Abstract
The objective was to characterize the transcriptome profile of in vivo-derived female embryos competent to establish and maintain gestation. Blastocysts from superovulated heifers were bisected to generate two demi-embryos. One demi-embryo was transferred into a synchronized recipient and the other part was used for RNA-seq analysis. Data on transcript abundance was analyzed for 4 demi-embryos that established and maintained pregnancy to day 60 (designated as PP) and 3 that did not result in a pregnancy at day 30 (designated as NP). Using a FDR of P < 0.10 as cutoff, a total of 155 genes were differentially expressed between PP and NP embryos, of which 73 genes were upregulated and 82 genes were downregulated in PP group. The functional cluster with the greatest enrichment score for embryos that survived, representing 28 genes (48% of the annotated genes), was related to membrane proteins, particualry those related to olfaction and neural development and function. The functional cluster with the greatest enrichment score for downregulated genes in embryos that survived included terms related to oxidative phosphorylation, mitochondrial function, and transmembrane proteins. In conclusion, competence of in vivo-derived female bovine embryos to survive after transfer is associated with increased expression of genes encoding transmembrane proteins, perhaps indicative of differentiation of the inner cell mass to epiblast, and decreased expression of genes involved in oxidative phosphorylation, perhaps indicative of reduced metabolic activity.
Keywords: Preimplantation embryo, embryo survival, pregnancy, blastocyst, transcriptome
Introduction
One of the characteristics of pregnancy is its propensity for failure. Death of the conceptus can occur at any point during pregnancy, with the greatest incidence in the early stages of development. In dairy cattle, for example, it has been estimated that about 23 to 30% of embryos alive at day 5 to 7 of gestation die before day 27 to 32, and about 12 to 13% of embryos alive at day 28 die by day 45 to 60 of gestation (Hansen 2011, Wiltbank et al. 2016). About 2 to 6% of fetuses die between day 40 and day 80, and about 4% die thereafter (Jousan et al. 2005, Wiltbank et al. 2016). A similar phenomenon has been described in the human (Boklage 1990, Ammon Avalos et al. 2012) and pig (Vonnahme et al. 2002, Langendijk et al. 2016).
The competence of the embryo to maintain pregnancy to term has been partly determined by the time it reaches the blastocyst stage; development of embryos to this stage depends on genetic inheritance (Ledoux et al. 2015), occurrence of chromosomal abnormalities (Griffin and Ogur 2018) and non-genetic characteristics of the oocyte (Hansen et al. 2010) and sperm (Trasler et al. 1985, Esakky et al. 2016) from which the embryo is derived.. Identifying the expression of genes that are associated with embryonic competence for development after the blastocyst stage could lead to insights into genetic pathways important for embryonic development, identification of genetic loci important for genetic variation in fertility and implementation of approaches to enhance embryonic survival in agricultural or clinical settings.
In the cow, the association between gene expression and competence of the blastocyst for sustained development has been examined in a model whereby one portion of a bisected blastocyst is transferred into a recipient female and the other half is used for transcriptome analysis. These experiments, initially using microarray analysis of gene expression, have resulted in identification of gene sets that differ in expression between embryos competent to develop after term as compared to those that are not for embryos produced in vitro (El-Sayed et al. 2016) and in vivo (Salilew-Wondim et al. 2010, Ghanem et al. 2011). More recently, the same model has been used in conjunction with high-throughput RNA sequencing to assess gene expression in female blastocysts produced in vitro (Zolini et al., 2020). In that study, there were a total of 617 and 470 genes that differed in expression between embryos that survived to day 30 or 60 of gestation, respectively, versus those that did not. Many differentially-expressed genes (DEG) identified in the in vitro produced embryos that sustained pregnancies were related to cellular responses to stress.
The transcriptome profile of the blastocyst produced in vitro is different compared to that of the blastocyst produced in vivo (Smith et al. 2005, Corocoran et al. 2007, Smith et al. 2009, Driver et al. 2012). The objective of the current experiment was to identify differentially-expressed genes for in vivo produced, female blastocysts that survive after embryo transfer compared to those that fail to establish pregnancy. There are large differences in gene expression between male and female blastocysts (Bermejo-Alvarez et al. 2010). Accordingly, female embryos were used to reduce variation due to sex. Results revealed that the genes and gene pathways associated with survival of the blastocyst were largely distinct from those found earlier for the embryo produced in vitro (Zolini et al., 2020). In particular, competence of in vivo produced embryos to survive after transfer was associated with the upregulation of genes encoding transmembrane proteins and neural processes, and with downregulation of genes involved in oxidative phosphorylation.
Materials and methods
Animals
All animal procedures were approved by the University of Florida Institutional Animal Care and Use Committee. The experiment was conducted on a commercial farm located in Battle Creek MI, USA (42°25”N, 85°14” W). A total of 18 virgin Holstein heifers, 13 to 14 months of age, were used as embryo donors (n = 6) or recipients (n = 12). Donors and recipients were housed in free-stall barns with sand bedding and had ad libitum access to a total mixed ration formulated for Holstein heifers.
Production of embryos
Donor heifers were subjected to a protocol for induction of superovulation initiated at random stages of the estrous cycle. To synchonize the emergence of a new follicle wave, all visible follicles ≥ 5 mm in diameter were removed by transvaginal ultrasound-guided follicle ablation. Briefly, heifers were administered an epidural block consisting of 5 mL of 2% (w/v) lidocaine. Ovarian follicles were visualized using an ECM ExaPad ultrasound unit equipped with a 7.5 mHz microconvex probe (IMV Imaging, Rochester, MN, USA) enclosed in a plastic needle guide. Follicle ablation was performed using an 18-gauge, 5.5 cm needle (Watanabe Tecnologia Aplicada, Campinas, Brazil) attached to a metal rod. Immediately after follicle ablation, each heifer received a controlled internal drug releasing device (CIDR - Eazi-BreedTM CIDR®, Zoetis, Kalamazoo, MI, USA) placed intravaginally. Follicle superstimulation was initiated 48 h after follicular ablation and CIDR insertion with 8 i.m. injections of decreasing amounts of follicle stimulating hormone (FSH; Folltropin-V, Vetoquinol, Fort Worth, TX, USA), with 50, 40, 40, 30, 30, 20, 20 and 10 mg administered at 12 h intervals. To induce luteolysis, heifers received 25 mg of prostaglandin F2α (Lutalyse, Zoetis, Kalamazoo, MI, USA), i.m., in conjunction with each of the last two administrations of FSH. The CIDR was removed 12 h after the last injection of FSH and 100 μg of gonadorelin hydrochloride (GnRH, Factrel, Zoetis) was injected, i.m., 24 h after CIDR removal. All heifers were inseminated 12 and 24 h after GnRH with one straw of X-sexed sorted semen from a single Holstein bull (Modesty 507HO12600 - ST Genetics, Navasota, TX, USA).
Embryos were recovered by flushing the uterus 8 days after GnRH administration. Heifers were restrained and administered an epidural block consisting of 5 mL of 2% (w/v) lidocaine. A 16 Fr Foley catheter was inserted into one of the uterine horns with the cuff positioned at the base. Using gravity flow, commercial embryo recovery medium (Vigro Complete Flush, Vetoquinol, Fort Worth, TX, USA) was infused into the uterine horn and then expelled out of the uterine horn and collected into an embryo collection filter (Maxi filter, SPI™, Canton, TX, USA) attached via Y-tubing to the Foley catheter. The process was then repeated for the opposite uterine horn. Up to 500 mL of embryo recovery medium was used to flush each uterine horn. Blastocysts and expanded blastocysts were harvested and subjected to bisection.
Embryo bisection
Bisection of Grade 1 blastocysts (Robertson and Nelson, 1998) was performed as described elsewhere (Zolini et al. 2020) except that a scratch was made on the culture dish to facilitate positioning of the embryo. Briefly, a micro-blade fixed to a micromanipulator was gently pressed against the middle of the blastocyst so that trophectoderm (TE) and inner cell mass (ICM) cells were evenly distributed to both demi-embryos. The micro-blade was slowly moved forward and backward until the blastocyst was completely bisected and the zona pellucida was removed. One portion of the bisected blastocyst (the smaller or less intact portion if bisection was not uniform) was directly placed into a 0.2 mL tube and stored at −80°C until processing for RNAseq analysis. The other demi-embryo was placed into a 25 μL drop of holding medium (ViGro™ Holding Plus Media, Vetoquinol) and then loaded into a 0.25 mL embryo transfer straw and transported for 30 minutes to the farm at room temperature.
Embryo transfer and pregnancy diagnosis
Recipient heifers were subjected to an ovulation synchronization protocol consisting of 100 μg GnRH (Factrel, Zoetis), i.m., and intravaginal insertion of a CIDR on day −9 (Day 0 = day of anticipated ovulation), 25 mg prostaglandin F2α (Lutalyse, Zoetis), i.m., and CIDR removal on day −4, 25 mg prostaglandin F2α on day −3, and 100 μg GnRH on day −1. At day 7 after anticipated ovulation (8 d after the last GnRH injection), each recipient was subjected to examination of the ovaries by transrectal ultrasonography (ECM ExaPad, 7.5 MHz linear transducer) to confirm the presence of a corpus luteum. Animals received an epidural block consisting of 5 mL of 2% (w/v) lidocaine and then, randomly, a single demi-embryo was transferred into the ipsilateral uterine horn relative to the corpus luteum. Pregnancy diagnosis was carried out by transrectal ultrasonography on day 30 and 60 of gestation relative to the time of anticipated ovulation. There was no pregnancy loss between day 30 and 60. Based on pregnancy diagnosis, each embryo was classified as PP (survived to day 30 of pregnancy and continued to maintain pregnancy until at least day 60) or NP (not pregnant at day 30).
RNA extraction, cDNA library preparation and transcriptome sequencing
Total RNA from each demi-embryo was isolated using the RNeasy Micro Kit (Qiagen, Germantown, MD, USA) according to the manufacturer’s protocol. Barcoded fragment libraries were constructed using the Bovine Custom AnyDeplete Ovation SoLo RNA-Seq System from NuGEN (San Carlos, CA, USA) following the manufacturer’s protocol. DNA integrity of the libraries was assessed using the High Sensitivity D1000 ScreenTape on the Agilent 4200 TapeStation (Santa Clara, CA, USA) and DNA concentration was measured using the KAPA qPCR kit (Boston, MA, USA) following the manufacturer’s protocol. Samples were then multiplexed and sequenced with 1 × 75 bp single-end reads on an Illumina NEXT-seq (Illumina, Inc., San Diego, CA, USA) according to the manufacturer’s protocol. Raw counts were obtained using CLC Genomics 1 × 75 bp kit Workbench software (Qiagen).
Bioinformatics
Analytic procedures were performed with the limma (Ritchie et al., 2015) and edgeR (Robinson et al., 2010) packages for R software. RNA-seq data was modified with the voom function of the limma package. Three samples were considered outliers based on distribution in a multidimensional scaling plot and because the samples had the greatest proportions of zero counts. Data from these samples were excluded from the analyses. Another sample was also excluded because it was determined to represent a male embryo based on the expression of the Y-linked genes DEAD-box helicase 3 Y-linked (DDX3Y), eukaryotic translation initiation factor 1A, Y-chromosomal (EIF1AY), sex determining region Y (SRY), and ubiquitously transcribed tetratricopeptide repeat containing, Y-linked (UTY) (Chang et al. 2013). All of the retained samples expressed genes characteristically expressed in both TE [actin alpha 2, smooth muscle (ACTA2), caudal type homeobox 2 (CDX2), GATA binding protein 2 (GATA2) and GATA binding protein 3 (GATA3)] and ICM [DNA methyltransferase 3 alpha (DNMT3A), H2A.Z variant histone 1 (H2AFZ), and Nanog homeobox (NANOG)] (Negrón-Pérez et al. 2017). Genes with low expression counts (less than 1 count per million reads, n = 3536) were filtered out before normalization and a total of 21,080 transcripts were eligigble for downstream analysis. For samples remaining in the analysis, embryos were classified as PP (n = 4) or NP (n = 3) and as derived from one of four donor cows (3742, 1 pregnant cow; 3844, 1 pregnant and 2 non-pregnant; 3891, 1 pregnant and 1 non-pregnant; 3901, 1 pregnant). Number of samples meets the requirement for minimum number for RNA-Seq experiments discussed by Conesa et al. (2016).
Gene expression data were normalized through the weighted trimmed mean of M-values method, implemented with the edgeR package. Normalization factors for all samples had a mean of 1, with a minimum of 0.72 and a maximum of 1.2. The voom function of the limma package was employed to transform the data for linear modeling. This function uses variances of the model residuals, and thus the model to be fitted was specified as a previous step, considering the effect of the group (PP and NP) and the donor cow. After data transformation, a linear model using weighted least squares for each gene was fit and a contrast between PP and NP groups was performed, in order to calculate estimated coefficients and standard errors. Finally, moderated t- and F- statistics and log-odds of differential expression were computed by empirical Bayes moderation of the standard errors towards a common value. P-values were adjusted by the Benjamini & Hochberg method, which controls for the false discovery rate (FDR). The differentially-expressed genes (DEG) were defined in two ways: as those with a P-value adjusted for FDR of < 0.05 and those with a FDR of < 0.10. The P-values and the magnitude of fold change (log2FC) were visualized in a volcano plot, highlighting the up- and down-regulated DEG.
Voom-transformed expression values from the DEG with FDR < 0.10 were subjected to hierarchical clustering of the genes and samples, and to construct a heatmap for expression levels. Genes and samples were clustered using the Euclidean distance as similarity metric and centroid linkage as the clustering method, with the Cluster 3.0 software (de Hoon et al., 2004). The clusters’ tree and heatmap were visualized with with Java Treeview (Saldanha, 2004).
Functional annotation and clustering of the DEGs were determined using DAVID Bioinformatics Resources 6.8 (Dennis et al. 2003). A total of 108 of the 155 DEG were annotated. Data were analyzed using the list of all expressed transcripts from the embryos as the background. Functional annotation clusters containing one or more terms with Benjamini-Hochberg adjusted P-values < 0.05 are reported. Selected pathways showing DEG were visualized using Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (Kanehisa and Goto, 2000) and the Pathview program (Luo et al., 2017). In addition, Ingenuity Pathway Analysis (IPA, Qiagen) was used to indentify canonical pathways predicted to be activated (z ≥ 2.0) or inhibited (z ≤ 2.0), prediction of molecular, cellular or physiological functions (exclusive of disease) predicted to be be activated (z ≥ 2.0) or inhibited (z ≤ 2.0), and identification of putative upstream regulators predicted to be activated (z ≥ 2.0) or inhibited (z ≤ 2.0). The classes of upstream regulators considered from the analysis output were endogenous mammalian chemicals, cytokine, G-protein coupled receptor, growth factor, ligand-dependent nuclear receptor, transcription regulator, translational regulator, and transmembrane receptor. A total of 91 of the 155 DEG were annotated by IPA and subjected to analysis.
Results
Identification of genes differentially expressed between embryos that survived and maintained pregnancy versus those that did not
A total of 12 demi-embryos were transferred into synchronized recipients as described above. Of these, 5 demi-embryos survived and maintained gestation until day 60 (41.7%) and the remaining 7 demi-embryos did not survive until day 30 of gestation (61.3%). One demi-embryo was lost during processing. After excluding three other embryos that were considered outliers and one embryo that was male based on expression of Y-linked genes, transcript abundance was analyzed in 4 female demi-embryos that survived and maintained pregnancy to day 60 (PP) and 3 female demi-embryos that did not result in a pregnancy at day 30 (NP).
Data on effects of pregnancy status on abundance of individual transcripts are presented in Supplementary File 1 Table S1. Using a FDR < 0.10, a total of 155 DEGs between PP and NP embryos were identified of which 73 genes were upregulated and 82 genes were downregulated in the PP group (Figure 1). When a more stringent cutoff was applied ( FDR < 0.05), the total number of DEGs was reduced to 99 DEG, of which 37 genes were upregulated and 62 genes were downregulated in embryos that survived (Figure 1). Clustering of samples based on expression of DEG with FDR < 0.10 indicated a cluster of PP embryos in one group and NP embryo in the other group (Figure 2).
Fig. 1.
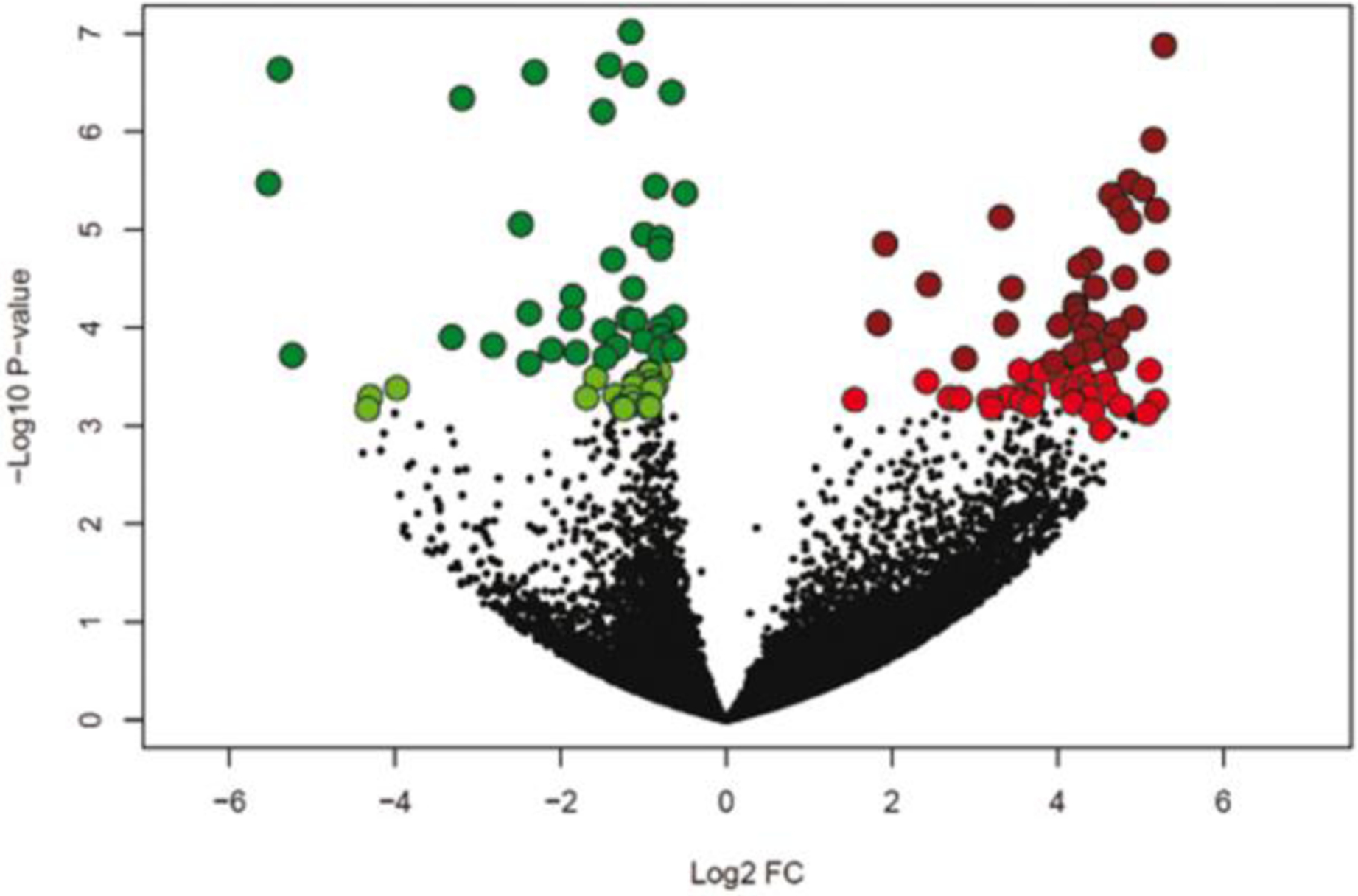
Volcano plot illustrating differentially-expressed genes using a false-discovery rate of 0.10 (light green and light red) and 0.05 (dark green and dark red). The x-axis represents the log2 fold-change of expression for blastocysts that survived to day 30 and 60 compared to those that did not survive to day 30. Upregulated genes are shown in red and downregulated genes in green.
Fig. 2.
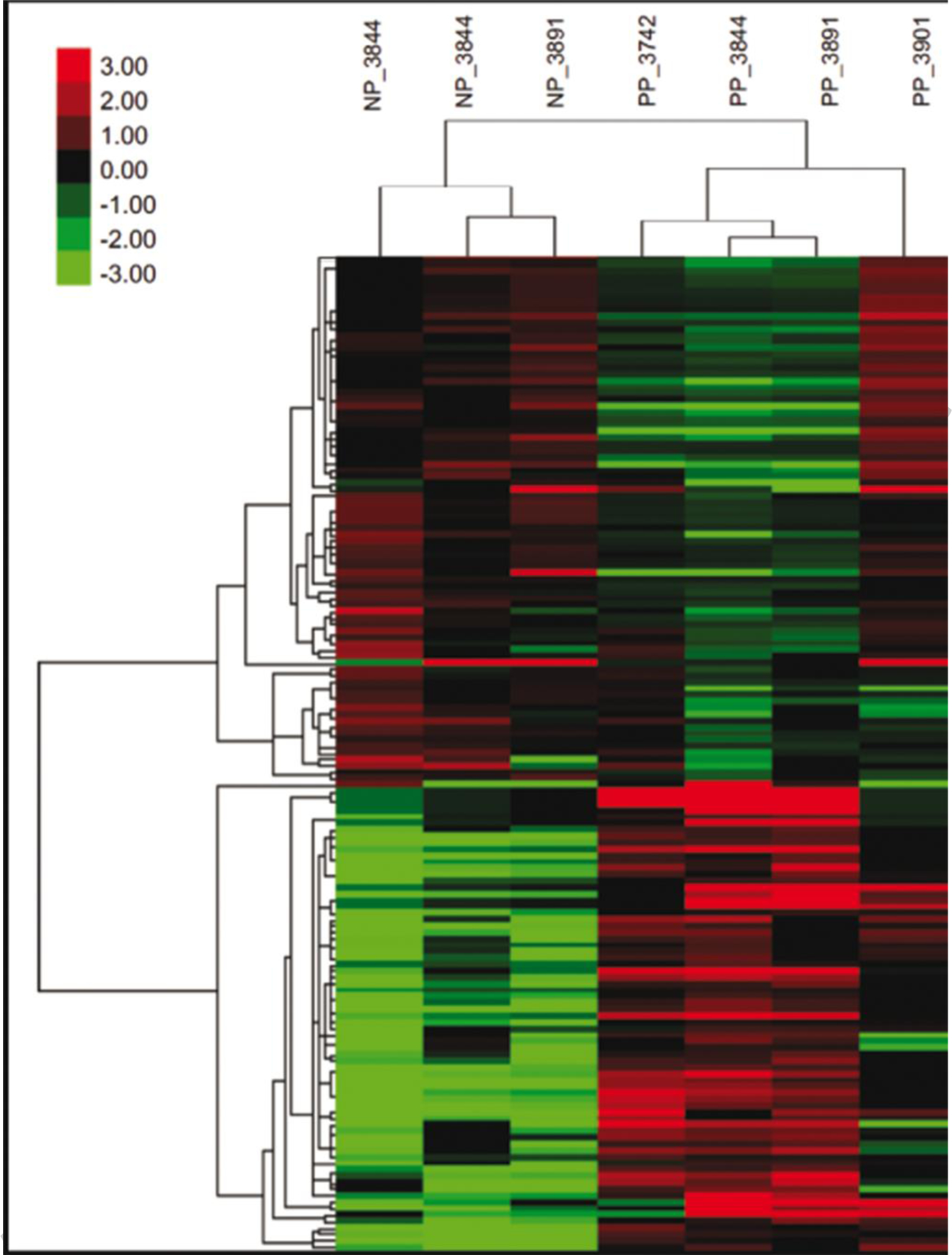
Heat map showing the expression pattern of genes that were differentially expressed between blastocysts that survived to day 30 and 60 (PP) compared to blastocysts that did not survive to day 30 (NP). Shown are the transformed and normalized expression values for each differentially-expressed gene (false discovery rate < 0.10) for each sample. The number following the PP and NP designation is the number of the donor from which the embryo was derived.
Functional annotation of DEG
Functional annotation of upregulated and downregulated DEGs (FDR < 0.10 and FDR < 0.05) was perfomed using the DAVID bioinfomatic tool.
Upregulated genes with FDR < 0.10 (58 annotated genes) were enriched in three functional clusters (Supplementary File 1, Table S2). The first annotation cluster (enrichment score = 3.19), representing 28 genes (48% of the annotated genes), was enriched for terms related to transmembrane proteins including olfactory receptors, ion-channel proteins, and cell adhesion proteins. The list of genes in this annotation cluster is provided in Figure 3. A total of 13 of the 28 genes in this cluster (46% of annotated genes) were olfactory receptor genes. Other genes, including ADGRV1, EPHA5, SCN1A, SCN2A, SORCS1, MFSD6, and UNC5C, are involved in neural development of function. Another annotation cluster was enriched for terms related to transmembrane proteins (enrichment score = 3.16) and the third term (enrichment score = 1.90) contained terms related to ion transport. There were no functional annotation terms with Benjamini-Hochberg scores with P < 0.05 when the set of upregulated genes for FDR < 0.05 (27 annotated genes) was analyzed. Nontheless, the top two clusters were for terms related to transmembrane helix and receptors and signal transduction (Supplementary File 1 Table S3).
Fig. 3.
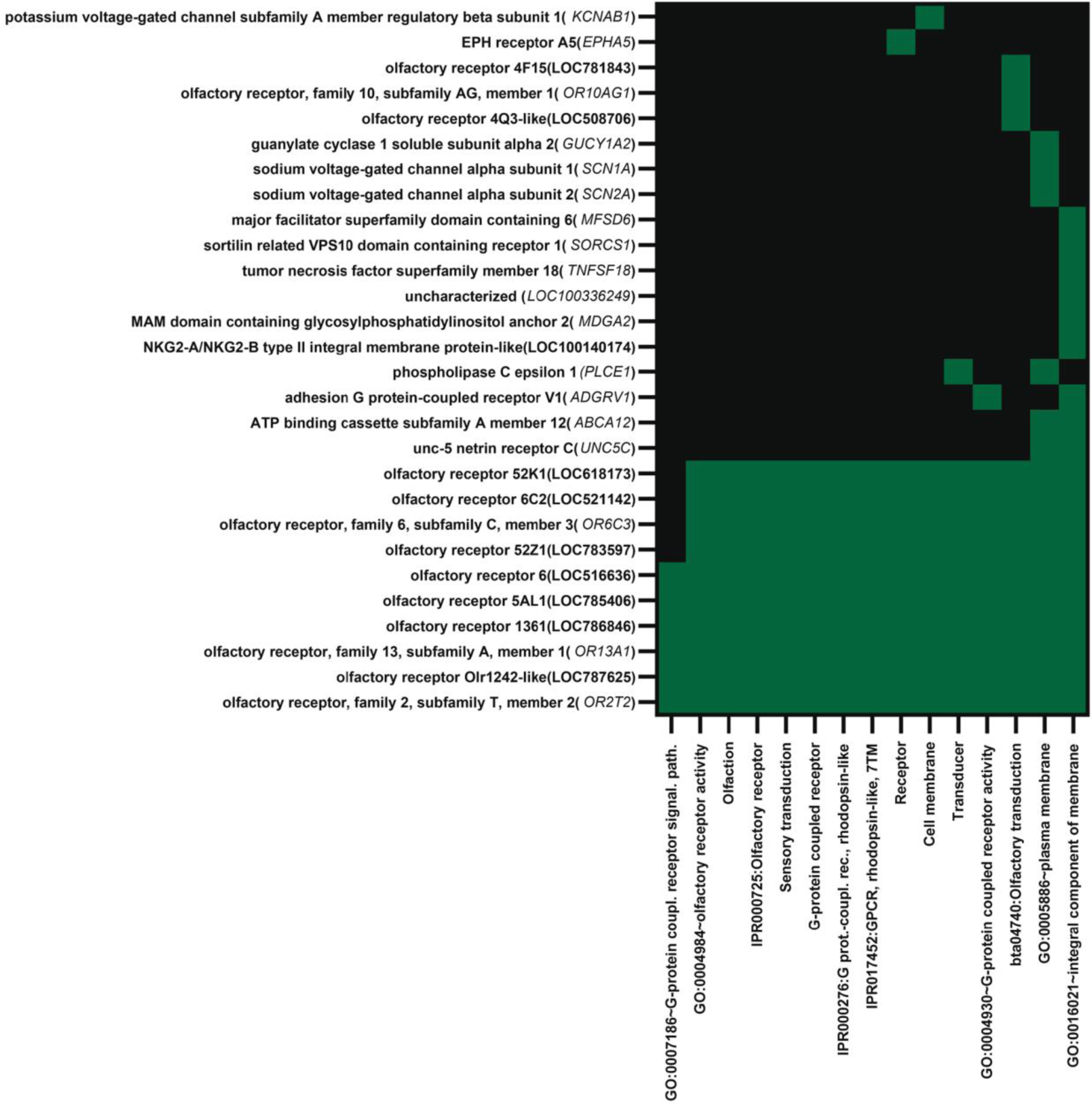
Functional annotation cluster 1 for genes upregulated in blastocysts that survived to day 30 and 60 of pregnancy as determined by analysis using DAVID. The cluster was enriched for terms related to olfaction, receptors and signal transduction. Genes located in a specific annotation are indicated by green. Note that 13 of the 28 genes in the cluster were olfactory receptors.
The downregulated genes with FDR < 0.10 (50 annotated genes) were enriched in six functional clusters (Supplementary File 1, Table S4). The first annotation cluster (enrichment score = 6.12) contained 26 genes (52% of the annotated genes) and was enriched for terms related to oxidative phosphorylation, mitochondrial function, and transmembrane proteins. The list of genes in this annotation cluster (Supplementary File 1 Table S5) include 7 genes for NADH dehydrogenase subunits, two subunits of ATP synthase F0, two cytochrome c subunits, and cytochrome B (Figure 4). The next greatest annotation cluster was enriched for terms related to ribosomal proteins (enrichment score = 2.0). When the DAVID tool was used to analyze the smaller set of downregulated genes, those with FDR < 0.05 (39 annotated genes), the functional annotation cluster related to oxidative phosphorylation and mitochondrial function had the greatest enrichment score (enrichment score = 6.76) and included 23 functional terms with Benjamini-Hochberg adjusted P-values < 0.05 (Supplementary File 1 Table S6).
Fig. 4.
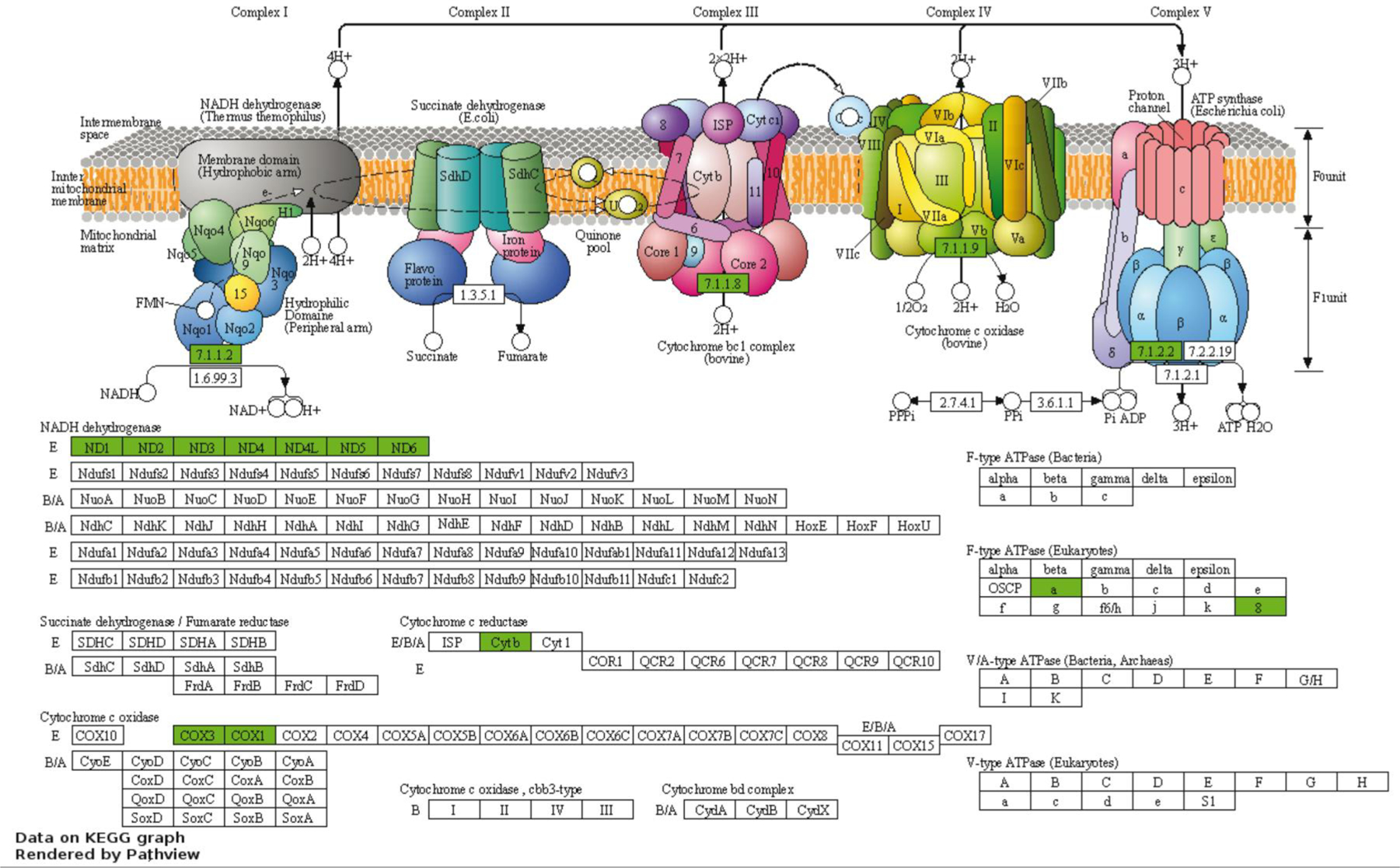
Genes present in the oxidative phosphorylation pathway (KEGG bta:00190) that were downregulated in embryos that survived transfer. Data were visualized with Pathview and are reproduced with permission of Kyoto Encyclopedia of Genes and Genomes. Genes that were downregulated in embryos that survived are shown in green. There were no upregulated genes in the pathway.
The functional annotations enriched in the set of DEG with FDR < 0.10 were also identfied by IPA. A total of 91 of the 155 genes in this dataset were annotated (Supplementary File 1 Table S7). Of these, 28 encoded for plasma membrane proteins (32% of annotated genes) including 8 classified as G-protein coupled receptors (7 olfactory receptors and ADGRV1), 4 transmembrane receptors (CLDN4, MFSD6, KLRC, and UNC5C), 5 ion channels and 9 transporters. One of the other plasma membrane genes also encodes for a receptor (EPHA5). The other major type of protein encoded for by DEG was enzyme (19 genes; 21% of annotated DEG). There were five genes encoding for transcriptional regulators, with 4 upregulated in embryos that survived after transfer (DMRT2, MXI1, RFX3, and ZBTB37) and one that was downregulated (NPM1). There were two genes for cell signaling moleceules that were both upregulated in embryos that survived, ANGPTL3 and TNFSF18.
Using IPA, the top molecular and cellular functions in which DEG were overrepresented (based on Bonferonni-Hochberg adjusted P-value) were cell-to-cell signaling and interaction (28 genes), cell death and survival (33 genes), cell morphology (12 genes), cellular assembly and organization (24 genes) and cellular compromise (12 genes). There were two canonical pathways whose activity was predicted to be changed (z ≥ 2.0 or ≤ −2.0) in embryos that survived and maintained pregnancy. Oxidative phosphorylation was predicted to be inhibited in embryos that survived to day 30 and 60 (z = −3.32). There were 11 genes involved in oxidative phosphorylation that were downregulated in embryos that survived including two cytochrome C subunits (COX1 and COX3), six genes encoding for proteins in complex I (ND1, ND2, ND3, ND4, ND4L and ND5), one gene in complex III (CYTB), two genes in complex IV (COX1 and COX3), and one gene in complex V (ATP6). The sirtuin pathway was predicted to be activated (z = 2.65; 9 genes) in embryos that survived to day 30 and 60 because of downregulation of expression of 9 genes that were largely part of the same set of genes associated with decreased oxidative phosphorylation (ATP6, CYTB, ND1, ND2, ND3, ND4, ND4L, ND5, and ND6). Exclusive of functions associated with diseases, it was predicted that DEG would lead to one change in cellular function - decreased activation of cells for embryos that survived after transfer (Figure 5).
Fig 5.
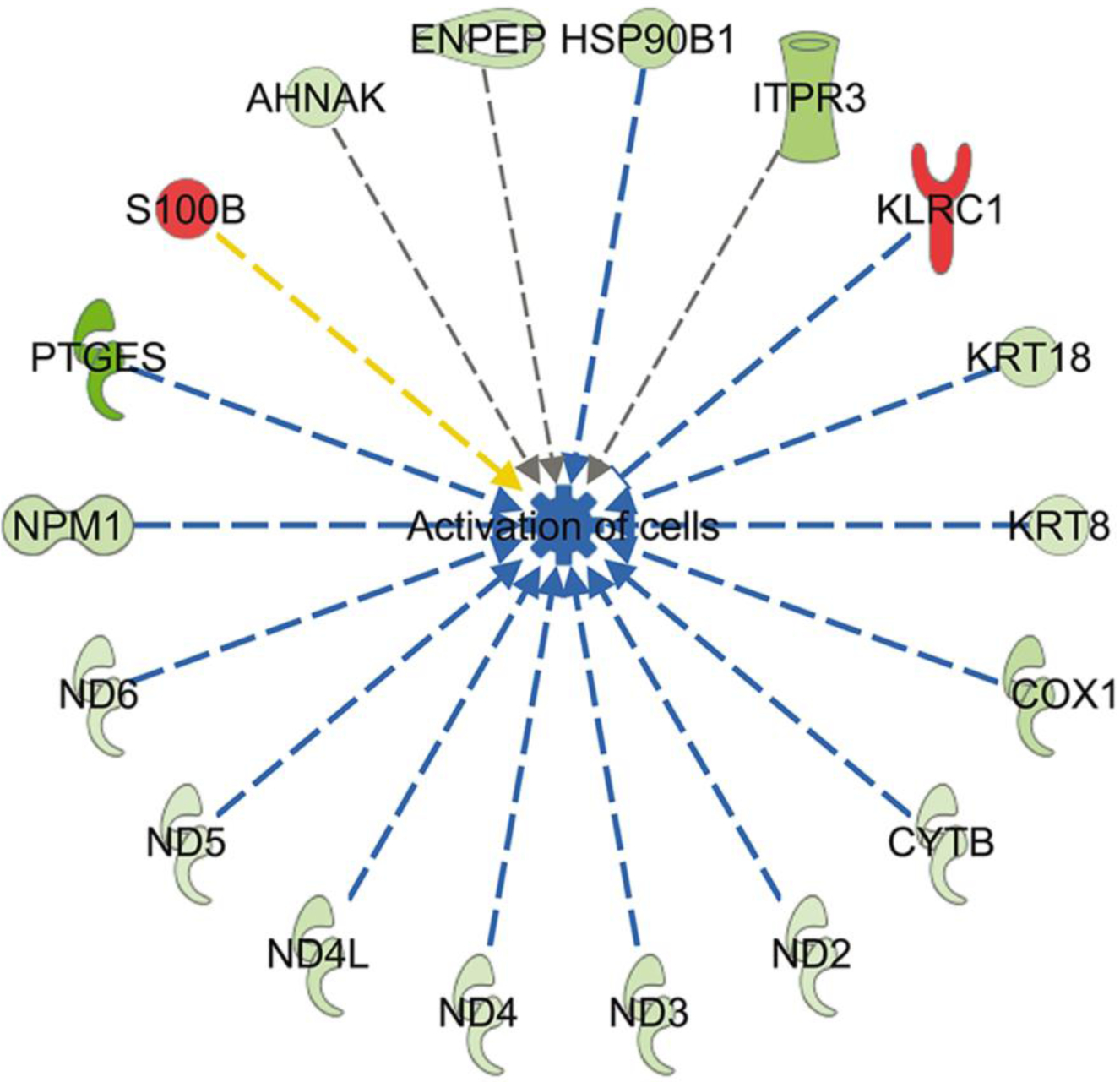
Predicted inhibition of cell activation in blastocysts that survived to day 30 and day 60 based on changes in gene expression. Genes in red were upregulated and genes in green were downregulated in embryos that survived. Blue lines represent relationships that lead to inhibition of cell activation, the yellow line represents a relationship inconsistent with inhibition of cell activation, and gray lines represent situations where the effect is not predicted.
DEG associated with ICM, TE, or hypoblast
The list of DEG were screened to identify genes associated with ICM, TE or hypoblast as described elsewhere (Negrón-Pérez et al. 2017). This analysis was performed to evaluate whether embryos that survived to day 30 and 60 exhibit characteristics of gene expression consistent with a different ratio of ICM, TE or hypoblast cells than embryos that did not survive to day 30. The only such gene that was differentially expressed was the TE gene KRT8 (FDR = 0.037), which was expressed 1.6-fold less for PP embryos. Other markers of ICM, TE, or hypoblast were not significantly different between embryo types. The fold-change difference between PP and NP embryos for genes considered epiblast markers was −1.28 for HNF4A, −1.25 for NANOG, and −1.10 for POU5F1. The fold-change difference in expression between PP and NP for genes considered TE markers was −3.92 for CDX2, 4.79 for ELF5, −1.40 for GATA2, and −1.44 for GATA3. Fold-change differences for hypoblast markers was 1.10 for GATA6, −1.94 for FGFR2 1.88 for PDGFRA, and 3.20 for SOX17.
Comparison of DEG with other studies relating gene expression to embryo survival
Using the list of DEG with FDR < 0.10 from the present study, there were no genes that were regulated in the same direction as any of the 617 DEG identified for in vitro produced embryos that established pregnancy at day 30 compared to those that did not (Zolini et al. 2020). The study of Zolini et al. (2020) used broader inclusion criteria for DEG than the current study (P<0.01 and a 2-fold or greater change in expression). If the same broad criteria were applied to the current experiment, there were only 2 genes expressed in the same direction as the 617 DEG identified by Zolini et al. (2020). In both experiments, expression of PGHDH was upregulated in embryos that established pregnancy and the unannotated gene ENSBTAG00000043649 was downregulated.
The list of expressed genes in the current experiment was also compared to a list of 34 genes that were found to be differentially expressed among in vivo derived embryos that survived to term after transfer vs those that did not (Salilew-Wondim et al., 2010). Only two genes were significantly associated with pregnancy outcome in the current experiment in the same direction as in the earlier experiment, namely EEFA1A and KRT8.
To assess the concordance between all experiments performed to assess the relationship between gene expression and embryo survival, we evaluated effects of pregnancy outcome in this experiment as well as others (El-Sayed et al., 2006; Salilew-Wondim et al., 2010; Ghanem et al., 2011; Zolini et al., 2020) on expression of 19 DEG that were found to be in common by Ghanem et al. (2011) when comparing their results to that of El-Sayed et al. (2006). Results are summarized in Figure 6. Expression of EEF1A1 was downregulated in blastocysts that survived transfer in four of five experiments and expression of ND1 was downregulated in blastocysts that survived transfer in three of five experiments with blastocysts produced in vivo. There were 16 other genes in which there was a significant effect of pregnancy outcome in the same direction in two experiments, although the most common result for these genes was similarity between results of El-Sayed et al. (2006) and Ghanem et al. (2011), who both used the same microarray platform to assess gene expression.
Fig. 6.
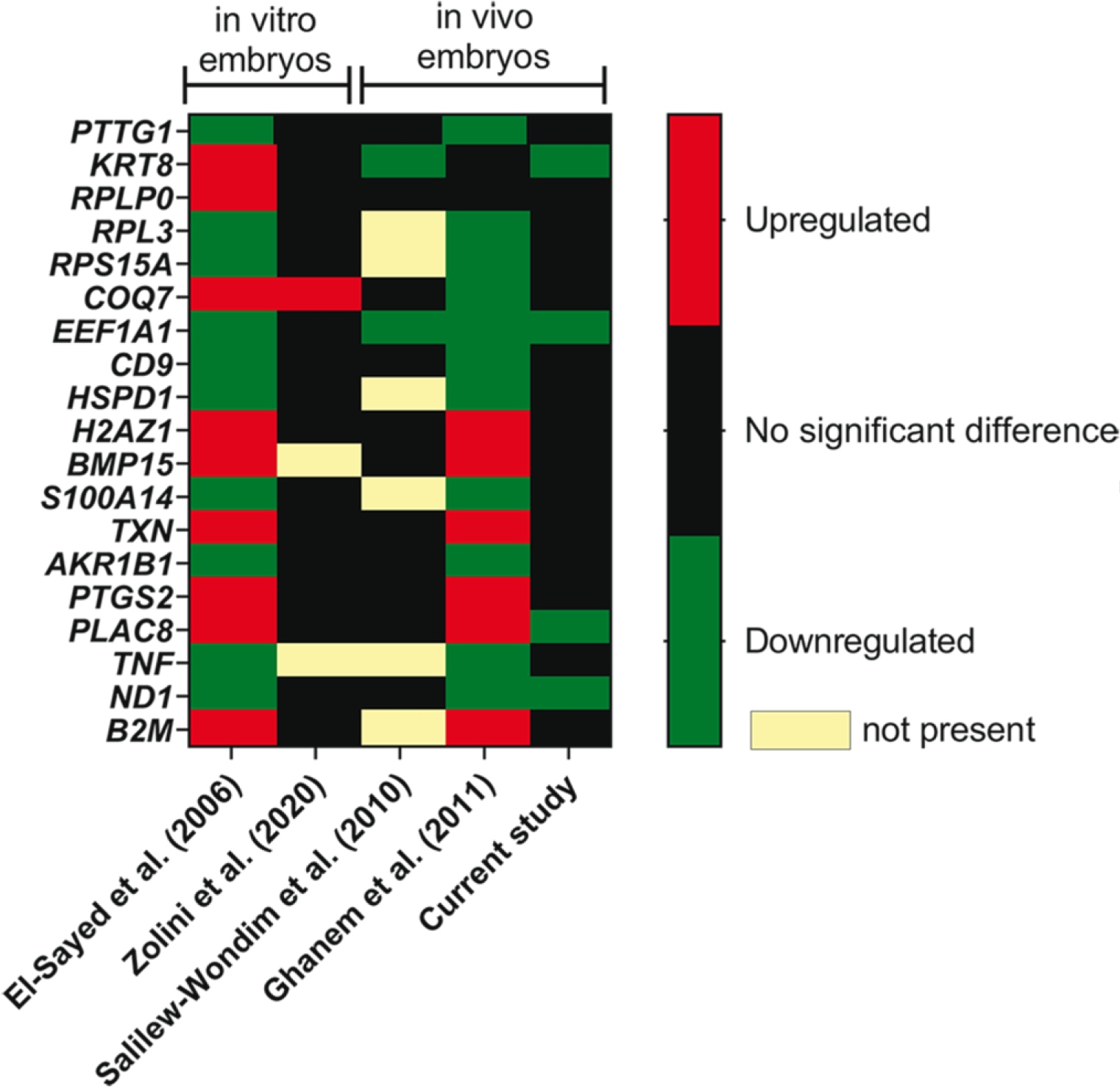
Concordance between differentially-expressed genes related to embryo survival in the present experiment with two earlier experiments that related gene expression of the blastocyst to survival of transferred embryos to term. The color code indicates whether the gene was upregulated, downregulated or not significantly associated with either calf delivery (El-Sayed et al. 2006; Ghanem et al. 2011), survival to day 30 (Zolini et al. 2020) or survival to day 30 and 60 (current study).
Colony stimulating factor 2 (CSF2) can increase the competence of embryos produced in vitro to establish pregnancy after transfer (Loureiro et al. 2019, Denicol et al. 2014). Accordingly, the set of DEG in the present experiment (FDR < 0.10) was also compared with a list of 635 genes regulated by CSF2 (P<0.01 and fold-change > 2) in embryos produced in vitro (Zolini et al. 2020). There were three genes in common between the two datasets. In particular, two non-annotated genes, ENSBTAG00000039456 and ENSBTAG00000043565, as well as EPS8L1 were downregulated in PP embryos in the current study and were also downregulated by CSF2 in the earlier experiment. When the set of genes where PP differed from NP at P< 0.01 was evaluated, there was one additional gene that was also regulated by CSF2 in the same direction. Specifically, NKAPL was upregulated in PP embryos and by CSF2.
Upstream regulators of differentially-expressed genes
Several categories of upstream regulators of DEG were evaluated by IPA. As summarized in Supplementary File 1, Table S8, a total of 7 transcriptional regulators (STAT3, PPARGC1B, MLXIPL, TFE3, PDX1, MYC and the ligand-dependent nuclear receptor AHR were predicted to be suppressed and one translational regulator ( AGO2) was predicted to be activated in embryos that survived after transfer . There was a total of 5 hormones, cytokines and growth factors predicted to be inhibited in embryos that survived after transfer, including angiotensinogen, leptin, leukemia inhibitory factor, estradiol and testosterone.
Discussion
Like for recent observations with female embryos produced in vitro (Zolini et al. 2020), the transcriptomal profile of in vivo-derived, female blastocysts competent to survive after embryo transfer was found to be different from blastocysts that did not establish pregnancy after transfer. Importantly, however, the set of genes associated with embryonic survival, and the biological functions associated with those genes, are largely dissimilar for embryos produced in vivo versus those produced in vitro. For in vivo-derived embryos, as represented schematically in Fig. 7, most of the DEG associated with embryo survival were either upregulated genes associated with transmembrane proteins, notably olfactory receptors and proteins involved in neural function and development, or were downregulated genes involved in oxidative phosphorylation. For embryos produced in vitro, in contrast, there was greater variety in the types and functions of DEG and many of the pathways associated with the DEG were related to cellular stress and DNA damage (Zolini et al. 2020). Thus, while the ability to survive the stress of cell culture is an important determinant of the subsequent fate of the embryo produced in vitro, other aspects of embryonic function and development are important for survival of the embryo that develops in vivo.
Fig. 7.
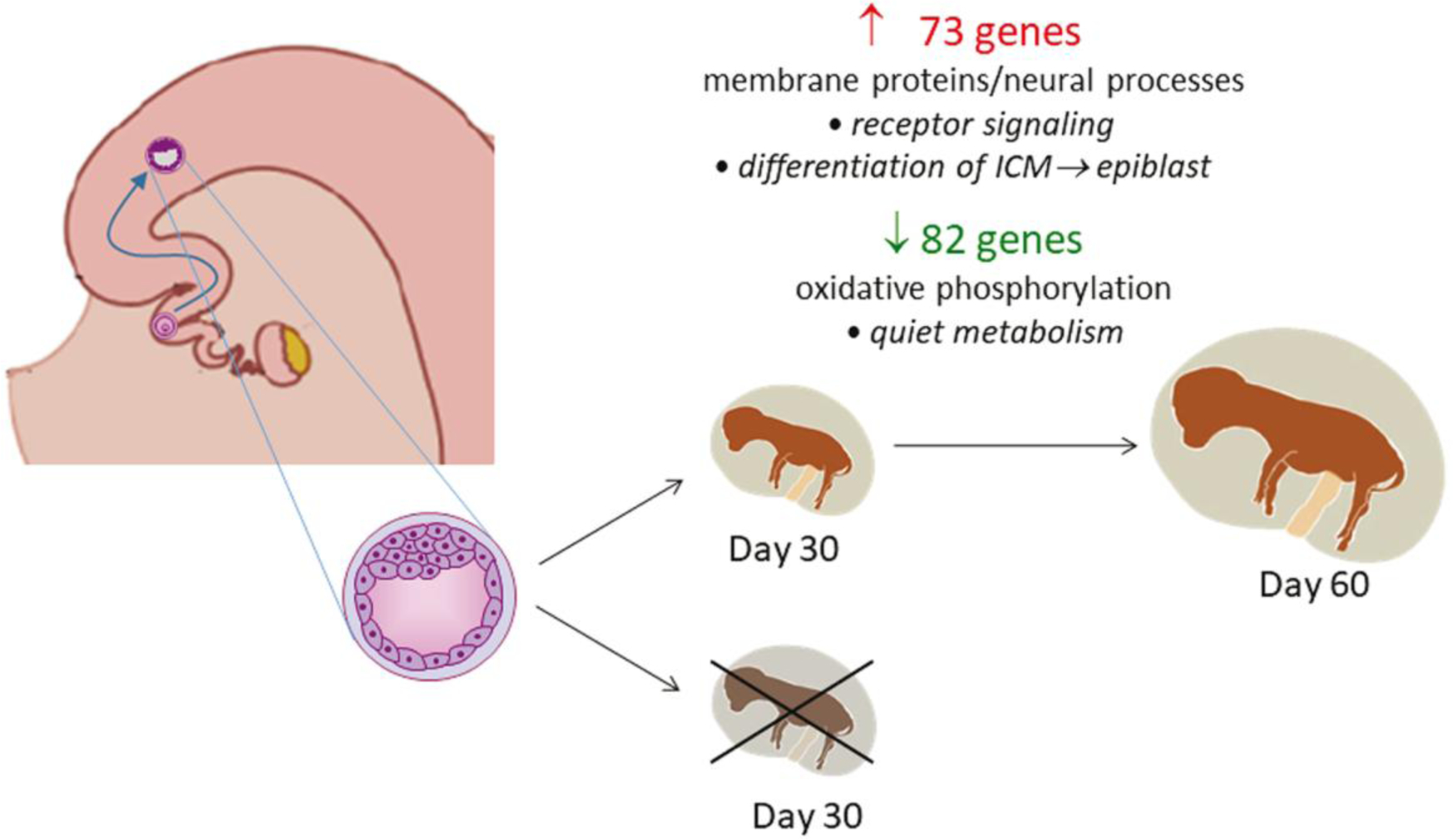
Summary of changes in gene expression in the blastocyst produced in vivo related to subsequent survival and maintenance of pregnancy to day 60. The number of upregulated (red) and downregulated (green) genes for embryos that survived after transfer are illustrated. The major functional categories represented by differentially-expressed genes is indicated under the number of genes and possible functional implications of those changes in expression are listed in italics.
In the current study, almost 50% of the genes upregulated in embryos that survived after transfer were genes encoding for transmembrane proteins, particularly olfactory receptors and proteins involved in neural development and function. Two explanations for why increased expression of these genes could facilitate successful development of the embryo are postulated. One possibility is that upregulation of specific receptor genes makes the embryo better able to participate in cell-cell interactions with the endometrium or itself or to respond to regulatory signals produced by the mother or embryo. One upregulated gene, CLDN4, is necessary for blastocyst formation in the mouse because of its role in tight junction formation (Moriwaki et al. 2007). Another upregulated gene was EPHA5. Ephrin ligands and receptors have been implicated in trophoblast attachment to the endometrium (Fujii et al. 2006). The largest cluster of upregulated DEG (13 genes) was for those involved in olfactory transduction. One receptor gene, OR52K1, probably recognizes n-aliphatic dicarboxylic acids (Lee et al. 2013) which can be detected in uterine fluid of the cow (Tríbulo et al. 2019). Olfactory receptors are not exclusively expressed in olfactory sensory neurons but are expressed in a variety of mammalian tissues where they have been implicated in the regulation of cell growth, differentiation, apoptosis, cell-cell recognition, migration and secretion (Maßberg and Hatt 2018).
The second possibility is that the abundance of upregulated genes associated with neural development and function is indicative that differentiation of epiblast from ICM is an important determinant of embryo survival. Many of the genes encoding for transmembrane proteins upregulated in embryos that survived to day 30 and 60 are involved in neural function or development. Besides the olfactory receptor genes, these include ADGRV1 (Myers et al. 2018), EPHA5 (Das et al. 2016), SCNA1A and SCNA2A (Kwong and Carr 2015), MFSD6 (Bagchi et al. 2020), SORCS1 (Ribeiro et al. 2019), and UNC5C (Poliak et al. 2015). Upregulation of genes involved in neural processes is a characteristic of differentiation of the ICM to epiblast in cynomologous monkey (Nakamura et al. 2016). In the cow, too, genes upregulated in the ICM include several associated with neural functions (Ozawa et al. 2012). It is possible that the upregulation of large numbers of genes associated with neural tissue for embryos that survive after transfer is a reflection that these embryos have more extensive differentiation of the ICM than embryos that did not survive transfer. Another observation consistent with this idea is the finding that embryos that survived to day 30 had reduced expression of NPM1. The protein encoded by this gene interacts with the pluripotency factors OCT4, SOX2 and NANOG and its downregulation in mouse embryonic stem cells can cause differentiation of mesoderm and ectoderm (Johansson and Simonsson, 2010).
As found earlier for embryos produced in vitro (Zolini et al. 2020), the genes associated with embryonic survival did not include many genes considered as markers of ICM, TE and hypoblast lineages in the blastocyst. Thus, there was no evidence that allocation of cells into these first three lineages of the blastocyst is a major determinant of embryo competence for subsequent development.
Another characteristic of embryos that survived after transfer was downregulation of genes involved in energy metabolism. There were 11 downregulated genes involved in oxidative phosphorylation in embryos that survived including CYTC, six genes encoding for proteins in complex I (ND1, ND2, ND3, ND4, ND4L and ND5), one gene in complex III (CYTB), two genes in complex IV (COX1 and COX3), and one gene in complex V (ATP6). These changes in gene expression, as well as decreased expression of genes such as the chaperone genes HSP90B1 and NPM1, the prostaglandin synthase gene PTGES, and the intermediate filament genes KRT8 and KRT18 resulted in the IPA prediction that cell activation was reduced in embryos that survived after transfer. Predicted changes in upstream regulators of gene expression, including upregulation of AGO2 involved in RNA interference (Herrera-Carrillo and Berkhout, 2017) and downregulation of a variety of transcription factors and extracellular cell signaling ligands, is also consistent with reduced metabolism in embryos that survived after transfer.
Leese and colleagues have proposed the “quiet embryo” hypothesis that states that “thrifty” embryos that do not need to have high rates of metabolism are more likely to be successful in sustaining development than embryos that require a high rate of metabolism (Leese, 2002, 2012). Such embryos would enjoy lower oxygen consumption, diminished demand for energy and nutrients, and reduced exposure to reactive oxygen species. There are data from embryo transfer experiments that are inconsistent with this hypothesis. For human embryos produced in vitro, those that established pregnancy after transfer exhibited higher consumption of glucose (Gardner et al. 2011) and oxygen in culture (Tejera et al. 2012). In the cow, there was a great deal of variation in oxygen consumption among blastocysts that developed in vivo but there was a non-significant tendency for pregnancy rate to be higher for blastocysts with higher oxygen consumption (Lopes et al. 2007). One difficulty in interpreting results of experiments measuring oxygen consumption is that embryo metabolism has never been measured in situ in the reproductive tract. Metabolism of mouse embryos changes within 3 h after removal from the reproductive tract and placement in culture (Lane and Gardner, 1998).
One must be careful when interpreting results of gene expression because changes in transcript abundance may not reflect oxidative phosphorylation activity. With that caveat, results are consistent with the idea that low expression of oxidative phosphorylation genes by the blastocyst is beneficial for its subsequent long-term survival. Moreover, present results are similar to those of earlier studies examining the relationship between gene expression and embryo survival in cattle. The gene ND1, which encodes for one the NADH dehydrogenase subunits of complex I in the electron transport chain, was less expressed for blastocysts that survived after transfer not only in the current experiment but also for earlier experiments with embryos produced in vitro (El-Sayed et al. 2006) and in vivo (Ghanem et al. 2011). In those two earlier studies, embryos that resulted in calf delivery also exhibited reduced expression of two mitochondrial transcripts that were not assessed in the current experiment.
Comparison of results between all of the experiments that identified genes whose expression in the blastocyst is related to embryo survival revealed only two that were consistently regulated differentially between blastocysts that survived as compared to those that did not. Such a result is not surprising considering the considerable variation between studies in the breed and sex of the blastocysts studied as well as in the platforms used to measure gene expression. Besides ND1, the other gene whose expression was consistently differentially regulated was EEF1A1, which was downregulated in blastocysts that survived transfer in four of the five studies. The repeatability in identification of ND1 and EEF1A1 among experiments is suggestive that these two genes are involved in processes important for embryo survival. As mentioned in the previous paragraph, ND1 may be one of the oxidative phosphorylation genes whose downregulation facilitates a metabolically-quiet phenotype. The same may be true for EEF1A1, which encodes for the alpha subunit of the elongation factor-1 complex that plays diverse roles in cell including delivery of amino-acyl tRNA to the ribsome for protein synthesis, signal transduction, organization of the cytoskeleton, and export of nuclear proteins (Sasikumar et al, 2012).
It is possible that the biopsy procedure damaged some embryos to the extent that subsequent survival was compromised. In fact, it was observed that embryos produced in vivo were more difficult to bisect than embryos produced in vitro. Thus, some of the embryos classified as not surviving to day 30 may have been competent to survive after transfer except for damage associated with biopsy. As a result, the power of the experiment to detect genes whose expression was different for embryos that survived or not may have been reduced. Probably, there are more genes associated with embryonic survival than were detected here.
In conclusion, results reported here show that the survival of the in vivo produced, female blastocyst to day 60 of pregnancy is associated with increased expression of genes encoding for transmembrane proteins, particularly those related to neural development, and decreased expression of genes involved in oxidative phosphorylation.
Supplementary Material
Table S1. Association of gene expression with pregnancy outcome.
Table S2. Functional annotation clusters for genes upregulated in embryos that established pregnancy (FDR < 0.10). Shown are annotation clusters where there was at least one term with Benjamini-Hochberg value of P < 0.05.
Table S3. Top two functional annotation clusters for genes upregulated in embryos that established pregnancy (FDR < 0.05).
Table S4. Functional annotation clusters for genes downregulated in embryos that established pregnancy (FDR < 0.10). Shown are annotation clusters where there was at least one term with Benjamini-Hochberg value of P < 0.05.
Table S5. Genes in functional annotation cluster 1 for genes downregulated in embryos that established pregnancy (FDR < 0.10).
Table S6. Functional annotation clusters for genes downregulated in embryos that established pregnancy (FDR < 0.05). Shown are annotation clusters where there was at least one term with Benjamini-Hochberg value of P < 0.05.
Table S7. Characteristics of differentially-expressed genes that were annotated by Ingenuity Pathway Analysis.
Table S8. Upstream activators predicted to be activated or inhibited in embryos that survived to day 30 or 60 as determined using Ingenuity Pathway Analysis.
Acknowledgments
The authors thank the owners and staff of Halbert Dairy Farm, Battle Creek, MI, for the use of heifers. The authors also thank Franca Rings of the University of Bonn for training with embryo bisection.
Funding
Research was supported by Zoetis Inc., NIH R01 HD088352 and the L.E. “Red” Larson Endowment.
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Compliance with ethical standards
The authors declare there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported. All animal procedures were approved by the University of Florida Institutional Animal Care and Use Committee. The owner of the animals provided written consent for their use in the experiment. There were no human subjects so informed consent was not required.
References
- Ammon Avalos L, Galindo C, Li D-K (2012) A systematic review to calculate background miscarriage rates using life table analysis. Birth Defects Res A Clin Mol Teratol 94:417–423 doi: 10.1002/bdra.23014. [DOI] [PubMed] [Google Scholar]
- Bagchi S, Perland E, Hosseini K, Lundgren J, Al-Walai N, Kheder S, Fredriksson R. (2020) Probable role for major facilitator superfamily domain containing 6 (MFSD6) in the brain during variable energy consumption. Int J Neurosci, in press. doi: 10.1080/00207454.2019.1694020. [DOI] [PubMed] [Google Scholar]
- Bermejo-Alvarez P, Rizos D, Rath D, Lonergan P & Gutierrez-Adan A (2010) Sex determines the expression level of one third of the actively expressed genes in bovine blastocysts. Proc Natl Acad Sci USA 107:3394–3399. (doi: 10.1073/pnas.0913843107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boklage CE (1990) Survival probability of human conceptions from fertilization to term. Int J Fertil 35:75–94. [PubMed] [Google Scholar]
- Chang TC, Yang Y, Retzel EF, Liu WS (2013) Male-specific region of the bovine Y chromosome is gene rich with a high transcriptomic activity in testis development. Proc Natl Acad Sci USA 110:12373–12378. doi: 10.1073/pnas.1221104110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, Madrigal P, Tarazona S, Gomez-Cabrero D, Cervera A, McPherson A, Szcześniak MW, Gaffney DJ, Elo LL, Zhang X, Mortazavi A. A survey of best practices for RNA-seq data analysis. Genome Biol 2016;17:13. doi: 10.1186/s13059-016-0881-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran D, Rizos D, Fair T, Evans ACO, Lonergan P (2007) Temporal expression of transcripts related to embryo quality in bovine embryos cultured from the two-cell to blastocyst stage in vitro or in vivo. Mol Reprod Dev 74:972–977. doi: 10.1002/mrd.20677. [DOI] [PubMed] [Google Scholar]
- Das G, Yu Q, Hui R, Reuhl K, Gale NW, Zhou R. (2016) EphA5 and EphA6: regulation of neuronal and spine morphology. Cell Biosci 6:48. doi: 10.1186/s13578-016-0115-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon MJL, Imoto S, Nolan J, Miyano S (2004) Open source clustering software. Bioinformatics 20:1453–1454. [DOI] [PubMed] [Google Scholar]
- Denicol AC, Block J, Kelley DE, Pohler KG, Dobbs KB, Mortensen CJ, Ortega MS, Hansen PJ (2014) The WNT signaling antagonist Dickkopf-1 directs lineage commitment and promotes survival of the preimplantation embryo. FASEB J 28:3975–3986. doi: 10.1096/fj.14-253112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G Jr., Shermna BT, Hosack DA, Yang J, Baseler MW, Lane HC, Lempicki RA (2003) DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4:P3. [PubMed] [Google Scholar]
- Driver AM, Peñagaricano F, Huang W, Ahmad KR, Hackbart KS, Wiltbank MC, Khatib H (2012) RNA-Seq analysis uncovers transcriptomic variations between morphologically similar in vivo- and in vitro-derived bovine blastocysts. BMC Genomics 13:118. doi: 10.1186/1471-2164-13-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sayed A, Hoelker M, Rings F, Salilew D, Jennen D, Tholen E, Sirard MA, Schellander K, Tesfaye D (2006) Large-scale transcriptional analysis of bovine embryo biopsies in relation to pregnancy success after transfer to recipients. Physiol Genomics 28:84–96. doi: 10.1152/physiolgenomics.00111.2006. [DOI] [PubMed] [Google Scholar]
- Esakky P, Hansen DA, Drury AM, Felder P, Cusumano A, Moley KH (2016) Paternal exposure to cigarette smoke condensate leads to reproductive sequelae and developmental abnormalities in the offspring of mice. Reprod Toxicol 65:283–294. doi: 10.1016/j.reprotox.2016.08.017. [DOI] [PubMed] [Google Scholar]
- Fujii H, Tatsumi K, Kosaka K, Yoshioka S, Fujiwara H, Fujii S. (2006) Eph-ephrin A system regulates murine blastocyst attachment and spreading. Dev Dyn 235:3250–3258. [DOI] [PubMed] [Google Scholar]
- Gardner DK, Wale PL, Collins R, Lane M. (2011) Glucose consumption of single post-compaction human embryos is predictive of embryo sex and live birth outcome. Hum Reprod 26:1981–1986. doi: 10.1093/humrep/der143. [DOI] [PubMed] [Google Scholar]
- Ghanem N, Salilew-Wondim D, Gad A, Tesfaye D, Phatsara C, Tholen E, Looft C, Schellander K, Hoelker M (2011) Bovine blastocysts with developmental competence to term share similar expression of developmentally important genes although derived from different culture environments. Reproduction 142:551–564. doi: 10.1530/REP-10-0476. [DOI] [PubMed] [Google Scholar]
- Griffin DK, Ogur C (2018) Chromosomal analysis in IVF: just how useful is it? Reproduction 156:F29–F50. doi: 10.1530/REP-17-0683. [DOI] [PubMed] [Google Scholar]
- Hansen PJ (2011) Challenges to fertility in dairy cattle: from ovulation to the fetal stage of pregnancy. Rev Brasil Reprod Anim 35:229–238. [Google Scholar]
- Hansen PJ, Block J, Loureiro B, Bonilla L, Hendricks KEM (2010) Effects of gamete source and culture conditions on the competence of in vitro-produced embryos for post-transfer survival in cattle. Reprod Fertil Dev 22:59–66. doi: 10.1071/RD09212. [DOI] [PubMed] [Google Scholar]
- Herrera-Carrillo E, Berkhout B. (2017) Dicer-independent processing of small RNA duplexes: mechanistic insights and applications. Nucleic Acids Res 45:10369–10379. doi: 10.1093/nar/gkx779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson H, Simonsson S (2010) Core transcription factors, Oct4, Sox2 and Nanog, individually form complexes with nucleophosmin (Npm1) to control embryonic stem (ES) cell fate determination. Aging 2:815–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jousan FD, Drost M, Hansen PJ (2005) Factors associated with early and mid-to-late fetal loss in lactating and nonlactating Holstein cattle in a hot climate. J Anim Sci 83:1017–1022. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S.(2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong K, Carr MJ. (2015) Voltage-gated sodium channels. Curr Opin Pharmacol 22:131–139. doi: 10.1016/j.coph.2015.04.007. [DOI] [PubMed] [Google Scholar]
- Lane M, Gardner DK. (1998) Amino acids and vitamins prevent culture-induced metabolic perturbations and associated loss of viability of mouse blastocysts. Hum Reprod 13:991–977. [DOI] [PubMed] [Google Scholar]
- Langendijk P, Chen TY, Athorn RZ, Bouwman EG (2016) Embryonic survival at day 9, 21 and 35 of pregnancy in intact and unilaterally oviduct ligated multiparous sows. Animal 10: 1336–1341. doi: 10.1017/S175173111600029X. [DOI] [PubMed] [Google Scholar]
- Ledoux D, Ponsart C, Grimard B, Gatien J, Deloche MC, Fritz S, Lefebvre R, Humblot P (2015) Sire effect on early and late embryonic death in French Holstein cattle. Animal 9:766–774. doi: 10.1017/S1751731114003140. [DOI] [PubMed] [Google Scholar]
- Lee K, Nguyen DT, Choi M, Cha SY, Kim JH, Dadi H, Seo HG, Seo K, Chun T, Park C (2013) Analysis of cattle olfactory subgenome: the first detail study on the characteristics of the complete olfactory receptor repertoire of a ruminant. BMC Genomics 14:596. doi: 10.1186/1471-2164-14-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leese HJ (2002) Quiet please: do not disturb. A hypothesis of embryo metabolism and viability. BioEssays 24:845–849. [DOI] [PubMed] [Google Scholar]
- Leese HJ. (2012) Metabolism of the preimplantation embryo: 40 years on. Reproduction 143:417–427. doi: 10.1530/REP-11-0484. [DOI] [PubMed] [Google Scholar]
- Lopes AS, Madsen SE, Ramsing NB, Løvendahl P, Greve T, Callesen H. (2007) Investigation of respiration of individual bovine embryos produced in vivo and in vitro and correlation with viability following transfer. Hum Reprod 22:558–566. [DOI] [PubMed] [Google Scholar]
- Loureiro B, Bonilla L, Block J, Fear JM, Bonilla AQS, Hansen PJ (2009) Colony-stimulating factor 2 (CSF-2) improves development and posttransfer survival of bovine embryos produced in vitro. Endocrinology 150:5046–5054. doi: 10.1210/en.2009-0481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Pant G, Bhavnasi YK, Blanchard SG, Brouwer C. (2017) Pathview Web: user friendly pathway visualization and data integration. Nucleic Acids Res, Web Server issue, doi: 10.1093/nar/gkx372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maßberg D, Hatt H (2018) Human olfactory receptors: novel cellular functions outside of the nose. Physiol Rev 98:1739–1763. doi: 10.1152/physrev.00013.2017. [DOI] [PubMed] [Google Scholar]
- Moriwaki K, Tsukita S, Furuse M (2007) Tight junctions containing claudin 4 and 6 are essential for blastocyst formation in preimplantation mouse embryos. Dev Biol 312:509–522. [DOI] [PubMed] [Google Scholar]
- Myers KA, Nasioulas S, Boys A, McMahon JM, Slater H, Lockhart P, Sart DD, Scheffer IE (2018) ADGRV1 is implicated in myoclonic epilepsy. Epilepsia 59:381–388. doi: 10.1111/epi.13980. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Okamoto I, Sasaki K, Yabuta Y, Iwatani C, Tsuchiya H, Seita Y, Nakamura S, Yamamoto T, Saitou M (2016) A developmental coordinate of pluripotency among mice, monkeys and humans. Nature 537:57–562. doi: 10.1038/nature19096. [DOI] [PubMed] [Google Scholar]
- Negrón-Pérez VM, Zhang Y, Hansen PJ (2017) Single-cell gene expression of the bovine blastocyst. Reproduction 154:627–644. doi: 10.1530/REP-17-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa M, Sakatani M, Yao J, Shanker S, Yu F, Yamashita R, Wakabayashi S, Nakai K, Dobbs KB, Sudano MJ, Farmerie WG, Hansen PJ (2012) Global gene expression of the inner cell mass and trophectoderm of the bovine blastocyst. BMC Dev Biol 12:33. doi: 10.1186/1471-213X-12-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliak S, Morales D, Croteau LP, Krawchuk D, Palmesino E, Morton S, Cloutier JF, Charron F, Dalva MB, Ackerman SL, Kao TJ, Kania A (2015) Synergistic integration of Netrin and ephrin axon guidance signals by spinal motor neurons. Elife 4:pii: e10841. doi: 10.7554/eLife.10841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro LF, Verpoort B, Nys J, Vennekens KM, Wierda KD, de Wit J (2019) SorCS1-mediated sorting in dendrites maintains neurexin axonal surface polarization required for synaptic function. PLoS Biol 17:e3000466. doi: 10.1371/journal.pbio.3000466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47. doi: 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson I, Nelson RE (1998) Certification of the embryos In: Stringfellow DA, Seidel SM (eds), Manual of the International Embryo Transfer Society. Savoy, IL, USA: pp. 103–134. [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha AJ (2004) Java Treeview – extensible visualization of microarray data. Bioinformatics 20:3246–3248. [DOI] [PubMed] [Google Scholar]
- Salilew-Wondim D, Hölker M, Rings F, Ghanem N, Ulas-Cinar M, Peippo J, Tholen E, Looft C, Schellander K, Tesfaye D (2010) Bovine pretransfer endometrium and embryo transcriptome fingerprints as predictors of pregnancy success after embryo transfer. Physiol Genomics 42:201–218. doi: 10.1152/physiolgenomics.00047.2010. [DOI] [PubMed] [Google Scholar]
- Sasikumar AN, Perez WB, Kinzy TG. (2012) The many roles of the eukaryotic elongation factor 1 complex. Wiley Interdiscip Rev RNA 3:543–555. doi: 10.1002/wrna.1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SL, Everts RE, Sung LY, Du F, Page RL, Henderson B, Rodriguez-Zas SL, Nedambale TL, Renard JP, Lewin HA, Yang X, Tian XC (2009) Gene expression profiling of single bovine embryos uncovers significant effects of in vitro maturation, fertilization and culture. Mol Reprod Dev 76:38–47. doi: 10.1002/mrd.20927. [DOI] [PubMed] [Google Scholar]
- Smith SL, Everts RE, Tian XC, Du F, Sung LY, Rodriguez-Zas SL, Jeong BS, Renard JP, Lewin HA, Yang X (2005) Global gene expression profiles reveal significant nuclear reprogramming by the blastocyst stage after cloning. Proc Natl Acad Sci USA 102:17582–17587. doi: 10.1073/pnas.0508952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tejera A, Herrero J, Viloria T, Romero JL, Gamiz P, Meseguer M (2012) Time-dependent O2 consumption patterns determined optimal time ranges for selecting viable human embryos. Fertil Steril 98:849–857.e1–3. doi: 10.1016/j.fertnstert.2012.06.040. [DOI] [PubMed] [Google Scholar]
- Trasler JM, Hales BF, Robaire B (1985) Paternal cyclophosphamide treatment of rats causes fetal loss and malformations without affecting male fertility. Nature 316:144–146. [DOI] [PubMed] [Google Scholar]
- Tríbulo P, Balzano-Nogueira L, Conesa A, Siqueira LG, Hansen PJ (2019) Changes in the uterine metabolome of the cow during the first 7 days after estrus. Mol Reprod Dev 86:75–87. doi: 10.1002/mrd.23082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonnahme KA, Wilson ME, Foxcroft GR, Ford SP (2002) Impacts on conceptus survival in a commercial swine herd. J Anim Sci 80:553–559. [DOI] [PubMed] [Google Scholar]
- Wiltbank MC, Baez GM, Garcia-Guerra A, Toledo MZ, Monteiro PLJ, Melo LF, Ochoa JC, Santos JEP, Sartori R (2016) Pivotal periods for pregnancy loss during the first trimester of gestation in lactating dairy cows. Theriogenology 86:239–253. doi: 10.1016/j.theriogenology.2016.04.037. [DOI] [PubMed] [Google Scholar]
- Zolini AM, Block J, Rabaglino MB, Tribulo P, Hoelker M, Rincon G, Bromfield JJ, Hansen PJ (2020) Molecular fingerprint of female bovine embryos produced in vitro with high competence to establish and maintain pregnancy. Biology of Reproduction 102:292–305. doi: 10.1093/biolre/ioz190. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Association of gene expression with pregnancy outcome.
Table S2. Functional annotation clusters for genes upregulated in embryos that established pregnancy (FDR < 0.10). Shown are annotation clusters where there was at least one term with Benjamini-Hochberg value of P < 0.05.
Table S3. Top two functional annotation clusters for genes upregulated in embryos that established pregnancy (FDR < 0.05).
Table S4. Functional annotation clusters for genes downregulated in embryos that established pregnancy (FDR < 0.10). Shown are annotation clusters where there was at least one term with Benjamini-Hochberg value of P < 0.05.
Table S5. Genes in functional annotation cluster 1 for genes downregulated in embryos that established pregnancy (FDR < 0.10).
Table S6. Functional annotation clusters for genes downregulated in embryos that established pregnancy (FDR < 0.05). Shown are annotation clusters where there was at least one term with Benjamini-Hochberg value of P < 0.05.
Table S7. Characteristics of differentially-expressed genes that were annotated by Ingenuity Pathway Analysis.
Table S8. Upstream activators predicted to be activated or inhibited in embryos that survived to day 30 or 60 as determined using Ingenuity Pathway Analysis.


