinery (ribosomes). Of note, several of these conserved uORFs have been shown to function as nascent peptides in the ribosome and confer regulation in response to specific metabolites.
Bacterial uORFs
Trp control of TnaC
The E. coli tna (tryptophanase) operon encodes two structural genes, tnaA and tnaB. tnaA specifies tryptophanase and tnaB specifies a tryptophan transporter. The operon is under catabolite control and control by levels of Trp. Specifically, operon expression is induced when preferred carbon sources are not available and Trp levels are high. In 1985, Stewart and Yanofsky reported the isolation of cis-acting mutants in the tna operon that caused high levels of expression independent of the presence of Trp. The mutations mapped to the mRNA 5’-leader in a sequence specifying a 24-residue leader peptide they named tnaC (111). Subsequent work showed that Trp acts with nascent TnaC to cause ribosome stalling with the TnaC termination codon in the ribosome A site. This stalling causes release from transcription attenuation, resulting in transcription of the full-length mRNA and expression of the structural genes (27). In the model for tna regulation, catabolite control causes transcription initiation to increase when rich carbon sources are not available, and translation of TnaC in the presence of Trp inhibits Rho-dependent termination in the mRNA 5’leader (14). (Fig. 1)
The requirements for TnaC-mediated stalling have been investigated by mutational analyses of the TnaC peptide and the ribosome (13; 75; 76; 138). The C-terminal half of the peptide is most important for regulation and residues critical for regulation are evolutionarily conserved (15). Regulation by TnaC is impacted by the stop codon that terminates its synthesis (61). The explanation for this is that TnaC stalling is accentuated when RF2 is the release factor (UGA stop codon) compared to when RF1 is the release factor (UAG stop codon) (21). The inherently different activities of RF2 and RF1 – RF1 is a more efficient release factor than RF2 – could explain why TnaC and Trp do not act as efficiently to inhibit RF1 activity. This hypothesis is supported by mutational analysis of RF2; mutations that increase its activity also decrease the effect of TnaC and Trp on stalling in a reconstituted cell-free translation system (21). The structural model for TnaC regulation, based on cryo-EM data, indicates that TnaC and Trp cause changes in the conformation of the ribosome that result in the large ribosomal subunit rRNA nucleotide A2602 shifting to a position that does not enable RF2 function (7), consistent with the mutational data. Here, and for the other bacterial stalling peptides, because of space considerations, we do not discuss the extensive work on the consequences of ribosome mutations for nascent peptide stalling mechanisms.
Recently, TnaC control was examined from a systems perspective by using flow cytometry to measure reporter expression from more than a thousand TnaC mutations (124). The analyses corroborated the importance of specific residues in the TnaC C-terminal half in Trp-mediated ribosome stalling. They also uncovered mutations in the less-conserved amino-terminal half of the coding region that alter the regulation of downstream reporter expression. These N-terminal mutations are not crucial at the amino acid coding level but are important at the nucleotide level. They appear to be relevant for forming RNA structures that govern translation initiation rates at the TnaC start codon. Thus, the sequences encoding the amino-terminal region (5’-half) modulate translation initiation to maintain levels that allow coupling of translation to transcription. Such coupling is critical so that the timing of ribosome stalling can affect the action of RNA polymerase at the Rho-dependent termination site in the mRNA 5’-leader.
Ornithine control of SpeFL/Orf34
E. coli and Salmonella typhimurium encode an inducible ornithine decarboxylase (ODC) named SpeF, which is induced by high ornithine levels (4; 37). The mechanism through which induction occurs has been determined: ornithine causes ribosome stalling during the translation of the 34-residue SpeFL leader peptide (37). Stalling in response to ornithine results in masking of a Rho utilization site (rut site) and transcription antitermination, enabling transcription to continue into the structural gene for SpeF. A structure of the ribosome containing the stalled peptide and ornithine gave insight into the stalling mechanism. Notably and excitingly, this is the first high resolution structure revealing how a nascent stalling peptide interacts with a metabolite in the peptide exit tunnel of the ribosome. In the structure, ornithine fits into a pocket created by both the ribosome and SpeFL resulting in a conformational change of the nascent peptide and subsequent interference with translation termination mediated by RF1 (37).
The SpeFL structure can be considered as two domains: an N-terminal sensor domain and a C-terminal effector domain. The junction of these domains, residues H10IRRXXH16, forms the tight ornithine binding pocket with the ribosome. Tight ornithine binding is proposed to compact the nascent peptide’s effector domain, and as a consequence, to change the structure of the ribosome and interfere with RF1 activity through steric clash (37).
SpeFL (named Orf34 in S. typhimurium) is conserved in γ-proteobacteria (4; 37). How ribosome stalling within SpeFL is coupled to transcription termination appears to be complex and may not yet be fully understood. In E. coli, the rare Arg12 and Arg13 codons of SpeFL are proposed to be important for fully exposing the Rho-utilization (rut) site so that, in the absence of ornithine, basal levels of SpeF are low. Ornithine-mediated stalling of the lead ribosome at the termination codon would then create a situation in which ribosomes accumulate behind the paused ribosome and occlude the rut site (37). In S. typhimurium, these conserved tandem Arg codons mediate regulation of downstream SpeF by Arg availability (4); this makes metabolic sense because Arg is also a precursor for polyamine synthesis. Limited Arg-availability is proposed to stabilize the mRNA in S. typhimurium by slowing translation at these tandem codons and enabling different RNA structures to form. The alternative secondary structures that form as a consequence of leader peptide translation in S. typhimurium also appear to affect translation initiation at the downstream SpeF start codon by impacting the accessibility of the Shine-Delgarno site (4).
Macrolide control of ErmC
Macrolide antibiotics, which interfere with protein synthesis in complex ways (59; 60), bind to ribosomes within the peptide exit tunnel (20; 112). Resistance to the macrolide erythromycin (ERY) is conferred by methylation of 23S rRNA (65) at A2058 (105) through the Erm family of methyltransferases; methylation of A2058 reduces macrolide binding to the ribosome (131).
Staphylococcus aureus ermC encodes this methyltransferase and its expression is induced by subinhibitory concentrations of ERY. The mechanism of induction was inferred early on to be at the level of translation. Specifically, in 1980, antibiotic-induced stalling of synthesis of the ErmCL leader peptide was proposed as the mechanism to increase ErmC translation (28; 42). Subsequent work established that the peptide sequence, not the mRNA sequence, of the leader peptide was important, and that ribosome stalling occurred during elongation near the C-terminus of the short (10-residue) peptide, with codon-9 in the P site and codon-10 in the A site (123). Stalling changes the mRNA structure so that translation of ErmC can be efficiently initiated (Fig. 1). Analyses of the translation of Erm leader peptides with different primary amino acid sequences in a variety of Erm-encoding mRNAs, and a cryo-EM structure of Erm-stalled ribosomes, showed that stalling was used generally as a regulatory mechanism to induce this resistance mechanism (3; 30). Understanding how different macrolide-related antibiotics trigger or do not trigger ribosome stalling and thus would or would not induce expression of distinct Erm methyltransferases is important to consider for the clinical treatment of resistant bacteria (30). Finally, translation of specific leader peptides is also proposed to translationally control operons that confer resistance to another antibiotic, chloramphenicol, that binds at the peptidyl transferase center of the bacterial ribosome, not in the exit tunnel (20; 112), but the mechanisms remain unclear (47).
SecM control of secretory capacity
E. coli SecM (secretion monitor), named by Nakatogawa and Ito (82) who discovered its function as a stalling peptide, was initially identified as a 170-residue upstream coding region called Gene X in the 5’-leader of the mRNA specifying E. coli SecA (103). SecA encodes a translocation ATPase important for targeting secreted proteins to the SecYEG channel (110). Observations, including the identification of mutations in the Gene X secretion sequence that caused SecA to be constitutively expressed (85), led Nakatogawa and Ito to examine SecM function in detail (82). They established that SecM protein synthesis stalls during elongation near the C-terminus of the peptide, and that insertion of SecM into the membrane releases the stall. They obtained data that the nascent peptide itself was critical for stalling, and they proposed that, if the stall was not released, then the secondary structure of the mRNA would be affected, and SecA translation would increase. These predictions were borne out by subsequent work (reviewed in 47; 48). Thus SecM-mediated ribosome stalling, which arises when secretory stress results in failure of SecM to get pulled into membrane, results in a stalled ribosome that changes the mRNA structure to expose a downstream Shine-Dalgarno sequence and to enable SecA translation (Fig. 1).
Elongation stalling in SecM occurs with a prolyl-tRNA in the stalled ribosome A site; this Pro is not incorporated into the nascent peptide (81). While variation appears tolerated in many residues of the functionally conserved region of the nascent peptide (140), the Pro residue is critical; one possible reason is that Pro in the A site of the stalled ribosome inhibits quality control mechanisms that would otherwise degrade mRNAs on which a ribosome is stalled (26). Cryo-EM studies of SecM-stalled ribosomes, which may not fully reflect the dynamic situation in vivo (48) and which are at variance with a biophysical in-solution study (135), nevertheless indicate structural changes that would interfere with peptidyl transferase activity (5; 29; 145).
MifM regulation and membrane insertion capacity
Bacillus subtilis MifM (membrane protein insertion and folding monitor) was identified initially as YqzJ, a 95-residue uORF peptide with a membrane insertion sequence that is encoded in the 5’-leader of the yidC2 operon (12). YidC2 is an insertase whose expression increases when the activity of the major insertase YidC1 appears inadequate to meet cellular needs. Genetic selection for YidC2-LacZ reporter overexpression yielded mutations affecting the sequence of this uORF peptide. Subsequent analyses revealed that the MifM peptide stalls ribosomes when cellular membrane insertion capacity is low, and the stalled ribosome affects RNA structure to expose the Shine-Dalgarno sequence important to initiate YidC2 translation (Fig. 1) and increase this capacity (12). When insertion capacity increases, arrest is relieved and YidC2 expression is reduced. Thus, regulation of YidC2 by MifM in response to insertion capacity appears analogous to regulation of SecA by SecM in response to secretory capacity. However, there is little similarity at the amino acid sequence level between MifM and SecM peptides, and in reconstituted systems, MifM is specific for B. subtilis ribosomes and SecM for E. coli ribosomes (11). A third class of peptide that monitors protein export through elongation arrest has been described in Vibrio (45). The mechanism of MifM action has been reviewed in detail (47; 48).
MifM stalls ribosomes engaged in elongation, and the stalls occur, not at a specific site, but in a region near a highly acidic amino acid cluster (10). Cryo-EM structural analyses of stalled ribosomes indicate that the nascent peptide interacts with the rRNA to reduce activity at the peptidyl transferase center (106). Additional N-terminal MifM residues, located far from the stalling site and that appear to be outside of the exit tunnel when arrest occurs, also have roles in arrest, indicating that the mechanism by which the conformational changes that lead to stalling are triggered by peptide-ribosome interactions is not fully understood (23).
Fungal uORFs
Arg control by the fungal AAP
The Arg biosynthetic pathway in fungi begins with the production of carbamoyl phosphate. Arginine-specific carbamoyl phosphate synthetase (CPS-A, which differs from CPS-P, which is used for pyrimidine synthesis) is a two subunit enzyme in these organisms (16). The CPS-A small subunit is subject to negative regulation by Arg in both organisms. The gene specifying the small subunit in Neurospora crassa, arg-2, was among the first amino acid biosynthetic genes identified by mutation in any organism (109), and cis-dominant mutations that removed Arg-specific regulation of the Saccharomyces cerevisiae small subunit gene, CPA1, were identified in the early 1970s (117). In 1987, these and other cis-acting mutations in CPA1 were shown to change the amino acid sequence of a 25-codon uORF in the CPA1 mRNA 5’-leader and a model for translational control through the uORF was proposed to account for the observations that the amino acid sequence of the uORF appeared important for regulation (133).
The sequences of the N. crassa arg-2 gene and cDNA revealed that a 24-codon uORF, conserved at the amino acid sequence level, was also present in the mRNA (88). Direct biochemical evidence for translational control by the arg-2 uORF in vivo was obtained; classical genetic selection for mutations that lost arg-2 uORF regulatory function identified the critical residue Asp12 in the peptide as was also found in the S. cerevisiae CPA1 uORF (22; 71; 72). An N. crassa cell-free translation system was developed that recapitulated Arg-specific regulation by the uORF (127). Primer-extension inhibition analyses (toeprinting) showed that Arg causes ribosomes to stall with the uORF stop codon in the ribosome A site (128) and that conserved residues in the peptide were crucial for stalling.
Further work established that the arg-2 uORF-encoded peptide could cause stalling of ribosomes involved in translation elongation in addition to ribosomes involved in termination, and the peptide was named the arginine attenuator peptide (AAP) (125). That Arg, or closely related compounds, were important for regulation, but levels of aminoacylated tRNA were not, was shown (126). The proposed mechanism of translational regulation by the uORF-encoded AAP (whose start codon is in a weak initiation context), with AAP-mediated stalling controlling leaky scanning of ribosomes (125) (Fig. 2, scheme 7), was confirmed in both N. crassa and S. cerevisiae cell-free translation systems with both the arg-2 and CPA1 AAPs (25).
A combination of a weak-context start codon, and a strong Arg-dependent stall at its termination codon, result in AAP-mediated stalling triggering NMD of the S. cerevisiae CPA1 and N. crassa arg-2 mRNAs (24; 146), destabilizing these transcripts. Thus, stalling by this eukaryotic uORF impacts mRNA levels. The early classical genetic studies of CPA1 regulation (117), accomplished before any of these processes were known, prefigured these results. The cis-acting CPA1 mutations altered the AAP, making it unable to stall ribosomes as discussed above, and a trans-acting gene identified to increase CPA1 gene expression reduced the function of UPF1, which is essential for NMD (78).
The AAP is conserved through a billion years of fungal evolution, and the evolutionarily conserved region is critical for regulation (108). The relative positions of highly conserved amino acids in the nascent AAP with respect to the translating ribosome are critical for the AAP to exert its stalling function; however, there is no evidence that a specific amino acid must be in the P site to trigger stalling (129), unlike what is observed for some of the other nascent peptides described here. In the presence of Arg, the AAP’s positioning in the ribosome exit tunnel changes (136), and Arg-regulation is then exerted by the AAP interfering with the activity of the ribosome peptidyl transferase center (129). The effect of Arg to change the relative position of the AAP nascent peptide can occur before the optimal position for stalling is reached (136).
The position of the AAP in the ribosome has been determined by cryo-EM at low-resolution (6). Recently, mutations affecting the plant ribosome tunnel at positions predicted from the structure to interact with the AAP were found to impact stalling by the AAP (115). Importantly, the same tunnel mutations affected other stalling peptides in different ways, showing that there is not one specific interaction with the ribosome by which these peptides, which have different lengths and sequences, cause stalling. Based on these data, it appears that while the end-result (ribosome arrest) can be the same, different eukaryotic stalling peptides have different ways of interacting with the ribosome to trigger an arrest.
Mammalian uORFs
Bioinformatic analyses have identified over one million uORFs in humans; however, the number of these that are translated and that regulate mORF translation is much lower (54; 74; 77). The number of uORFs encoding conserved peptides has not been determined conclusively for mammals. Rather than cataloging examples of mRNAs containing uORFs with conserved peptide sequences, we will focus on a few exemplar uORFs that have been shown to confer regulation in response to a metabolite in a sequence-dependent manner or whose mechanism of translational control has been investigated in depth.
Polyamine-regulated uORFs
The small organic polycations putrescine, spermidine, and spermine have been implicated in a variety of cellular processes, with prominent functions in transcription, translation, and ion channels (93). The positively charged polyamines interact with negatively charged nucleic acids (DNA or RNA), membranes and proteins, and stimulate the processes involving these molecules, in some cases functioning like Mg2+ ions. While the precise molecular functions of polyamines have not been elucidated, the regulation of polyamine biosynthesis is under tight homeostatic control, with much of the control occurring at the level of translation (18).
The enzyme ornithine decarboxylase (ODC) converts ornithine to putrescine in the first step of polyamine biosynthesis. Under conditions of polyamine excess, the protein antizyme (OAZ1), whose synthesis is induced by polyamine-regulated +1 ribosomal frameshifting, binds to ODC and targets it for degradation (92). When polyamine levels drop, OAZ1 synthesis is repressed and synthesis of antizyme inhibitor (AZIN1), a non-functional ortholog of ODC, is induced (58). The AZIN1 binds and sequesters OAZ1, enabling ODC levels to increase and catalyze the synthesis of more polyamines. To generate the higher-order polyamines, a molecule of decarboxylated S-adenosylmethionine (dcSAM) donates an aminopropyl moiety to convert putrescine to spermidine and another dcSAM is used to convert spermidine to spermine. dcSAM is generated from SAM by the enzyme S-adenosylmethionine decarboxylase (AdoMetDC or AMD1) (92). uORFs encoding evolutionarily conserved peptide sequences mediate polyamine-controlled translation of both the AZIN1 and AMD1 mRNAs. Conserved uORFs in genes of the polyamine biosynthetic pathway are a recurrent feature of eukaryotic evolution (49).
AMD1 (AdoMetDC)
In vertebrates, the AMD1 mRNA contains a uORF encoding the peptide MAGDIS. The uORF is located 13-14 nts downstream of the 5’ cap (100; 101). As ribosomes are inefficient at initiating translation close to the 5’ cap, a fraction of the scanning ribosomes scans past the uORF start codon without initiating (leaky scanning) (Fig. 2, scheme 2). These ribosomes that leaky scan past the uORF can translate the AMD1 mORF. On occasion, a scanning ribosome initiates translation of the uORF, terminates, and then either disengages from the mRNA or resumes scanning (as a 40S subunit). These subunits that resume scanning will be able to reinitiate translation at the AMD1 mORF. Under conditions of high polyamines, a ribosome that translates the uORF stalls at termination (67; 96; 97) (Fig. 2, scheme 7). The peptidyl-tRNA remains bound to the ribosome paused on the stop codon. This stalled ribosome, while presumably not preventing the mRNA cap-binding protein eIF4E from associating with the cap, will, through steric occlusion, prevent additional ribosomes from loading on the mRNA and thus repress translation.
While the mechanism by which polyamines interfere with translation termination on the AMD1 uORF is not clear, insights have been obtained into the uORF determinants for translational control. First, as stated above, the amino acid, but not the nucleotide, sequence of the uORF is perfectly conserved in vertebrates. Consistent with the idea that the uORF peptide sequence is critical for polyamine regulation, missense mutations in the uORF coding region, but not synonymous codon substitutions, impair regulation both in mammalian cells (80) and in heterologous yeast (S. cerevisiae) cells (79; 80). While the MAGDIS uORF confers regulation in yeast, indicating that regulation is mediated by conserved elements within the translational apparatus, the yeast homolog of AMD1, SPE2, lacks a MAGDIS uORF in its transcript. Site-directed mutational studies revealed that the three C-terminal uORF residues, DIS, are the most critical for regulation (38), and saturation mutagenesis studies of these residues revealed an absolute requirement for Asp at position 4 and either Ile or Val at position 5 (80). Nonsense mutations at the fourth, fifth or sixth position that shorten the uORF or mutations that lengthen the uORF by one or three residues at the C-terminus or by three residues at the N-terminus weaken the inhibitory effect of the uORF on mRNA translation (38; 80). Ribosome stalling on the MAGDIS uORF is greater than on the mutant uORFs, even in the absence of added polyamines, indicating that the MAGDIS uORF is intrinsically inefficient at termination (67). As the critical C-terminal residues of the MAGDIS peptide will be in the peptidyl transferase center and the short nascent peptide will be contained within the beginning of the exit tunnel of the ribosome, these findings suggest that the nascent peptide contributes to the decreased efficiency of translation termination. Polyamines might further exacerbate the termination defect by interfering with the function of the peptide release factors (eRF1, eRF3) or other factors or ribosome constituents that contribute to translation termination.
AZIN1
Comparative sequence analyses of AZIN1 mRNAs and a subset of eukaryotic ODC mRNAs revealed the presence of a conserved upstream Conserved Coding region (uCC) in these distinct transcripts (50). In vertebrate AZIN1 mRNAs, the uCC is approximately 50 codons in length; these uCCs are, by definition, uORFs; however, they lack in-frame AUG start codons and instead initiate at the near-cognate start codon AUU. Whereas mutation of the AUU start codon of the uCC in a human AZINl-luciferase reporter mRNA to noncognate UUU resulted in constitutive derepression of luciferase repression, replacement of the AUU codon by a conventional AUG start codon lead to constitutive repression of AZIN1 mRNA translation (50). Thus, translation of the uORF impairs AZIN1 production. The wild type uCC regulates translation of the AZIN1 mORF in response to polyamines with high translation in cells depleted of polyamines and translational repression in the presence of polyamines. Multi-sequence alignments of uCCs from vertebrate AZIN1 mRNAs as well as from some invertebrate and fungal ODC mRNAs revealed that the uCC sequence conservation is highest near the C-terminus of the encoded peptide with strong conservation of the motif EPPWxPS* (* denotes a stop codon), though in many fungi this C-terminal motif is PP* (50). Frameshift mutations that shift the reading frame for the last ten codons of the uCC lead to derepression of AZIN1 reporter expression and block polyamine regulation, indicating that the amino acid sequence conservation is critical for polyamine regulation (50).
Ribosome profiling (ribo-seq) can be used to determine the positions of ribosomes on translated mRNAs and, in optimal cases, can pinpoint ribosome pauses during initiation, elongation, and termination. Applying this technique to cells depleted for polyamines or supplemented with high levels of polyamines revealed that polyamines trigger ribosome pausing on the PPW motif within the uCC with the Trp codon in the A site of the paused ribosome; an additional pause under high polyamine conditions was observed at the stop codon of the uCC (52). Mutating the PP or W of the PPW motif abolished polyamine induction of uCC translation and derepressed AZIN1 synthesis (52). These studies also provided a new connection to previous studies that showed that translation factor eIF5A was important for translation elongation through polyproline motifs (31). Specifically, eIF5A was found to be required for PPW peptide synthesis, and in addition, high polyamine levels interfered with the ability of eIF5A to stimulate PPW peptide synthesis (52).
Consistent with the findings that polyamines inhibit AZIN1 synthesis, and that translation of the uCC inhibits AZIN1 mORF translation, polyamines stimulate translation of the uCC (50). Supporting the notion that polyamines are acting through inhibition of eIF5A to regulate AZIN1 expression, inhibition of eIF5A function mimicked high polyamines and stimulated uCC translation. Notably, the ability of polyamines (or inhibition of eIF5A) stimulate uCC translation was dependent on the C-terminal amino acid sequence of the uCC, including the translation pause site (52). This paradoxical finding that the specific amino acid sequence of an uORF can stimulate translation initiation of that ORF from its upstream near-cognate start codon provided experimental evidence for a model of ribosome queuing to control translation initiation from uORFs that have weak initiation codons (Fig. 2, scheme 9).
These findings, taken together, suggest a model in which, under low polyamine conditions, most scanning ribosomes leaky scan past the near-cognate start codon of the uCC without initiating and then translate the AZIN1 mORF (Fig. 2, scheme 2). The occasional ribosome that translates the uCC under low polyamine conditions is likely to disengage from the mRNA at termination owing to the length of the uCC (~50 codons; Fig. 2, scheme 4). Under high polyamine conditions, any ribosome that initiates translation of the uCC will encounter the PPW motif. The polyamines will interfere with eIF5A function and cause the elongating ribosome to pause on the PPW motif (Fig. 2, scheme 6). Subsequent scanning ribosomes, most of which will leaky scan past the uCC start codon, will form a queue behind the elongating ribosome paused on the PPW motif. As the queue lengthens, a scanning ribosome will eventually be poised in the vicinity of the uCC start codon, increasing initiation events at that start codon (Fig. 2, scheme 9). The increased translation of the uCC will reinforce the ribosome pause resulting in a positive feedback loop that enhances uCC translation and leads to reduced AZIN1 synthesis. In support of this ribosome queuing model, inhibition of ribosome loading, which should impair queue formation, derepressed AZIN1 expression (52), presumably because the elongation pause is only temporary and once it dissipates the queued ribosomes can access the mORF.
The key features important for translational control of AZIN1, a uORF with a weak start codon and highly conserved peptide sequence that sensitizes translation to a metabolite (in this case, polyamines) is reminiscent of the fungal AAP (see above). In the latter case, Arg and the nascent uORF peptide provoke a stall that enhances initiation at the weak (poor context) start codon of the AAP, leading to further reduction in downstream translation initiation for the Arg biosynthetic enzyme. We hypothesize that similar mechanisms might account for translational control of mRNAs containing sequence conserved uORFs that initiate with weak start codons (107) and encode stalling peptides.
Mg2+-regulated uORFs in PRL-2/PTP4A2 and TRPM7 mRNAs
Two mammalian mRNAs encoding proteins linked to Mg2+ transport have been found to contain conserved uORFs that regulate translation in response to Mg2+ levels. The mRNA encoding the phosphatase of regenerating liver-2 (PRL-2, also known as PTP4A2), which associates with cyclin M (CNNM) magnesium regulators to coordinate Mg2+ homeostasis, contains an evolutionarily conserved uORF encoding a peptide of either 39 or 21 amino acids depending on whether translation initiates at the first or an internal AUG start codon (34). Mutational studies indicate that the internal AUG codon, though in poor context with a U at position −3, is the more likely start site for translation. Mutation of the second AUG codon derepresses expression of a PRL-2 reporter and also impairs Mg2+ regulation (34). Further supporting the idea that the 21 amino acid uORF mediates Mg2+ control of PRL-2 mRNA translation, mutations that scramble the amino acid sequence encoded by the uORF without affecting the start and stop codons nearly abolish Mg2+ repression of PRL-2 reporter expression, whereas synonymous codon substitutions within the uORF maintain regulation (34). Thus, the amino acid sequence of the uORF is critical for translational control. As aggregate ribosome profiling data from many studies revealed prominent ribosome occupancy near the 3’ end of the uORF (34), perhaps the Mg2+-induced pause of elongating ribosomes on the uORF controls ribosome access to the mORF. However, additional studies are needed to identify the critical residues within the uORF required to mediate Mg2+ responsiveness and to define how the uORF controls ribosome access to the PRL-2 mORF.
Like the PRL-2 mRNA, the mRNA encoding the transient receptor potential melastatin channel TRPM7, an Mg2+ transporter, contains highly conserved uORFs that confer Mg2+-regulated translation to the mORF (83). The complex mRNA structure consists of a long uORF (~ 130 codons in humans) that overlaps the mORF plus a second uORF (~20-32 codons) that is upstream of the mORF, but entirely embedded within the longer first uORF. While this uORF structure is conserved among fish, frogs, birds, and mammals, amino acid sequence conservation is greatest in the mORF and lesser in the uORFs – though high amino acid sequence conservation in the uORFs is found within mammals (83). In vitro translation assays in mammalian cell extracts revealed that the longer uORFl represses mORF translation, while uORF2 alters the Mg2+ optimum for mORF translation. Mg2+ was also found to repress TRPM7 reporter expression in vivo (83). While aggregate ribosome profiling data reveal extensive translation of the longer uORFl (for example, see https://gwips.ucc.ie/), additional studies are needed to determine whether the conserved uORF residues contribute to Mg2+ regulation of translation. As there is extensive overlap between the functions of Mg2+ and polyamines in cells, especially in translation (reviewed in 18), it is tempting to speculate that uORFs controlling synthesis of proteins involved in Mg2+ and polyamine homeostasis might show cross-regulation. Accordingly, it will be of interest to assess the role, if any, of proline-containing motifs in the TRPM7 uORFs. Moreover, it is intriguing that the biochemically similar molecules ornithine, arginine and polyamines have been linked to the uORF nascent peptide-dependent mode of translational control in bacteria, fungi, mammals and plants, perhaps revealing a propensity for these metabolites to interact with nascent peptides in the exit tunnel and inhibit ribosome peptidyl transferase activity.
eIF2α–P regulated uORFs in CHOP (DDIT3) and GADD34 (PPP1R15A) mRNAs
The integrated stress response (ISR) couples phosphorylation of the translation initiation factor eIF2α with (1) inhibition of global translation and (2) upregulated translation of a few mRNAs including those encoding stress-related transcription factors ATF4 and CHOP (DDIT3) and the phosphatase subunit GADD34 (PPP1R15A) (91; 142). Translational induction of ATF4 expression by eIF2α phosphorylation involves regulated reinitiation (reviewed in 91; 142). The ATF4 mRNA contains two uORFs, the second of which overlaps the beginning of the ATF4 mORF in an alternate reading frame. Following translation of uORF1, ribosomes resume scanning and reinitiate translation at either the inhibitory uORF2 or at the ATF4 mORF. Phosphorylation of eIF2α, by indirectly lowering eIF2 function, promotes scanning past uORF2 and thereby enhances ATF4 synthesis (regulation is not nascent peptide sequence-dependent). In contrast, the CHOP and GADD34 mRNAs each contain uORFs whose amino acid sequences are important for regulation.
In the human CHOP mRNA, the uORF contains two in-frame AUG start codons in good but not perfect contexts. The longer uORF encodes a peptide of 34 amino acids with striking amino acid sequence conservation among vertebrates over the C-terminal half of the peptide, including the near-perfect conservation of the C-terminal HHHT* motif (56; 141). Whereas mutations of the uORF start codon or missense mutations that alter the encoded peptide sequence of the uORF cause derepression of CHOP expression, synonymous codon substitutions retain regulation (56; 90). The poor context of the uORF start codon is critical for induction by eIF2α phosphorylation and ribosomal toeprinting experiments revealed that the ribosome stalls near the 3’ end of the uORF (141). Taken together, these findings have suggested a model (141; 142) in which phosphorylation of eIF2α contributes to enhanced leaky scanning of the weak uORF start codon enhancing CHOP expression, but for those ribosomes that translate the uORF the ribosomal stall contributes to the inhibitory effect of the uORF on CHOP synthesis. Whether the efficiency of stalling is itself regulated remains unknown.
The mammalian GADD34 mRNAs contain two uORFs; uORF2 is more highly conserved at the amino acid level than is uORF1, and uORF2 terminates with the nearly perfectly conserved motif PPG*. Mutating the uORF2 start codon, which is in good but not perfect context, mimicked the regulatory effect of eIF2α phosphorylation on GADD34 and caused derepression of expression (68; 143). Mutating the uORF1 start codon did not significantly impact GADD34 basal or induced expression (143). These results indicate that the conserved uORF2 principally confers eIF2α phosphorylation control of GADD34 mRNA translation. Mutations that delete or change the amino acid sequence of the conserved C-terminal half of uORF2 cause depression of GADD34 expression, while strengthening the start codon context for uORF2 represses GADD34 expression (143). Toeprinting experiments revealed a modest level of ribosome pausing near the uORF2 stop codon that was partially alleviated by mutating the C-terminal PPG* motif (143).
The common features of GADD34 and CHOP mRNA translation, including the imperfect start codon contexts that enable leaky scanning and the conserved peptide sequences that provoke elongation or termination pauses, suggest that eIF2α phosphorylation controls the translation of these mRNAs via similar mechanisms. Interestingly, a related mechanism might also be relevant for translational control of the mRNA encoding human hemojuvelin (HJV), a bone morphogenetic protein (BMP) co-receptor whose dysfunction is linked to hereditary hemochromatosis. The HJV mRNA contains a 28-codon uORF whose inhibitory function is dependent on the encoded peptide sequence and overcome by phosphorylation of eIF2α in response to iron overload (86).
Human cytomegalovirus (HCMV) gp48/UL4 uORF2
While several additional mammalian mRNAs have been found to contain conserved uORFs that impair mORF translation, we will focus on one final example from human cytomegalovirus (HCMV). The mRNA for the UL4 gene encoding the glycoprotein gp48 contains three uORFs; translation of the conserved 22-codon uORF2 is clearly responsible for repression of gp48 synthesis. Mutational analyses demonstrated that the amino acid sequence of uORF2 is critical for its inhibitory function, especially the C-terminal PP* motif (17); some internal residues also contribute to the inhibitory function of this uORF (2). Biochemical studies revealed that the ribosome stalls at the uORF2 stop codon with bound peptidyl-tRNAPro and termination factor eRF1 (8; 53), indicating that the nascent peptide impairs termination at the uORF2 stop codon. Structural studies indicated that the nascent peptide adopts an elongated conformation in the ribosome peptide exit tunnel with conserved rRNA nucleotides contacting the critical residues Pro21 and Pro22. Interestingly, a loop in ribosomal protein uL4 forms part of the constriction in the peptide exit tunnel, and the cryo-EM structures indicate that the uORF2 residue Ser12 interacts with the uL4 loop in the stalled nascent chain complex (6). Recent studies using in vitro translation assays revealed that mutations in the corresponding loop of Arabidopsis uL4 relieve ribosome stalling at the uORF2 stop codon, but not at the AMD1 MAGDIS uORF stop codon (115). These findings are consistent with a model in which the nascent peptide interactions with residues in the peptide exit tunnel can induce a conformation in the nascent peptide that exacerbates the inherently poor termination efficiency of the C-terminal PP* motif. The lack of effect of the uL4 mutation on the MAGDIS uORF is consistent with this peptide being too short to interact with the altered tunnel constriction residues during translation termination. The importance of the interaction between uL4 and the uORF2 peptide is further supported by mutations at Ser12 of the uORF2 peptide that partially relieve the repressive function of uORF2 in mammalian cell assays (2). Finally, it is noteworthy that the AUG start codon for uORF2 is in conserved poor context, which is critical for leaky scanning and the constitutively low levels of gp48 expression (8). The attributes of uORF2 (weak translation start codon context, conserved amino acid sequence, termination stall) share features with uORF2 in the CHOP mRNA described above as well with the uCC in the AZIN1 mRNA. It is tempting to speculate that cellular conditions, as yet undetermined, impact eIF2α phosphorylation, impair ribosome queuing, or modulate stalling and could lead to enhanced leaky scanning and derepression of gp48 expression.
Plant uORFs
Searches for conserved uORFs in plants
Some of the first systematic searches for uORFs conserved at the amino acid level were conducted in plants. Hayden and Jorgensen identified 58 conserved uORF containing genes belonging to 26 homology groups (35). The same group was able to add to the list and to link conserved uORFs to specific classes of plant genes. Conserved uORFs are overrepresented in transcripts for transcription factors and other regulatory genes (55). Other studies using similar techniques have identified additional conserved uORFs (113; 114; 119). The most careful and comprehensive analysis to date was performed by van der Horst et al., who identified 29 novel conserved uORFs in Arabidopsis (122). Importantly, 15 of these 29 novel uORFs are predicted to initiate from near-cognate start codons. Therefore, there are now over 100 different plant uORFs with conserved peptide sequences.
Sucrose-regulated uORFs in select bZIP mRNAs
Arguably the first and best-studied example of metabolite-controlled conserved uORF peptide-dependent translational regulation in plants is that of the Arabidopsis thaliana C/S1-group bZIP transcription factors (bZIP1, bZIP2/GBP5, ATB2/bZIP11, bZIP44, and bZIP53). These bZIP transcription factors control general metabolism as well as amino acid and sugar metabolism. Translation of their mRNAs is regulated by sucrose concentrations (134). Sucrose is the sugar most frequently transported between cells in plants (118). The 5’-leader of the ATB2/bZIP11 mRNA contains four uORFs that partially overlap with each other (99), and this 5’ leader conferred sucrose-dependent regulation to reporters. Deletion of the region including all four uORFs resulted in loss of sucrose repression (98). Subsequently, it was shown that one of the four ATB2/bZIP11 uORFs is highly conserved at the amino acid level throughout plants. The conservation is most pronounced in the 3’ region of the uORF, including its stop codon. This single conserved uORF is sufficient for sucrose regulation of ATB2/bZIP11 mRNA (134). Similarly conserved homologous uORFs are present in the 5’-leaders of bZIP1, bZIP2/GBP5, bZIP44, and bZIP53 mRNAs, and these uORFs confer sucrose regulation of the other four bZIP paralogs (132).
The ATB2/bZIP 11 regulatory uORF is initiated by an AUG codon in a conserved suboptimal context. Improving the context of the start codon makes the uORF more repressive but does not abolish sucrose regulation. Alanine-scanning mutagenesis revealed that several of the most highly conserved 14 C-terminal residues of the uORF nascent peptide are critical for sucrose regulation. The last amino acid of the uORF, a nearly universally conserved Ser, can be changed to Ala without abolishing regulation; however, changing the stop codon to a sense codon to create a 3’-extended uORF leads to loss of regulation. These findings strongly suggest that the conserved peptide affects termination in a sucrose-dependent fashion, but only if the peptide occupies a specific position in the exit tunnel of the ribosome. The same series of experiments also showed that the conserved uORF, when inserted in a 5’ leader, can confer sucrose-dependent regulation on a heterologous mRNA (94).
In vitro translation experiments have provided more detailed investigations of the mechanism of sucrose-dependent regulation by the conserved uORF of ATB2/bZIP11 mRNA (137). Sucrose was found to induce ribosome stalling during translation of the uORF. The stall is specific to sucrose as other sugars, including D-fructose, D-glucose, UDP-D-glucose, and sucrose-6-phosphate, did not induce stalling. These cell-free experiments also showed that the pause must be at the stop codon. The same study identified the sequence SFSVxFLxxLYYV near the C-terminus of the uORF nascent peptide as necessary for sucrose-induced ribosome stalling. Additionally, the stalled ribosomes in the presence of sucrose are less sensitive to release by puromycin, suggesting that the combination of the nascent peptide positioned correctly in the exit tunnel and the presence of sucrose inhibits the activity of the peptidyl transferase center during termination of uORF translation.
Ribosome profiling of A. thaliana grown under hypoxia and normoxia indicated that the ratio of ribosome footprints on the uORF and mORF of all five of the sucrose-regulated bZIP mRNAs is different under these two conditions. Under normoxia, the uORF footprint densities are relatively high and the mORF densities are relatively low (repressive conditions). This pattern is reversed under hypoxic conditions, consistent with uORF-dependent derepression of the mORF. Profiling under normoxic conditions also indicated increased ribosome density near the termination codon of the uORFs. These findings suggest that, under repressive conditions, ribosomes pause at or near the termination codon of the uORF, and that this pause is correlated with repression of mORF translation (57). However, intracellular sucrose concentrations were not measured inside cells in these experiments, so it is not clear if the derepression under hypoxia is due to lower sucrose levels in these cells or due to a different mechanism.
Polyamine regulation of plant AdoMetDC mRNA
In plants, polyamines facilitate stress tolerance. As both low and high cytoplasmic levels of polyamines have negative consequences for cells, polyamine homeostasis is tightly regulated in most eukaryotes, including plants. A key target for polyamine-regulated translational control in many eukaryotic systems is AdoMetDC (AMD1) (18). However, polyamine regulation of AdoMetDC mRNA translation in plants is distinct from the mechanism involving the MAGDIS uORF that is employed in mammalian cells as described above. The 5’-leader of plant AdoMetDC mRNAs usually contains two conserved overlapping uORFs, one “tiny” and the other “small.” The tiny uORF is typically only three codons. The small uORF is in the range of 40-65 codons and shows higher conservation at the amino acid level. The C-terminus of the small uORF always ends with the sequence PS* (49). Overexpression of the wild-type AdoMetDC mRNA in plants does not lead to the accumulation of polyamines or elevated AdoMetDC activity, presumably due to homeostatic translational autoregulation (33). By contrast, overexpression of AdoMetDC mRNA, using a construct in which the two uORFs are eliminated by mutating their respective start codons, led to a large increase of AdoMetDC enzymatic activity, disrupted polyamine homeostasis, and resulted in abnormal plant growth and development (33). Subsequent work showed that the function of the tiny uORF appears to be to prevent ribosomes from translating the small inhibitory uORF (32). Interestingly, when tested in a heterologous yeast system, the combination of removing the tiny uORF and altering the amino acid sequence of the small uORF results in derepression of the mORF (32).
The peptide encoded by the small uORF facilitates stalling at its stop codon in a polyamine and C-terminal sequence-dependent fashion in cell-free translation experiments (120). The exact mechanism through which the polyamine-induced pause at the small uORF termination codon is propagated to confer polyamine regulation is currently not understood.
Thermospermine regulation of SAC51
Thermospermine is a structural isomer of spermine. In Arabidopsis, the production of thermospermine is catalyzed by the enzyme thermospermine synthase, ACL5, that converts spermidine to thermospermine. Loss-of-function mutants of ACL5 display severe defects in stem elongation due to the overproliferation of xylem vessels (43). sac51-d is a dominant mutation in SAC51 that was identified in a screen for acl5 suppressors (44). SAC51 encodes a basic helix-loop-helix (bHLH) transcription factor. The mutation in sac51-d is a premature termination codon in one of several uORFs in the SAC51 mRNA. In the sac51-d mutant, translation of the mORF is upregulated compared to wild type, as determined by reporter experiments, suggesting that the uORF is inhibitory to downstream translation under normal conditions (44). The uORF mutated in sac51-d is highly conserved at the amino acid level in plants (35). Work in plants showed that thermospermine derepressed reporters containing the wild type SAC51 leader (46). The sac51-d mutation, as well as other mutations that inactivate the conserved uORF, caused depression even in the absence of thermospermine (46). Whether thermospermine exerts its effect directly through the uORF (during its translation) or if thermospermine works indirectly, remains to be determined. One interpretation of the results presented by Ishitsuka et al. (46) is that under low thermospermine conditions, translation of the conserved SAC51 uORF represses mORF expression. A high concentration of thermospermine might promote leaky scanning past the conserved uORF or might relieve an elongation/termination stall on the uORF.
Ascorbate-regulated uORF in GGP mRNA
The main roles of ascorbate (vitamin C) are to regulate the redox potential of cells and to serve as a cofactor in enzymes (89). The rate-limiting step in ascorbate biosynthesis in plants is catalyzed by GDP-L-Galactose phosphorylase (GGP); up- or down-regulation of GGP activity controls the amount of ascorbate in the cell. While some of the regulation of GGP is at the transcriptional level, the GGP mRNA is also translationally regulated by ascorbate in an autoregulatory loop, with high ascorbate levels resulting in repression of GGP mRNA translation. This translational regulation is mediated by a uORF that is initiated by a predicted near-cognate ACG start codon. Interestingly, the peptide encoded by the uORF is conserved throughout plants and eukaryotic algae. Ribosome profiling experiments in cells under homeostatic conditions revealed a peak of protected fragments, suggesting a ribosome pause at conserved sequences near the uORF’s C-terminus (66). These conserved sequences consist of several distinct amino acid motifs and span a distance of approximately 30 amino acids, ending with the amino acids AGGG. The position of this last motif relative to the stop codon of the GGP uCC varies, suggesting that the ribosome pauses during elongation rather than termination.
The features of the GGP uORF – a near cognate start codon, conserved peptide sequence, and translation stalling activity – resemble the uCC elements in the mammalian AZIN1 mRNA described above, raising the possibility that the GGP uORF is an ascorbate-sensing uCC. Experiments with reporters and transient transformations of A. thaliana cells indicate that the GGP uORF represses expression of the mORF and that the repression is enhanced in the presence of ascorbate. These same experiments also identified a conserved His residue, located 20 amino acids upstream of the AGGG motif, as required for both repression and ascorbate regulation. Mutating the near-cognate ACG start codon of the uORF to an AUG codon leads to constitutive repression of mORF expression (66), indicating that leaky scanning past the uORF contributes to increased initiation at the GGP start codon. Inactivation of the GGP uORF in Arabidopsis thaliana by homozygous CRISPR driven modification resulted in plants that have >70% higher levels of ascorbate, confirming the involvement of the uORF in vitamin C homeostasis (144).
Laing et al. (66) suggested a model for the mechanism of GGP regulation by the uORF. In this model, ascorbate globally lowers the stringency of start codon selection, resulting in enhanced translation of the inhibitory uORF and reduced GGP synthesis. Alternatively, the GGP uORF might function in the same manner as the uCC of mammalian AZIN1. In this alternative model, ascorbate would cause an elongating ribosome to pause during translation of the uORF. The paused ribosome could block leaky scanning to the GGP start codon, and also trigger queuing of subsequent scanning ribosomes, eventually resulting in enhanced initiation on the suboptimal ACG start codon of the GGP uORF. To date, the published data are consistent with either model.
Boron-regulated Met-Stop uORF in NIP5;1 mRNA
Boron, an essential element for plants, is toxic at high concentrations. NIP5;1 is a diffusion facilitator of boron and is required for efficient uptake of boron by plant roots. The 5’-leader of the Arabidopsis thaliana NIP5;1 mRNA contains the tiniest uORF, AUGUAA, consisting only of a start and stop codon (116). This Met-stop uORF is conserved in the mRNAs encoding the boron facilitator, at least in flowering plants. Experiments with reporters using the 5’-leader sequence from A. thaliana showed that high concentrations of boron inhibited termination on this short uORF. Insertion of additional codons between the start and stop codons abolished the regulation, suggesting boron has a unique effect on termination events that immediately follow initiation events at least in these contexts, since the nucleotide sequences surrounding the uORF were also found to contribute to boron regulation.
The enhanced termination pause on the uORF appears to have two inhibitory effects on NIP5;1 expression. First, the termination pause destabilizes NIP5;1 mRNA. Second, it results in reduced reinitiation and translation of the downstream mORF. Placing the uORF and surrounding sequences in the leader of a heterologous mRNA conferred boron regulation. Furthermore, boron regulation was also recapitulated in mammalian cell-free translation systems (116), suggesting that boron is exerting its function through evolutionarily conserved components of the translation machinery.
Other plant uORFs with conserved peptide sequences and potential regulatory roles
Translational regulation via conserved uORFs has been reported for other plant mRNAs. The Arabidopsis XIPOTL1 gene encodes a phosphoethanolamine N-methyltransferase involved in phosphocholine biosynthesis via the methylation pathway. The mRNA encoding XIPOTL1 contains a uORF conserved at the amino acid level that has been suggested to mediate phosphocholine-dependent translational repression of XIPOTL1, and this regulation is dependent on translating the conserved peptide of the uORF (1). This and other examples of conserved uORF-regulated mRNAs in plants are described in more detail in a recent review (121). Finally, an internal peptide domain in A. thaliana cystathionine γ synthase, while not a uORF, is another example of a well-studied metabolite-controlled stalling peptide discovered early (87).
Conclusions
In this review, we have tried to highlight examples of uORFs encoding conserved peptides in bacteria, fungi, mammals, and plants that control mRNA translation or mRNA levels. Unfortunately, due to length restrictions, we were not able to discuss many additional examples of conserved uORFs or to more thoroughly discuss the uORFs we highlighted, and we apologize to those whose work we were not able to include in this review. With the enhanced efforts in bioinformatic analyses, and by combining these analyses with experimental insights obtained from ribosome profiling studies, we anticipate that the family of known metabolite-responsive uORFs will grow. While, in some cases, the identification of the controlling metabolite might be deducible from the function of the mORF, this may not hold for all uORFs. Along these lines, we wonder if some uORFs that are proposed to lead to constitutive ribosome stalling may, in fact, be responding to metabolites that are in abundance in the cell. New strategies, including chemical genetic approaches, should help shed light on how metabolites could control translation through such sequence-dependent uORFs.
Literature Cited
- 1.Alatorre-Cobos F, Cruz-Ramirez A, Hayden CA, Perez-Torres CA, Chauvin AL, et al. 2012. Translational regulation of Arabidopsis XIPOTL1 is modulated by phosphocholine levels via the phylogenetically conserved upstream open reading frame 30. J Exp Bot 63:5203–21 [DOI] [PubMed] [Google Scholar]
- 2.Alderete JP, Jarrahian S, Geballe AP. 1999. Translational effects of mutations and polymorphisms in a repressive upstream open reading frame of the human cytomegalovirus UL4 gene. J Virol 73:8330–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arenz S, Bock LV, Graf M, Innis CA, Beckmann R, et al. 2016. A combined cryo-EM and molecular dynamics approach reveals the mechanism of ErmBL-mediated translation arrest. Nat Commun 7:12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ben-Zvi T, Pushkarev A, Seri H, Elgrably-Weiss M, Papenfort K, Altuvia S. 2019. mRNA dynamics and alternative conformations adopted under low and high arginine concentrations control polyamine biosynthesis in Salmonella. PLoS Genet 15:e 1007646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhushan S, Hoffmann T, Seidelt B, Frauenfeld J, Mielke T, et al. 2011. SecM-stalled ribosomes adopt an altered geometry at the peptidyl transferase center. PLoS Biol 9:e1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhushan S, Meyer H, Starosta AL, Becker T, Mielke T, et al. 2010. Structural basis for translational stalling by human cytomegalovirus and fungal arginine attenuator peptide. Mol Cell 40:138–46 [DOI] [PubMed] [Google Scholar]
- 7.Bischoff L, Berninghausen O, Beckmann R. 2014. Molecular basis for the ribosome functioning as an L-tryptophan sensor. Cell Rep 9:469–75 [DOI] [PubMed] [Google Scholar]
- 8.Cao J, Geballe AP. 1995. Translational inhibition by a human cytomegalovirus upstream open reading frame despite inefficient utilization of its AUG codon. J Virol 69:1030–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao J, Geballe AP. 1996. Inhibition of nascent-peptide release at translation termination. Mol Cell Biol 16:7109–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiba S, Ito K. 2012. Multisite ribosomal stalling: a unique mode of regulatory nascent chain action revealed for MifM. Mol Cell 47:863–72 [DOI] [PubMed] [Google Scholar]
- 11.Chiba S, Kanamori T, Ueda T, Akiyama Y, Pogliano K, Ito K. 2011. Recruitment of a species-specific translational arrest module to monitor different cellular processes. Proc Natl Acad Sci USA 108:6073–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiba S, Lamsa A, Pogliano K. 2009. A ribosome-nascent chain sensor of membrane protein biogenesis in Bacillus subtilis. EMBO J 28:3461–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cruz-Vera LR, Rajagopal S, Squires C, Yanofsky C. 2005. Features of ribosome-peptidyl-tRNA interactions essential for tryptophan induction of tna operon expression. Mol Cell 19:333–43 [DOI] [PubMed] [Google Scholar]
- 14.Cruz-Vera LR, Sachs MS, Squires CL, Yanofsky C. 2011. Nascent polypeptide sequences that influence ribosome function. Curr Opin Microbiol 14:160–6 [DOI] [PubMed] [Google Scholar]
- 15.Cruz-Vera LR, Yanofsky C. 2008. Conserved residues Asp16 and Pro24 of TnaC-tRNAPro participate in tryptophan induction of Tna operon expression. J Bacteriol 190:4791–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis R 1986. Compartmental and regulatory mechanisms in the arginine pathways of Neurospora crassa and Saccharomyces cerevisiae. Microbiol Rev 50:280–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Degnin CR, Schleiss MR, Cao J, Geballe AP. 1993. Translational inhibition mediated by a short upstream open reading frame in the human cytomegalovirus gpUL4 (gp48) transcript. J Virol 67:5514–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dever TE, Ivanov IP. 2018. Roles of polyamines in translation. J Biol Chem 293:18719–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diaz de Arce AJ, Noderer WL, Wang CL. 2018. Complete motif analysis of sequence requirements for translation initiation at non-AUG start codons. Nucleic Acids Res 46:985–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunkle JA, Xiong L, Mankin AS, Cate JH. 2010. Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc Natl Acad Sci USA 107:17152–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emmanuel JS, Sengupta A, Gordon ER, Noble JT, Cruz-Vera LR. 2019. The regulatory TnaC nascent peptide preferentially inhibits release factor 2-mediated hydrolysis of peptidyl-tRNA. J Biol Chem 294:19224–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freitag M, Dighde N, Sachs MS. 1996. A UV-induced mutation in Neurospora that affects translational regulation in response to arginine. Genetics 142:117–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujiwara K, Ito K, Chiba S. 2018. MifM-instructed translation arrest involves nascent chain interactions with the exterior as well as the interior of the ribosome. Sci Rep 8:10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaba A, Jacobson A, Sachs MS. 2005. Ribosome occupancy of the yeast CPA1 upstream open reading frame termination codon modulates nonsense-mediated mRNA decay. Mol Cell 20:449–60 [DOI] [PubMed] [Google Scholar]
- 25.Gaba A, Wang Z, Krishnamoorthy T, Hinnebusch AG, Sachs MS. 2001. Physical evidence for distinct mechanisms of translational control by upstream open reading frames. EMBO J 20:6453–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garza-Sanchez F, Janssen BD, Hayes CS. 2006. Prolyl-tRNA(Pro) in the A-site of SecM-arrested ribosomes inhibits the recruitment of transfer-messenger RNA. J Biol Chem 281:34258–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gong M, Cruz-Vera LR, Yanofsky C. 2007. Ribosome recycling factor and release factor 3 action promotes TnaC-peptidyl-tRNA dropoff and relieves ribosome stalling during tryptophan induction of tna operon expression in Escherichia coli. J Bacteriol 189:3147–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gryczan TJ, Grandi G, Hahn J, Grandi R, Dubnau D. 1980. Conformational alteration of mRNA structure and the posttranscriptional regulation of erythromycin-induced drug resistance. Nucleic Acids Res 8:6081–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gumbart J, Schreiner E, Wilson DN, Beckmann R, Schulten K. 2012. Mechanisms of SecM-mediated stalling in the ribosome. Biophys J 103:331–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gupta P, Liu B, Klepacki D, Gupta V, Schulten K, et al. 2016. Nascent peptide assists the ribosome in recognizing chemically distinct small molecules. Nat Chem Biol 12:153–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gutierrez E, Shin BS, Woolstenhulme CJ, Kim JR, Saini P, et al. 2013. eIF5A promotes translation of polyproline motifs. Mol Cell 51:35–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanfrey C, Elliott KA, Franceschetti M, Mayer MJ, Illingworth C, Michael AJ. 2005. A dual upstream open reading frame-based autoregulatory circuit controlling polyamine-responsive translation. J Biol Chem 280:39229–37 [DOI] [PubMed] [Google Scholar]
- 33.Hanfrey C, Franceschetti M, Mayer MJ, Illingworth C, Michael AJ. 2002. Abrogation of upstream open reading frame-mediated translational control of a plant S-adenosylmethionine decarboxylase results in polyamine disruption and growth perturbations. J Biol Chem 277:44131–9 [DOI] [PubMed] [Google Scholar]
- 34.Hardy S, Kostantin E, Wang SJ, Hristova T, Galicia-Vazquez G, et al. 2019. Magnesium-sensitive upstream ORF controls PRL phosphatase expression to mediate energy metabolism. Proc Natl Acad Sci U S A 116:2925–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hayden CA, Jorgensen RA. 2007. Identification of novel conserved peptide uORF homology groups in Arabidopsis and rice reveals ancient eukaryotic origin of select groups and preferential association with transcription factor-encoding genes. BMC Biol 5:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He F, Jacobson A. 2015. Nonsense-Mediated mRNA Decay: Degradation of Defective Transcripts Is Only Part of the Story. Annu Rev Genet 49:339–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herrero Del Valle A, Seip B, Cervera-Marzal I, Sacheau G, Seefeldt AC, Innis CA. 2020. Ornithine capture by a translating ribosome controls bacterial polyamine synthesis. Nat Microbiol doi: 10.1038/s41564-020-0669-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hill JR, Morris DR. 1993. Cell-specific translational regulation of S-adenosylmethionine decarboxylase mRNA: dependence on translation and coding capacity of the cis-acting upstream open reading frame. J Biol Chem 269:726–31 [PubMed] [Google Scholar]
- 39.Hinnebusch AG. 2011. Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol Mol Biol Rev 75:434–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hinnebusch AG. 2014. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem 83:779–812 [DOI] [PubMed] [Google Scholar]
- 41.Hinnebusch AG, Ivanov IP, Sonenberg N. 2016. Translational control by 5’-untranslated regions of eukaryotic mRNAs. Science 352:1413–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horinouchi S, Weisblum B. 1980. Posttranscriptional modification of mRNA conformation: mechanism that regulates erythromycin-induced resistance. Proc Natl Acad Sci USA 77:7079–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Imai A, Akiyama T, Kato T, Sato S, Tabata S, et al. 2004. Spermine is not essential for survival of Arabidopsis. FEBS Lett 556:148–52 [DOI] [PubMed] [Google Scholar]
- 44.Imai A, Hanzawa Y, Komura M, Yamamoto KT, Komeda Y, Takahashi T. 2006. The dwarf phenotype of the Arabidopsis acl5 mutant is suppressed by a mutation in an upstream ORF of a bHLH gene. Development 133:3575–85 [DOI] [PubMed] [Google Scholar]
- 45.Ishii E, Chiba S, Hashimoto N, Kojima S, Homma M, et al. 2015. Nascent chain-monitored remodeling of the Sec machinery for salinity adaptation of marine bacteria. Proc Natl Acad Sci USA 112:E5513–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ishitsuka S, Yamamoto M, Miyamoto M, Imai A, Motose H, Takahashi T. 2019. Complexity and conservationof thermospermine-responsive uORFs of SAC51 family genes in angiosperms. Front Plant Sci 10:564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ito K, Chiba S. 2013. Arrest peptides: cis-acting modulators of translation. Annu Rev Biochem 82:171–202 [DOI] [PubMed] [Google Scholar]
- 48.Ito K, Mori H, Chiba S. 2018. Monitoring substrate enables real-time regulation of a protein localization pathway. FEMS Microbiol Lett 365. [DOI] [PubMed] [Google Scholar]
- 49.Ivanov IP, Atkins JF, Michael AJ. 2010. A profusion of upstream open reading frame mechanisms in polyamine-responsive translational regulation. Nucleic Acids Res 38:353–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ivanov IP, Loughran G, Atkins JF. 2008. uORFs with unusual translational start codons autoregulate expression of eukaryotic ornithine decarboxylase homologs. Proc Natl Acad Sci USA 105:10079–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ivanov IP, Loughran G, Sachs MS, Atkins JF. 2010. Initiation context modulates autoregulation of eukaryotic translation initiation factor 1 (eIF1). Proc Natl Acad Sci USA 107:18056–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ivanov IP, Shin BS, Loughran G, Tzani I, Young-Baird SK, et al. 2018. Polyamine control of translation elongation regulates start site selection on antizyme inhibitor mRNA via ribosome queuing. Mol Cell 70:254–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Janzen DM, Frolova L, Geballe AP. 2002. Inhibition of translation termination mediated by an interaction of eukaryotic release factor 1 with a nascent peptidyl-tRNA. Mol Cell Biol 22:8562–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ji Z, Song R, Regev A, Struhl K. 2015. Many lncRNAs, 5′UTRs, and pseudogenes are translated and some are likely to express functional proteins. Elife 4:e08890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jorgensen RA, Dorantes-Acosta AE. 2012. Conserved peptide upstream open reading frames are associated with regulatory genes in angiosperms. Front Plant Sci 3:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jousse C, Bruhat A, Carraro V, Urano F, Ferrara M, et al. 2001. Inhibition of CHOP translation by a peptide encoded by an open reading frame localized in the chop 5′UTR. Nucleic Acids Res. 29:4341–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Juntawong P, Girke T, Bazin J, Bailey-Serres J. 2014. Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc Natl Acad Sci USA 111:E203–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kahana C 2018. The antizyme family for regulating polyamines. J Biol Chem 293:18730–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kannan K, Kanabar P, Schryer D, Florin T, Oh E, et al. 2014. The general mode of translation inhibition by macrolide antibiotics. Proc Natl Acad Sci USA 111:15958–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kannan K, Vazquez-Laslop N, Mankin AS. 2012. Selective protein synthesis by ribosomes with a drug-obstructed exit tunnel. Cell 151:508–20 [DOI] [PubMed] [Google Scholar]
- 61.Konan KV, Yanofsky C. 1999. Role of ribosome release in regulation of tna operon expression in Escherichia coli. J Bacteriol 181:1530–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kozak M 1987. At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J Mol Biol 196:947–50 [DOI] [PubMed] [Google Scholar]
- 63.Kozak M 1989. Context effects and inefficient initiation at non-AUG codons in eukaryotic cell-free translation systems. Mol Cell Biol 9:5073–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kozak M 2001. Constraints on reinitiation of translation in mammals. Nucleic Acids Res 29:5226–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lai CJ, Weisblum B. 1971. Altered methylation of ribosomal RNA in an erythromycin-resistant strain of Staphylococcus aureus. Proc Natl Acad Sci USA 68:856–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Laing WA, Martinez-Sanchez M, Wright MA, Bulley SM, Brewster D, et al. 2015. An upstream open reading frame is essential for feedback regulation of ascorbate biosynthesis in Arabidopsis. Plant Cell 27:772–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Law GL, Raney A, Heusner C, Morris DR. 2001. Polyamine regulation of ribosome pausing at the upstream open reading frame of S-adenosylmethionine decarboxylase. J Biol Chem 276:38036–43 [DOI] [PubMed] [Google Scholar]
- 68.Lee YY, Cevallos RC, Jan E. 2009. An upstream open reading frame regulates translation of GADD34 during cellular stresses that induce eIF2α phosphorylation. J Biol Chem 284:6661–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lin Y, May GE, Kready H, Nazzaro L, Mao M, et al. 2019. Impacts of uORF codon identity and position on translation regulation. Nucleic Acids Res 47:9358–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Loughran G, Sachs MS, Atkins JF, Ivanov IP. 2012. Stringency of start codon selection modulates autoregulation of translation initiation factor eIF5. Nucleic Acids Res 40:2898–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Luo Z, Freitag M, Sachs MS. 1995. Translational regulation in response to changes in amino acid availability in Neurospora crassa. Mol Cell Biol 15:5235–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Luo Z, Sachs MS. 1996. Role of an upstream open reading frame in mediating arginine-specific translational control in Neurospora crassa. J Bacteriol 178:2172–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Luukkonen BG, Tan W, Schwartz S. 1995. Efficiency of reinitiation of translation on human immunodeficiency virus type 1 mRNAs is determined by the length of the upstream open reading frame and by intercistronic distance. J Virol 69:4086–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mackowiak SD, Zauber H, Bielow C, Thiel D, Kutz K, et al. 2015. Extensive identification and analysis of conserved small ORFs in animals. Genome Biol 16:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martinez AK, Gordon E, Sengupta A, Shirole N, Klepacki D, et al. 2014. Interactions of the TnaC nascent peptide with rRNA in the exit tunnel enable the ribosome to respond to free tryptophan. Nucleic Acids Res 42:1245–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Martinez AK, Shirole NH, Murakami S, Benedik MJ, Sachs MS, Cruz-Vera LR. 2012. Crucial elements that maintain the interactions between the regulatory TnaC peptide and the ribosome exit tunnel responsible for Trp inhibition of ribosome function. Nucleic Acids Res 40:2247–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McGillivray P, Ault R, Pawashe M, Kitchen R, Balasubramanian S, Gerstein M. 2018. A comprehensive catalog of predicted functional upstream open reading frames in humans. Nucleic Acids Res 46:3326–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Messenguy F, Vierendeels F, Pierard A, Delbecq P. 2002. Role of RNA surveillance proteins Upf1/CpaR, Upf2 and Upf3 in the translational regulation of yeast CPA1 gene. Curr Genet 41:224–31 [DOI] [PubMed] [Google Scholar]
- 79.Mize GJ, Morris DR. 2001. A mammalian sequence-dependent upstream open reading frame mediates polyamine-regulated translation in yeast. RNA 7:374–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mize GJ, Ruan H, Low JJ, Morris DR. 1998. The inhibitory upstream open reading frame from mammalian S-adenosylmethionine decarboxylase mRNA has a strict sequence specificity in critical positions. J Biol Chem 273:32500–5 [DOI] [PubMed] [Google Scholar]
- 81.Muto H, Nakatogawa H, Ito K. 2006. Genetically encoded but nonpolypeptide prolyl-tRNA functions in the A site for SecM-mediated ribosomal stall. Mol Cell 22:545–52 [DOI] [PubMed] [Google Scholar]
- 82.Nakatogawa H, Ito K. 2001. Secretion monitor, SecM, undergoes self-translation arrest in the cytosol. Mol Cell 7:185–92 [DOI] [PubMed] [Google Scholar]
- 83.Nikonorova IA, Kornakov NV, Dmitriev SE, Vassilenko KS, Ryazanov AG. 2014. Identification of a Mg2+-sensitive ORF in the 5′-leader of TRPM7 magnesium channel mRNA. Nucleic Acids Res 42:12779–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Noderer WL, Flockhart RJ, Bhaduri A, Diaz de Arce AJ, Zhang J, et al. 2014. Quantitative analysis of mammalian translation initiation sites by FACS-seq. Mol Syst Biol 10:748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Oliver D, Norman J, Sarker S. 1998. Regulation of Escherichia coli secA by cellular protein secretion proficiency requires an intact gene X signal sequence and an active translocon. J Bacteriol 180:5240–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Onofre C, Tome F, Barbosa C, Silva AL, Romao L. 2015. Expression of human Hemojuvelin (HJV) is tightly regulated by two upstream open reading frames in HJV mRNA that respond to iron overload in hepatic cells. Mol Cell Biol 35:1376–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Onouchi H, Nagami Y, Haraguchi Y, Nakamoto M, Nishimura Y, et al. 2005. Nascent peptide-mediated translation elongation arrest coupled with mRNA degradation in the CGS1 gene of Arabidopsis. Genes Dev 19:1799–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Orbach MJ, Sachs MS, Yanofsky C. 1990. The Neurospora crassa arg-2 locus: structure and expression of the gene encoding the small subunit of arginine-specific carbamoyl phosphate synthetase. J Biol Chem 265:10981–7 [PubMed] [Google Scholar]
- 89.Paciolla C, Fortunato S, Dipierro N, Paradiso A, De Leonardis S, et al. 2019. Vitamin C in plants: from functions to biofortification. Antioxidants 8:E519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Palam LR, Baird TD, Wek RC. 2011. Phosphorylation of eIF2 facilitates ribosomal bypass of an inhibitory upstream ORF to enhance CHOP translation. J Biol Chem 286:10939–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pavitt GD, Ron D. 2012. New insights into translational regulation in the endoplasmic reticulum unfolded protein response. Cold Spring Harb Perspect Biol 4:a012278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pegg AE. 2006. Regulation of ornithine decarboxylase. J Biol Chem 281:14529–32 [DOI] [PubMed] [Google Scholar]
- 93.Pegg AE. 2016. Functions of polyamines in mammals. J Biol Chem 291:14904–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rahmani F, Hummel M, Schuurmans J, Wiese-Klinkenberg A, Smeekens S, Hanson J. 2009. Sucrose control of translation mediated by an upstream open reading frame-encoded peptide. Plant Physiol 150:1356–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rajkowitsch L, Vilela C, Berthelot K, Ramirez CV, McCarthy JE. 2004. Reinitiation and recycling are distinct processes occurring downstream of translation termination in yeast. J Mol Biol 335:71–85 [DOI] [PubMed] [Google Scholar]
- 96.Raney A, Baron AC, Mize GJ, Law GL, Morris DR. 2000. In vitro translation of the upstream open reading frame in the mammalian mRNA encoding S-adenosylmethionine decarboxylase. J Biol Chem 275:24444–50 [DOI] [PubMed] [Google Scholar]
- 97.Raney A, Law GL, Mize GJ, Morris DR. 2002. Regulated translation termination at the upstream open reading frame in S-adenosylmethionine decarboxylase mRNA. J Biol Chem 277:5988–94 [DOI] [PubMed] [Google Scholar]
- 98.Rook F, Gerrits N, Kortstee A, van Kampen M, Borrias M, et al. 1998. Sucrose-specific signalling represses translation of the Arabidopsis ATB2 bZIP transcription factor gene. Plant J 15:253–63 [DOI] [PubMed] [Google Scholar]
- 99.Rook F, Weisbeek P, Smeekens S. 1998. The light-regulated Arabidopsis bZIP transcription factor gene ATB2 encodes a protein with an unusually long leucine zipper domain. Plant Mol Biol 37:171–8 [DOI] [PubMed] [Google Scholar]
- 100.Ruan H, Hill JR, Fatemie-Nainie S, Morris DR. 1994. Cell-specific translational regulation of S-adenosylmethionine decarboxylase mRNA: influence of the structure of the 5′ transcript leader on regulation by the upstream open reading frame. J Biol Chem 269:17905–10 [PubMed] [Google Scholar]
- 101.Ruan H, Shantz LM, Pegg AE, Morris DR. 1996. The upstream open reading frame of the mRNA encoding S-adenosylmethionine decarboxylase is a polyamine-responsive translational control element. J Biol Chem 271:29576–82 [DOI] [PubMed] [Google Scholar]
- 102.Saito K, Green R, Buskirk AR. 2020. Translational initiation in E. coli occurs at the correct sites genome-wide in the absence of mRNA-rRNA base-pairing. Elife 9:e55002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schmidt MG, Rollo EE, Grodberg J, Oliver DB. 1988. Nucleotide sequence of the secA gene and secA(Ts) mutations preventing protein export in Escherichia coli. J Bacteriol 170:3404–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Simms CL, Thomas EN, Zaher HS. 2017. Ribosome-based quality control of mRNA and nascent peptides. Wiley Interdiscip Rev RNA 8: 10.1002/wrna.366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Skinner R, Cundliffe E, Schmidt FJ. 1983. Site of action of a ribosomal RNA methylase responsible for resistance to erythromycin and other antibiotics. J Biol Chem 258:12702–6 [PubMed] [Google Scholar]
- 106.Sohmen D, Chiba S, Shimokawa-Chiba N, Innis CA, Berninghausen O, et al. 2015. Structure of the Bacillus subtilis 70S ribosome reveals the basis for species-specific stalling. Nat Commun 6:6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Spealman P, Naik AW, May GE, Kuersten S, Freeberg L, et al. 2018. Conserved non-AUG uORFs revealed by a novel regression analysis of ribosome profiling data. Genome Res 28:214–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Spevak CC, Ivanov IP, Sachs MS. 2010. Sequence requirements for ribosome stalling by the arginine attenuator peptide. J Biol Chem 285:40933–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Srb AM, Horowitz NH. 1944. The ornithine cycle in Neurospora and its genetic control. J Biol Chem 154:129–39 [Google Scholar]
- 110.Steinberg R, Knupffer L, Origi A, Asti R, Koch HG. 2018. Co-translational protein targeting in bacteria. FEMS Microbiol Lett 365:fny095. [DOI] [PubMed] [Google Scholar]
- 111.Stewart V, Yanofsky C. 1985. Evidence for transcription antitermination control of tryptophanase operon expression in Escherichia coli K-12. J Bacteriol 164:731–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Svetlov MS, Plessa E, Chen CW, Bougas A, Krokidis MG, et al. 2019. High-resolution crystal structures of ribosome-bound chloramphenicol and erythromycin provide the ultimate basis for their competition. RNA 25:600–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Takahashi H, Hayashi N, Yamashita Y, Naito S, Takahashi A, et al. 2019. Comprehensive genome-wide identification of angiosperm upstream ORFs with peptide sequences conserved in various taxonomic ranges using a novel pipeline, ESUCA. bioRxiv doi: 10.1101/524090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Takahashi H, Takahashi A, Naito S, Onouchi H. 2012. BAIUCAS: a novel BLAST-based algorithm for the identification of upstream open reading frames with conserved amino acid sequences and its application to the Arabidopsis thaliana genome. Bioinformatics 28:2231–41 [DOI] [PubMed] [Google Scholar]
- 115.Takamatsu S, Ohashi Y, Onoue N, Tajima Y, Imamichi T, et al. 2020. Reverse genetics-based biochemical studies of the ribosomal exit tunnel constriction region in eukaryotic ribosome stalling: spatial allocation of the regulatory nascent peptide at the constriction. Nucleic Acids Res 48:1985–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tanaka M, Sotta N, Yamazumi Y, Yamashita Y, Miwa K, et al. 2016. The Minimum Open Reading Frame, AUG-Stop, Induces Boron-Dependent Ribosome Stalling and mRNA Degradation. Plant Cell 28:2830–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Thuriaux P, Ramos F, Pierard A, Grenson M, Wiame JM. 1972. Regulation of the carbamoylphosphate synthetase belonging to the arginine biosynthetic pathway of }I}Saccharomyces cerevisiae}i}. J Mol Biol 67:277–87 [DOI] [PubMed] [Google Scholar]
- 118.Tognetti JA, Pontis HG, Martinez-Noel GM. 2013. Sucrose signaling in plants: a world yet to be explored. Plant Signal Behav 8:e23316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tran MK, Schultz CJ, Baumann U. 2008. Conserved upstream open reading frames in higher plants. BMC Genomics 9:361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Uchiyama-Kadokura N, Murakami K, Takemoto M, Koyanagi N, Murota K, et al. 2014. Polyamine-responsive ribosomal arrest at the stop codon of an upstream open reading frame of the AdoMetDC1 gene triggers nonsense-mediated mRNA decay in Arabidopsis thaliana. Plant Cell Physiol 55:1556–67 [DOI] [PubMed] [Google Scholar]
- 121.van der Horst S, Filipovska T, Hanson J, Smeekens S. 2020. Metabolite Control of Translation by Conserved Peptide uORFs: The Ribosome as a Metabolite Multisensor. Plant Physiol 182:110–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.van der Horst S, Snel B, Hanson J, Smeekens S. 2018. Novel pipeline identifies new upstream ORFs and non-AUG initiating main ORFs with conserved amino acid sequences in the 5′ leader of mRNAs in Arabidopsis thaliana. RNA [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Vazquez-Laslop N, Thum C, Mankin AS. 2008. Molecular mechanism of drug-dependent ribosome stalling. Mol Cell 30:190–202 [DOI] [PubMed] [Google Scholar]
- 124.Wang T, Zheng X, Ji H, Wang TL, Xing XH, Zhang C. 2019. Dynamics of transcription-translation coordination tune bacterial indole signaling. Nat Chem Biol [DOI] [PubMed] [Google Scholar]
- 125.Wang Z, Fang P, Sachs MS. 1998. The evolutionarily conserved eukaryotic arginine attenuator peptide regulates the movement of ribosomes that have translated it. Mol Cell Biol 18:7528–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang Z, Gaba A, Sachs MS. 1999. A highly conserved mechanism of regulated ribosome stalling mediated by fungal arginine attenuator peptides that appears independent of the charging status of arginyl-tRNAs. J Biol Chem 274:37565–74 [DOI] [PubMed] [Google Scholar]
- 127.Wang Z, Sachs MS. 1997. Arginine-specific regulation mediated by the Neurospora crassa arg-2 upstream open reading frame in a homologous, cell-free in vitro translation system. J Biol Chem 272:255–61 [DOI] [PubMed] [Google Scholar]
- 128.Wang Z, Sachs MS. 1997. Ribosome stalling is responsible for arginine-specific translational attenuation in Neurospora crassa. Mol Cell Biol 17:4904–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wei J, Wu C, Sachs MS. 2012. The arginine attenuator peptide interferes with the ribosome peptidyl transferase center. Mol Cell Biol 32:2396–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wei J, Zhang Y, Ivanov IP, Sachs MS. 2013. The stringency of start codon selection in the filamentous fungus Neurospora crassa. J Biol Chem 288:9549–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Weisblum B 1995. Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother 39:577–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Weltmeier F, Rahmani F, Ehlert A, Dietrich K, Schutze K, et al. 2009. Expression patterns within the Arabidopsis C/S1 bZIP transcription factor network: availability of heterodimerization partners controls gene expression during stress response and development. Plant Mol Biol 69:107–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Werner M, Feller A, Messenguy F, Pierard A. 1987. The leader peptide of yeast gene CPA1 is essential for the translational repression of its expression. Cell 49:805–13 [DOI] [PubMed] [Google Scholar]
- 134.Wiese A, Elzinga N, Wobbes B, Smeekens S. 2004. A conserved upstream open reading frame mediates sucrose-induced repression of translation. Plant Cell 16:1717–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Woolhead CA, Johnson AE, Bernstein HD. 2006. Translation arrest requires two-way communication between a nascent polypeptide and the ribosome. Mol Cell 22:587–98 [DOI] [PubMed] [Google Scholar]
- 136.Wu C, Wei J, Lin PJ, Tu L, Deutsch C, et al. 2012. Arginine changes the conformation of the arginine attenuator peptide relative to the ribosome tunnel. J Mol Biol 416:518–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yamashita Y, Takamatsu S, Glasbrenner M, Becker T, Naito S, Beckmann R. 2017. Sucrose sensing through nascent peptide-mediated ribosome stalling at the stop codon of Arabidopsis bZIP11 uORF2. FEBS Lett 591:1266–77 [DOI] [PubMed] [Google Scholar]
- 138.Yang R, Cruz-Vera LR, Yanofsky C. 2009. 23S rRNA nucleotides in the peptidyl transferase center are essential for tryptophanase operon induction. J Bacteriol 191:3445–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yanofsky C 1981. Attenuation in the control of expression of bacterial operons. Nature 289:751–8 [DOI] [PubMed] [Google Scholar]
- 140.Yap MN, Bernstein HD. 2009. The plasticity of a translation arrest motif yields insights into nascent polypeptide recognition inside the ribosome tunnel. Mol Cell 34:201–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Young SK, Palam LR, Wu C, Sachs MS, Wek RC. 2016. Ribosome elongation stall directs gene-specific translation in the integrated stress response. J Biol Chem 291:6546–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Young SK, Wek RC. 2016. Upstream open reading frames differentially regulate gene-specific translation in the integrated stress response. J Biol Chem 291:16927–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Young SK, Willy JA, Wu C, Sachs MS, Wek RC. 2015. Ribosome reinitiation directs gene-specific translation and regulates the integrated stress response. J Biol Chem 290:28257–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhang H, Si X, Ji X, Fan R, Liu J, et al. 2018. Genome editing of upstream open reading frames enables translational control in plants. Nat Biotechnol [DOI] [PubMed] [Google Scholar]
- 145.Zhang J, Pan X, Yan K, Sun S, Gao N, Sui SF. 2015. Mechanisms of ribosome stalling by SecM at multiple elongation steps. Elife 4:e09684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhang Y, Sachs MS. 2015. Control of mRNA stability in fungi by NMD, EJC and CBC factors through 3′UTR introns. Genetics 200:1133–48 [DOI] [PMC free article] [PubMed] [Google Scholar]


