Abstract
Breast cancer (BCa) proliferates within a complex, three-dimensional microenvironment amid heterogeneous biochemical and biophysical cues. Understanding how mechanical forces within the tumor microenvironment (TME) regulate BCa phenotype is of great interest. We demonstrate that mechanical strain enhanced the proliferation and migration of both estrogen receptor (ER)+ and triple-negative (TNBC) human and mouse BCa cells. Furthermore, a critical role for exosomes derived from cells subjected to mechanical strain in these pro-tumorigenic effects was identified. Exosome production by TNBC cells increased upon exposure to oscillatory strain (OS), which correlated with elevated cell proliferation. Using a syngeneic, orthotopic mouse model of TNBC, we identified that preconditioning BCa cells with OS significantly increased tumor growth and myeloid-derived suppressor cells (MDSCs) and M2 macrophages at the TME. This pro-tumorigenic myeloid cell enrichment also correlated with a decrease in CD8+ T cells. An increase in PD-L1+ exosome release from BCa cells following OS supported additive T cell inhibitory functions in the TME. The role of exosomes in MDSC and M2 macrophage was confirmed in vivo by cytotracking fluorescent exosomes, derived from labeled 4T1.2 cells, preconditioned with oscillatory strain. Additionally, in vivo internalization and intratumoral localization of tumor-cell derived exosomes was observed within MDSCs, M2 macrophages, and CD45-negative cell populations following direct injection of fluorescently-labeled exosomes. Our data demonstrate that exposure to mechanical strain promotes invasive and pro-tumorigenic phenotypes in BCa cells, indicating that mechanical strains can impact the growth and proliferation of cancer cell, alters exosome production by BCa, and induces immunosuppression in the TME by dampening anti-tumor immunity.
Keywords: mechanical strain, breast cancer cells, exosomes, immunosuppression, tumor microenvironment
Introduction
Triple-negative breast cancer (TNBC), characterized by the lack of estrogen and progesterone receptors and the absence of HER2 overexpression, represents a breast cancer (BCa) subset that demonstrates aggressive clinical behavior and poor prognosis with limited options for targeted intervention1. The lack of effective therapeutic approaches for the treatment of TNBC underscores the impending need for a deeper understanding of the complex molecular and biophysical processes involved in disease progression. Mechanical forces within the breast tumor microenvironment (TME) are potent regulators of cancer progression. As breast tumors grow, a variety of forces accumulate in the TME that create a complex scenario of elevated compression at the tumor interior, tension at the tumor periphery, and altered interstitial fluid flow throughout the tumor volume2, 3. These forces within and around tumors impact cancer progression by modulating invasion and the metastatic cascade4, 5. Furthermore, there is emerging evidence of biomechanical forces modulating the immune response through cancer cell-immune cell signaling6–8.
Although there are several mechanisms of cell-cell signaling within the TME, exosomes have emerged to play key roles in regulating tumor progression7. Exosomes mediate several biological processes associated with tumor initiation, progression and invasion, and can travel to distant sites to promote pre-metastatic niche formation through interaction with resident cell populations9, 10. Furthermore, exosomes can promote cancer cell immune escape by modulating immune cell activity, thereby promoting an environment prone to tumor development11. Cancer cells can directly or indirectly stimulate an inflammatory response through tumor cell-derived exosome-mediated intercellular signaling. BCa-derived exosomes have been shown to induce an inflammatory response in macrophages, which may promote metastatic tumor development12. Additionally, mechanical stretch in cardiomyocytes has been shown to enhance exosome secretion, and influence exosome cargo release13. Together these observations suggest that biomechanical forces may have the potential to directly or indirectly modulate cellular components through exosome-mediated signaling within the TME.
There are several biomimetic in vitro systems that are useful for determining the impact of mechanical forces on cancer cells. Microfluidic, and transwell systems have provided insights into flow-mediated BCa cell signaling through regulation of chemokines and protein expression14–16. Other in vitro systems employing compression have shown enhanced migration of BCa cells through cytoskeletal rearrangement in response to compressive force3. Application of constant tension to a TNBC cell line enhanced its proliferative and invasive potential through FAK-Rho-ERK–mediated signaling17. While these in vitro studies have addressed how constant flow or constant cell strain may directly influence cancer cells, studies to date have not addressed whether these mechanical forces may contribute to immune suppression in vivo. Additionally, a direct link between mechanical forces in malignant BCa cells, exosome release, and the immune response has not been investigated thus far.
We report here that oscillatory strain enhances both the proliferative and migratory potential of human and mouse TNBC cells as well as estrogen receptor (ER)+ BCa cells, while constant strain modulates proliferation of only TNBC cells. Further, oscillatory strain modulates overall exosome production by human and mouse TNBC cells and increases the PD-L1+ exosome production by murine TNBC cells. Importantly, exposure to oscillatory strain promotes mammary tumor growth in vivo and enhances tumor infiltration of immunosuppressive myeloid-lineage cells that internalize tumor-cell derived exosomes.
Materials and Methods
Cell culture
Human ER+ MCF-7 cells were obtained from American Type Culture Collection and subsequently transduced with GFP and luciferase (MCF-7-GFP/LUC) as previously described18. Human TNBC MDA-MB-231 cells were obtained from Dr. Danny Welch (University of Kansas) and subsequently transduced with GFP and luciferase (MDA-MB-231-GFP/LUC). MCF-7-GFP/LUC cells were maintained in Modified Eagle’s Medium (MEM, Corning, NY) supplemented with 10% Fetal Bovine Serum (FBS, Atlas Biologicals, Fort Collins, CO), 0.01 mg/ml insulin (Sigma Aldrich, St. Louis, MO), under the selection of 10 μg/ml puromycin (MP Biomedicals, Santa Ana, CA). MDA-MB-231-GFP/LUC cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Corning, NY) supplemented with 10% FBS under the selection of 10 μg/ml puromycin. The murine TNBC cell line 4T1.2 (an aggressive clone derived from 4T1) was obtained from Dr. Robin L. Anderson’s laboratory (Peter MacCallum Cancer Institute, Melbourne, Australia)19. 4T1.2 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% FBS and 10 mM HEPES (MP Biomedicals, Santa Ana, CA).
Exposure of BCa cells to strain
2.5 × 105 BCa cells (MCF-7-GFP/LUC, MDA-MB-231-GFP/LUC or 4T1.2) were seeded on collagen coated 6 well UniFlex culture plates (Flexcell International Corporation, Burlington, NC) and cultured to confluence in the growth medium appropriate for each cell line. Using a FlexCell FX-6000 or FX-5000 Tension System, plates were subjected to 10% uniaxial oscillatory strain at 0.3 Hz for 48 hours, 10% constant strain for 48 hours, or no strain for 48 hours with medium changed immediately prior to induction of strain.
Cell proliferation assay
Cell proliferation assays were performed using MTT uptake kit (Sigma Chemical, St. Louis, MO), per manufacturer’s instructions. 5 × 104 MCF-7-GFP/LUC cells, 2.5 × 104 MDA-MB-231-GFP/LUC cells or 2.5 × 104 4T1.2 cells (constant or oscillatory strained cells and unstrained cells) were seeded into flat-bottomed 96-well plates in 100 μl of growth medium/well and cultured for 24 to 72 hours. 10 μl of the MTT labeling reagent was added and incubated at 37° C for 4 hours. The purple formazan product was solubilized overnight at 37° C. The plate was read at 550 nm in a plate reader with reference wavelength at 690 nm. The cell proliferation rates were normalized to the controls.
Cell count by trypan blue
5 × 104 MCF-7-GFP/LUC cells, 2.5 × 104 MDA-MB-231-GFP/LUC cells or 2.5 × 104 4T1.2 cells (constant or oscillatory strained cells and unstrained cells) were seeded into flat-bottomed 96-well plates and cultured for 48 hours. Live cells were measured by trypan blue exclusion. Cell counts were normalized with those of unstrained control cells.
Cell migration assay
Strained or control BCa cells (MCF-7-GFP/LUC, MDA-MB-231-GFP/LUC or 4T1.2) were isolated from FlexCell culture plate membranes. Subsequently, 1 × 105 cells were plated in 24-well transwell inserts (8 μm pore size, Millicell; MilliporeSigma, Burlington, MA). Cells were incubated in serum free medium on the transwell inserts (i.e., top well), and medium containing 0.5% FBS + 80 μg/ml Collagen Type I (Advanced Biomatrix, San Diego, CA) was placed in the bottom well. After 24 hours, cells on the upper surface of the transwell filter were removed. Cell migration to the lower surface was evaluated via fluorescent microscopy. Cells that did not express a fluorescent protein (4T1.2) were stained with DAPI (5 μg/ml; Sigma-Aldrich, St. Louis, MO) prior to analysis. The number of cells that migrated through the filters was counted in 5, random 200× microscopic fields per filter.
Wound healing assay
Following 48 hours exposure to strain or control conditions cells were removed from flex plates and reseeded in complete growth media appropriate for the cell line in a 24 well plate. Upon confluence cells were scratched with a 200-μl pipette tip and a PBS wash was performed to remove detached cells. Serum free media were then utilized to halt proliferation for the evaluation of cell migration. Photomicrographs were taken immediately following wound formation (time 0) and 24 hours post wound formation using a digital inverted EVOS FL microscope (10x objective, Life Technologies, Grand Island, NY). The percent wound closure was measured using ImageJ software (National Institutes of Health).
Preparation of exosome-depleted media
Exosome-depleted medium was prepared as previously described20. Briefly, MEM supplemented with 20% FBS was centrifuged using an ultracentrifuge overnight at 100,000 × g at 4˚ C. Supernatant was filtered through a 0.2 μm cellulose acetate filter (Corning, NY). Exosome depleted media were then diluted 1:1 with MEM to make a final concentration of 10% FBS, to which 0.01 mg/ml insulin was added.
Isolation of Extracellular Vesicles and exosomes from conditioned media
Extracellular vesicles (EVs) were isolated using a previously described differential centrifugation method20, 21. Briefly, 96 ml of cell culture media were centrifuged at 300 × g for 10 minutes at 4˚ C. The supernatant was further centrifuged at 2,000 × g for 10 minutes at 4˚ C. The supernatant was spun again at 10,000 × g for 30 minutes at 4˚ C and filtered through a 0.2 μm cellulose acetate filter (Corning, NY). The filtrate was then centrifuged at 100,000 × g for 70 minutes at 4˚ C and the pellet was washed with fresh PBS to remove any contaminating proteins. Finally, the washed pellet was centrifuged at 100,000 × g for 70 minutes at 4˚ C. The pellet was re-suspended in 50 μl of fresh PBS and stored at −80˚ C.
Conditioned media were collected for exosome isolation following 48 hours exposure to strain or control conditions. Conditioned media were centrifuge at 2000 × g to remove any cell pellets and apoptotic bodies. The supernatant was then incubated with the Total Exosome Isolation Reagent for Cell Culture Media kit (ThermoFisher, Waltham, MA) per the manufacturer’s protocol. Purified exosomes were stored in 50 μl of PBS at −80° C.
NanoSight particle analysis for quantitation of exosome size and concentration
The concentrations and size distributions of purified exosomes were determined using a NanoSight NS300 (Cambridge, MA) as described previously. The instrument was calibrated using 100 nm polystyrene latex microspheres (Malvern Instruments Ltd., Malvern, UK). Exosomes were diluted 100-fold with PBS to make a final volume of 1 ml and loaded into a 1 ml syringe. The syringe was placed on a syringe pump attached to the NanoSight. The diluted exosomes were injected at a flow rate of 25 μl at room temperature. A total of 5 videos were acquired under the following settings. The camera level was set to 7, gain to 1, detection threshold to 5, and capture duration to 1 min per video.
Co-culture of exosomes with breast cancer cells
Exosomes were co-cultured with MCF-7 cells, MDA-MB-231 cells and 4T1.2 cells at a ratio of 10 exosomes per cell in 96-well plates for 48 hours. Cell proliferation was evaluated by MTT assay. The cell proliferation rates were normalized to those of the control cells cultured in the absence of exosomes. Live cells were measured by trypan blue exclusion. Cell counts were normalized with those of control cells cultured in the absence of exosomes.
ImageStream analysis of exosomes
ImageStream flow cytometry analysis of exosomes purified from cultured media was performed as previously described20. Exosomes were stained with the following antibodies: PE-conjugated anti-human CD54 (clone HA58; Life Technology, Grand Island, NY), eFlour450-conjugated anti-human CD63 (H5C6) and APC-conjugated anti-mouse CD63 (NVG-2) and PerCP-eFluor 710-conjugated anti-mouse CD274 (PD-L1, MIH5) antibodies were purchased from Life Technologies (Grand Island, NY). PE-conjugated anti-human CD9 (M-L13) antibody was purchased from BD Bioscience (San Jose, CA). PE-Cy7-conjugated anti-human CD81 (5A6), PE-conjugated anti-mouse CD81 (Eat-2), FITC-conjugated anti-mouse CD326 (EpCAM, G8.8), eFluor 450-conjugated anti-human CD63 (H5C6), PE-Cy7-conjugated anti-human CD81 (TAPA-1, 5A6), PE-Cy7-conjugated anti-mouse CD9 (MZ3) and Pacific blue-conjugated anti-mouse CD54 (YN1/1.7.4) antibodies were purchased from BioLegend (San Diego, CA). Alexa Fluor 647-conjugated anti-human TG101 (4A10) was purchased from Novus Biologicals (Centennial, CO). CD63, CD81, CD9 tetraspanins and endosomal sorting complexes required for transport (ESCRT)-I complex subunit TSG101 were used as positive markers to distinguish exosomes from other types of EVs, cellular debris, and calibration beads. Unstained control, single color staining, and calibration beads were used to calibrate the machine and adjust compensation. The stained samples were imaged at 60 × magnification with extended depth of field (EDF). The data were acquired on channels Ch01, Ch03, Ch06, Ch07, Ch09, Ch11 and Ch12. Ch01 and Ch09 were used as bright field channels whereas Ch12 was used for side-scatter. A total of 5,000 events were acquired for each sample, with three technical replicates per sample. The acquired data were analyzed using IDEAS software version 6.2 (EMD Millipore, Billerica, MA).
Syngeneic orthotopic mouse model of BCa
Female BALB/c mice at 6 to 8 week of age were purchased from The Jackson Laboratory (Bar Harbor, ME). Mice were kept in pathogen-free conditions and handled in accordance with the Guidelines for Animal Experiments at the University of Alabama at Birmingham. For some experiments, 4T1.2 cells were labeled with PKH67 Green Fluorescent Cell Linker (Sigma, St. Louis, MO), per manufactures instructions, before being subjected to oscillatory strain. 5 × 105 oscillatory strain-treated or untreated 4T1.2 cells in 60 μl PBS were injected into the fourth mammary fat pad of BALB/c mice. Tumor size was monitored by calipers at the indicated time points after injection. Tumor tissues were collected for analyses on day 14 after injection of 4T1.2 cells. In some experiments, 5 × 105 4T1.2 cells were injected into the fourth mammary fat pad of BALB/c mice on day 0. On day 6, 7.5 × 108 PKH67-labeled 4T1.2-derived exosomes or PBS were injected into the tumor nodule. On day 2 and day 8 after exosome injection, tumor tissues were harvested for analysis of the internalization of exosomes by immune cells and tumor cells.
Confocal microscopy
2 or 8 days following exosome or PBS control injection in vivo, tumors were resected and a portion of the tumor was preserved in O.C.T. Compound (Sakura Tissue-Tek, Torrance, CA) and stored at −80° C. 6 μm frozen sections were generated using a Microm HM 525 cryostat (MICROM International GmbH, Walldorf, Germany). Sections were stained with Hoechst (5 μg/mL, BD Biosciences, San Jose, CA) for 5 minutes at room temperature, following nuclei staining coverslips were mounted with Fluormount-G (SouthernBiotech, Birmingham, AL). Photomicrographs were acquired with a Nikon A1 confocal microscope with a 20x objective (6x digital zoom). Z-stacks were merged and channels were combined using ImageJ.
Flow cytometry
Tumor tissues were harvested and digested with collagenase B. Red blood cells were removed by ACK lysis buffer. Fc receptors were blocked with 3% BSA in PBS containing 2.4G2 antibody (anti-mouse CD16/CD32; BD Pharmingin), followed by staining with the antibodies below. Phycoerythrin (PE)–conjugated anti-mouse-MerTK (DSSMMER), anti-mouse-Gr-1 (RB6–8C5), allophycocyanin (APC)–conjugated anti-mouse-CD206 (MR6F3), anti-mouse-CD8 (53–6.7), PerCP-eFluor 710–conjugated anti-mouse-CD163 (TNKUPJ), PerCP-Cyanine (Cy) 5.5 conjugated anti-mouse-Ly6C (HK1.4), PE-Cy5-conjugated anti-mouse MHC-II (I-A/I-E, M5/114.15.2), PE-Cy7 conjugated anti-mouse-CD4 (GK1.5), and anti-streptavidin antibodies were purchased from Life Technologies (Grand Island, NY). V500-conjugated anti-mouse-CD45 (30-F11), BV605-conjugated anti-mouse-F4/80 (T45–2342), APC-Cy7–conjugated anti-mouse-CD11b (M1/70), anti-mouse-CD3 (145–2C11), and Alexa Fluor 700-conjugated anti-mouse-Ly6G (1A8) antibodies were purchased from BD Bioscience (San Jose, CA). Biotin-conjugated anti-mouse-CD64 (x54–5/7.1) and PE-Cy7-conjugated anti-mouse-CCR2 (SA203G11) antibodies were purchased from BioLegend (San Diego, CA). Data were collected with LSR-II flow cytometer (Becton Dickinson) and analyzed with FlowJo software (version 8.5.2; TreeStar, Ashland, OR).
Statistical analysis
One-way ANOVA with Tukey’s multiple comparisons test or two-way ANOVA with Sidak’s multiple comparisons test was used for analysis of multiple groups. Unpaired t test was used for the statistical analyses between two groups using GraphPad Prism 5. P < 0.05 was considered statistically significant. Pearson correlation analysis was performed to evaluate the correlation of exosome concentration and cell proliferation.
Results
Oscillatory strain promotes BCa cell proliferation and migration
Mechanical strain has been shown to regulate cancer progression and metastasis22–24. To determine whether mechanical strain modulates the proliferation of BCa cells in vitro, MCF-7 cells (human ER+), MDA-MB-231 cells (human TNBC), and 4T1.2 cells (murine TNBC) were exposed to constant or oscillatory strain for 48 hours, and cell proliferation was evaluated by MTT assay and cell count by trypan blue. MCF-7 cells exposed to oscillatory strain showed increased proliferation compared to control cells or cells exposed to constant strain (Fig. 1a and Supplemental Fig. S1a). Interestingly, proliferation of MDA-MB-231 (human) and 4T1.2 (murine) cells was increased following exposure to both constant and oscillatory strain compared to control cells (Fig. 1b–c and Supplemental Fig. S1b–c). Additionally, exposure to oscillatory strain enhanced the migratory potential of both ER+ and TNBC (both human and mouse) cells compared to control cells (Fig. 1d–f and Supplemental Fig. S1d–f). Exposure to constant strain increased transwell migration and wound closure of only human TNBC cells (Fig. 1e and Supplemental Fig. S1e), while the motility of ER+ cells exposed to constant strain was not altered compared to control cells (Fig. 1d and Supplemental Fig. S1d). Together, these data indicate that oscillatory strain promotes proliferation and migration of both ER+ and TNBC cells.
Fig. 1.
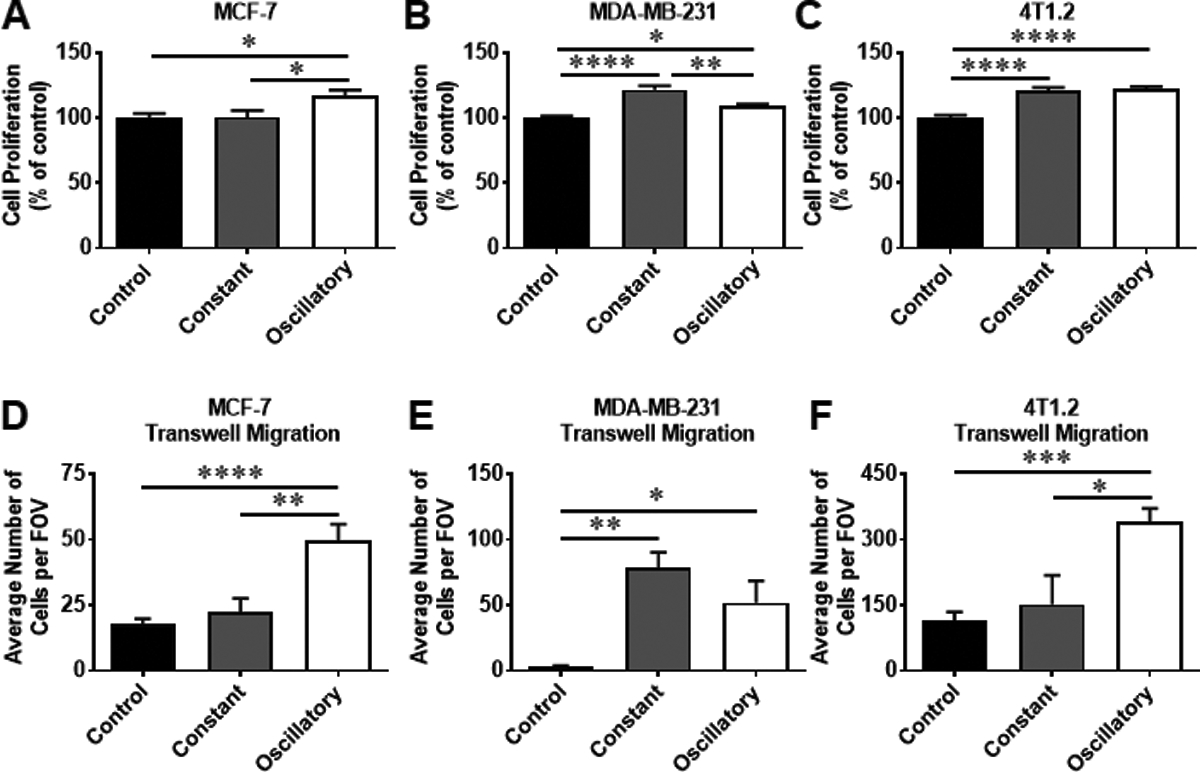
Proliferation and migration of BCa cells changes in response to strain. MCF-7 cells (Human ER+), MDA-MB-231 cells (Human TNBC), and 4T1.2 cells (Murine TNBC) were exposed to 10% uniaxial oscillatory or constant strain for 48 hours. 5 × 104 MCF-7 cells, 2.5 × 104 MDA-MB-231 cells, and 4T1.2 cells (constant or oscillatory strained cells and unstrained cells) were seeded into 96-well plates and cultured for 48 hours. Cell proliferation was evaluated by MTT assay. The cell proliferation rates were normalized to the unstrained control cells. a The cell proliferation rates of MCF-7 cells exposed to constant or oscillatory strain compared to control cells. b The cell proliferation rates of MDA-MB-231 cells exposed to constant or oscillatory strain compared to control cells. c The cell proliferation rates of 4T1.2 cells exposed to constant or oscillatory strain compared to control cells. 1 × 105 cells strained or unstrained MCF-7, MDA-MB-231, or 4T1.2 cells were plated in 24-well transwell inserts (8 μm pore size). After 24 hours, cell migration was evaluated via fluorescent microscope. The migration rates are shown as “average number of cells per field of view (FOV)”. d The transwell migration rates of MCF-7 cells exposed to constant or oscillatory strain compared to control cells. e The transwell migration rates of MDA-MB-231 cells exposed constant or oscillatory strain compared to control cells. f The transwell migration rates of 4T1.2 cells exposed constant or oscillatory strain compared to control cells. Statistical significance was evaluated using one-way ANOVA with Tukey’s multiple comparison testing. * P < 0.05, ** P < 0.01, *** P < 0.005, **** P < 0.001.
Mechanical strain alters exosome production by BCa cells
Tumor-derived exosomes (TEXs) are important mediators of intercellular communication and may have various roles in regulating cancer progression11, 25–27. Exosomes can induce immune suppression, promote the upregulation of inflammatory molecules, increase angiogenesis and vascular permeability, promote matrix remodeling, and determine organotropic metastasis in the pre-metastatic niche28. To investigate if mechanical strain regulates exosome production in BCa cells, MCF-7 cells, MDA-MB-231 cells, and 4T1.2 cells were exposed to oscillatory strain for 48 hours, and TEXs were isolated from the conditioned media. The exosome concentration in the conditioned media of oscillatory strain-exposed human and mouse TNBC cells was significantly higher compared to control cells with no exposure (Fig. 2a and 2e), while oscillatory strain did not alter TEX production by ER+ cells. Further, the exosome concentrations positively correlated with the increased proliferation observed in both human and mouse TNBC cells following exposure to oscillatory strain (Fig. 2b). In addition, no significant differences were observed in terms of mean sizes between exosomes from conditioned media following exposure to oscillatory strain and those from control condition (Fig. 2c–d).
Fig. 2.
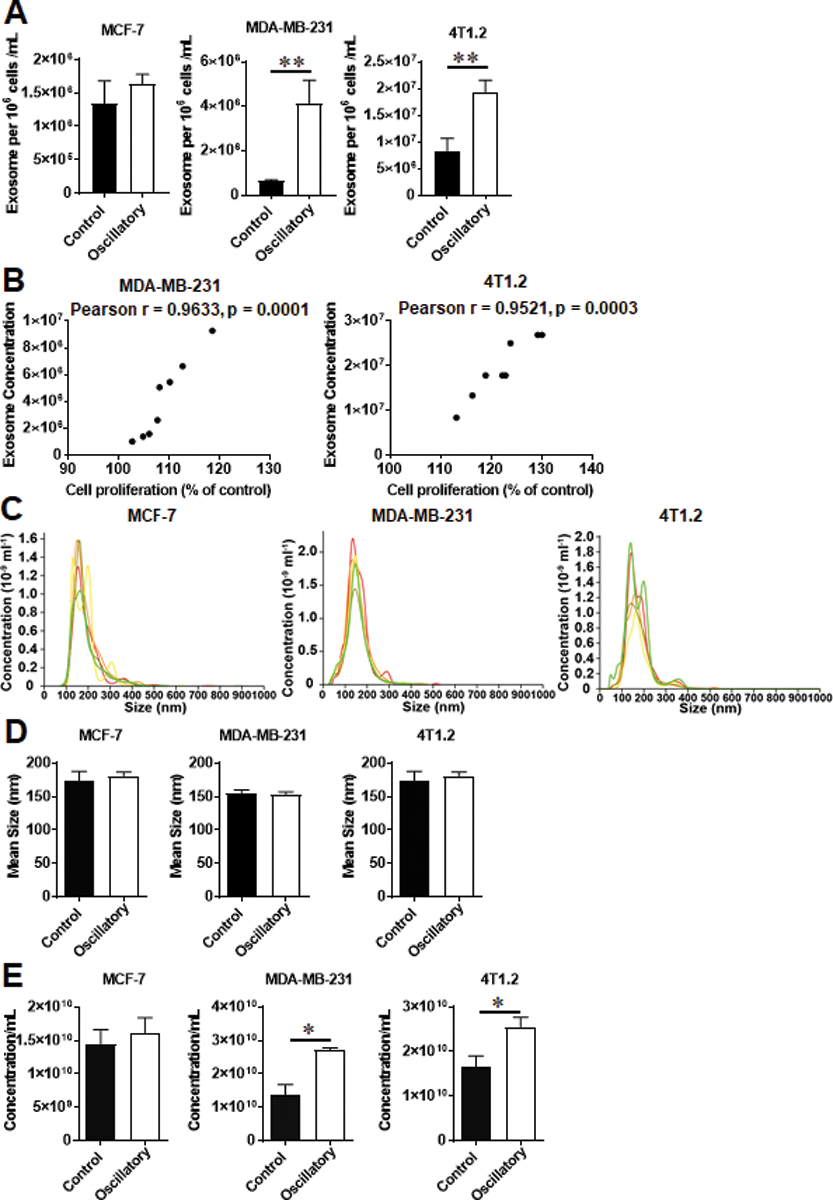
Mechanical strain regulates the concentration of exosomes released from TNBC cells. MCF-7 cells, MDA-MB-231 cells, and 4T1.2 cells were exposed to 10% uniaxial oscillatory strain for 48 hours. Exosomes were purified from conditioned media via differential centrifugation. The concentrations of purified exosomes were determined using ImageStream evaluation. a Exosome concentrations from conditioned media of MCF-7 cells, MDA-MB-231 cells, and 4T1.2 cells exposure to oscillatory strain compared to those of control cells were determined using ImageStream analysis. b Pearson correlation analysis of exosome concentration and cell proliferation of human TNBC MDA-MB-231 (n = 8) or murine TNBC 4T1.2 cells (n = 8). c NanoSight quantitation of one representative exosome isolation from conditioned media of MCF-7 cells, MDA-MB-231 or 4T1.2 cells by using the Total Exosome Isolation kit (ThermoFisher, Waltham, MA). Mean exosome sizes (d) and concentrations (e) quantified by NanoSight. Statistical significance was determined using unpaired t tests. * P < 0.05, ** P < 0.01.
It is unknown whether the increased exosomes production resulted in higher proliferation or if higher proliferation resulted in increased exosomes production. To address this question, we performed co-culture experiments as follows. MCF-7 cells, MDA-MB-231 cells and 4T1.2 cells were co-cultured in the absence or presence of exosomes derived from cancer cells with or without prior exposure to oscillatory strain at 10:1 ratio (10 exosomes per cell) for 48 hours. Cell proliferation was evaluated by both MTT assay and live cell count by trypan blue. We did not observe significant difference in cell proliferation of cancer cells between co-culture with exosomes derived from control cells and that with exosomes derived from cancer cells exposed to oscillatory strain (Supplemental Fig. S2). Thus, the increased exosomes production did not result in higher proliferation, indicating that higher proliferation may have resulted in increased exosome production noted in our study.
Oscillatory strain alters immunomodulatory exosome profiles of BCa cells
Tetraspanins, CD63, CD81, CD9, and the ESCRT-I complex subunit TSG101 are well-established markers that collectively identify exosomes from other types of extracellular vesicles (EVs)26, 29. Characterization of TEXs released from MCF-7 cells exposed to oscillatory strain showed increased frequency of CD63+ exosomes and decreased frequency of CD81+ exosomes compared to those released by cells not exposed to mechanical strain (control, Fig. 3a and d). No significant changes were noted in the tetraspanin profile of exosomes produced by human and mouse TNBC cells (Fig. 3b–c, 3e and f), as seen in representative image stream panels shown in Figure 3d–f. Cargos found in TEX are dependent on their cell-origin and can contain a variety of factors such as mRNA, miRNA, DNA, and lipids. Furthermore, the presence of immunomodulatory proteins such as immune check point PD-L1, and proteins that confer invasive potential such as EpCAM and CD54 on exosomes are beginning to be appreciated in facilitating immunosuppression, tumor invasion and metastasis30–32. When these markers were evaluated, the populations of CD81+PD-L1+ and CD63+PD-L1+ exosomes were increased while CD63+CD54+ exosomes were decreased when murine 4T1.2 cells were exposed to oscillatory strain and compared to control cells (Fig. 4a, d and f). No difference in CD81+EpCAM+ or CD63+EpCAM+ exosomes was noted between the two groups (Fig. 4b and e). Representative imagestream images of exosomes of these subsets are shown in Fig. 4g and h. Taken together, these findings indicate that oscillatory strain modulates PD-L1+ exosome production by TNBC cells.
Fig. 3.
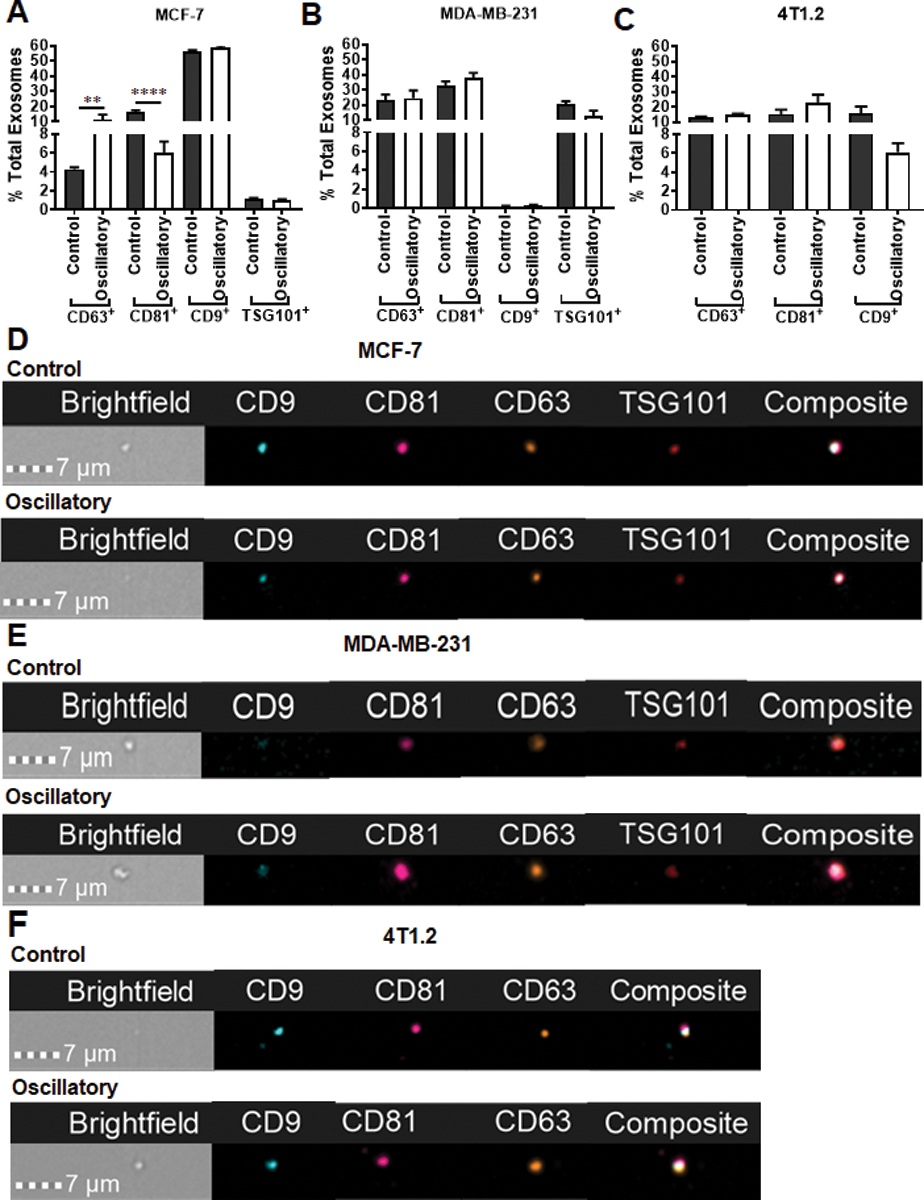
The exosome profile of BCa cells changes in response to oscillatory strain. MCF-7 cells, MDA-MB-231 cells, and 4T1.2 cells were exposed to 10% uniaxial oscillatory strain for 48 hours. Exosomes purified from conditional media were characterized by ImageStream. CD63, CD81, CD9 tetraspanins as well as ESCRT-I complex subunit TSG101 were used as positive markers of exosomes. a Frequency of CD63+, CD81+, CD9+ or TSG101+ exosomes from conditioned media of MCF-7 cells exposed to oscillatory strain compared to those from control cells. b Frequency of CD63+, CD81+, CD9+ or TSG101+ exosomes released from MDA-MB-231 cells exposed to oscillatory strain compared to those from control cells. c Frequency of CD63+, CD81+ or CD9+ exosomes from 4T1.2 cells exposed to oscillatory strain compared to those from control cells. d Image strips showing exosome marker profiles of exosomes released from MCF-7 cells exposed to oscillatory strain versus control. e Image strips showing exosome marker profiles of exosomes isolated from MDA-MB-231 cells exposed to oscillatory strain versus control. f Image strips showing exosome marker profiles of exosomes released from 4T1.2 cells exposed to oscillatory strain versus control. Statistical significance was evaluated using one-way ANOVA with Tukey’s multiple comparison testing. ** P < 0.01, **** P < 0.001.
Fig. 4.
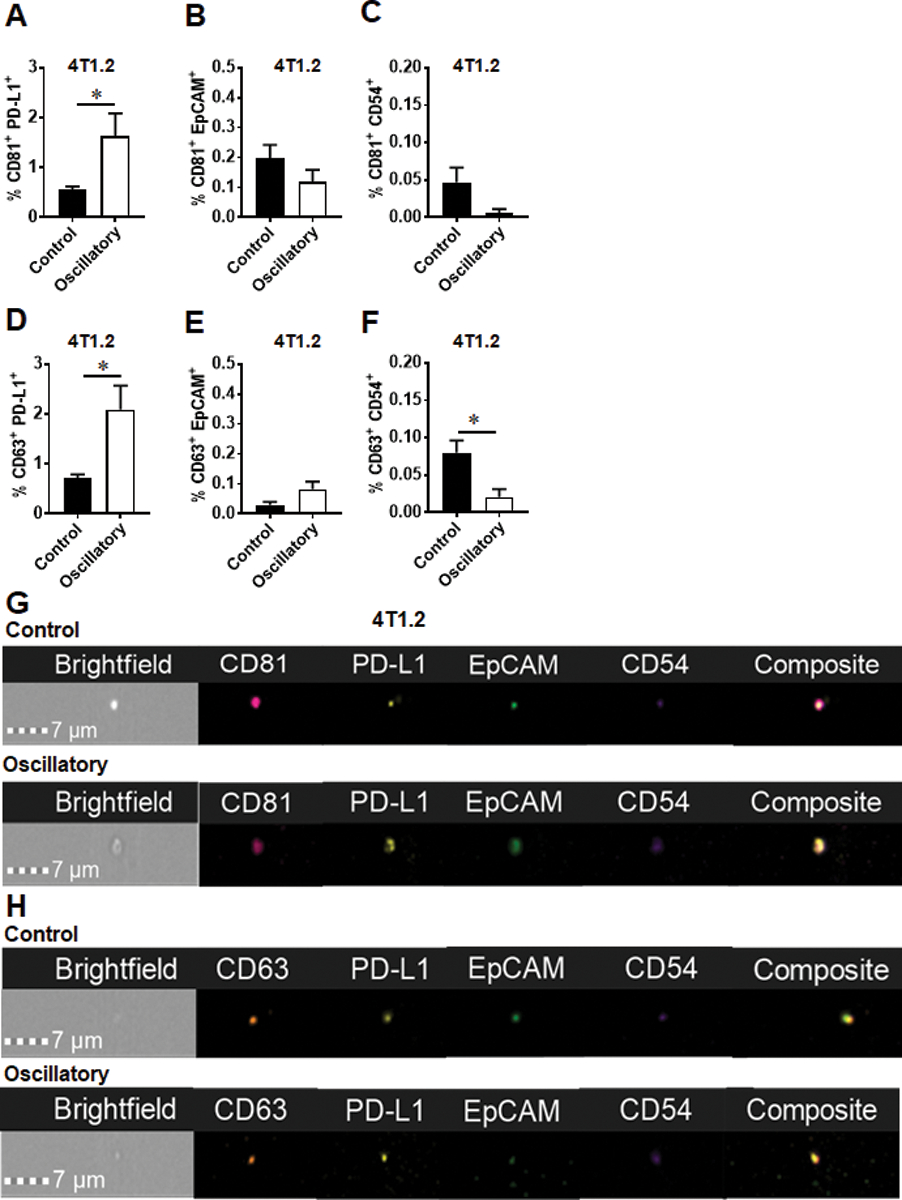
The exosome profile of murine TNBC 4T1.2 cells changes in response to oscillatory strain. 4T1.2 cells were exposed to 10% uniaxial oscillatory strain for 48 hours. Exosomes were purified from conditional media, profile was further characterized by ImageStream. a The population of CD81+PD-L1+ exosomes from 4T1.2 cells exposed to oscillatory strain compared to that from control cells. b The population of CD81+EpCAM+ exosomes from 4T1.2 cells exposed to oscillatory strain versus control. c The population of CD81+CD54+ exosomes from 4T1.2 cells exposed to oscillatory strain compared to control. d The population of CD63+PD-L1+ exosomes from 4T1.2 cells exposed to oscillatory strain compared to that from control cells. e The population of CD63+EpCAM+ exosomes from 4T1.2 cells exposed to oscillatory strain versus control. f The population of CD63+CD54+ exosomes from 4T1.2 cells exposed to oscillatory strain compared to control. Statistical significance was determined using unpaired t tests. * P < 0.05. g Image strips showing CD81+PD-L1+, CD81+EpCAM+ or CD81+CD54+ exosomes isolated from 4T1.2 cells exposed to oscillatory strain versus control. h Image strips showing CD63+PD-L1+, CD63+EpCAM+ or CD63+CD54+ exosomes isolated from 4T1.2 cells exposed to oscillatory strain versus control.
Oscillatory strain promotes tumor growth in an orthotopic model of TNBC
Next, to determine whether oscillatory strain promotes tumor growth in vivo, a syngeneic orthotopic mouse model of BCa was utilized to monitor the tumor sizes and the infiltration of immune cells. BALB/c mice were injected in the fourth mammary fat pad with aggressive TNBC cells (4T1.2) preconditioned with oscillatory strain for 48 hours. Tumor sizes were measured on days 5, 8, 11 after tumor implantation. As shown in Figure 5a, mice implanted with 4T1.2 cells exposed to oscillatory strain showed a significant increase in tumor growth on days 8 and 11 compared to control group implanted with unstrained cells. As heterogeneous myeloid-derived suppressor cells (MDSCs) and tumor-associated macrophages (TAMs) have tumor-promoting functions, and are drivers of tumor-associated immune suppression33–37, we investigated whether mechanical strain-induced changes in tumor cells altered the breast TME to modulate infiltration of these immune suppressive cells that could further contribute to enhanced tumor growth. Immune profiling of tumor tissues showed that the percentage of the monocytic (CD11b+Ly6G−Ly6Chigh) MDSC subset was significantly increased in the TME of mice implanted with 4T1.2 cells exposed to oscillatory strain on day 14, compared to that of mice transplanted with control 4T1.2 cells (Fig. 5b). There was no difference in the tumor-infiltrating granulocytic (CD11b+Ly6G+Ly6Clow) MDSC subset between the two groups (Fig. 5c). Additionally, we also observed increased infiltration of recruited macrophages (CD45+CD11b+F4/80+CD11c−CD206−CCR2+Ly6C+) in the TME of mice implanted with oscillatory strain-exposed 4T1.2 cells, compared to control cells (Fig. 5d). Notably, the percentage of CD8+ T cells showed a decreased trend in the TME of these mice (Fig. 5e). These results suggest that oscillatory strain promotes tumor growth via immune suppression in the TME.
Fig. 5.
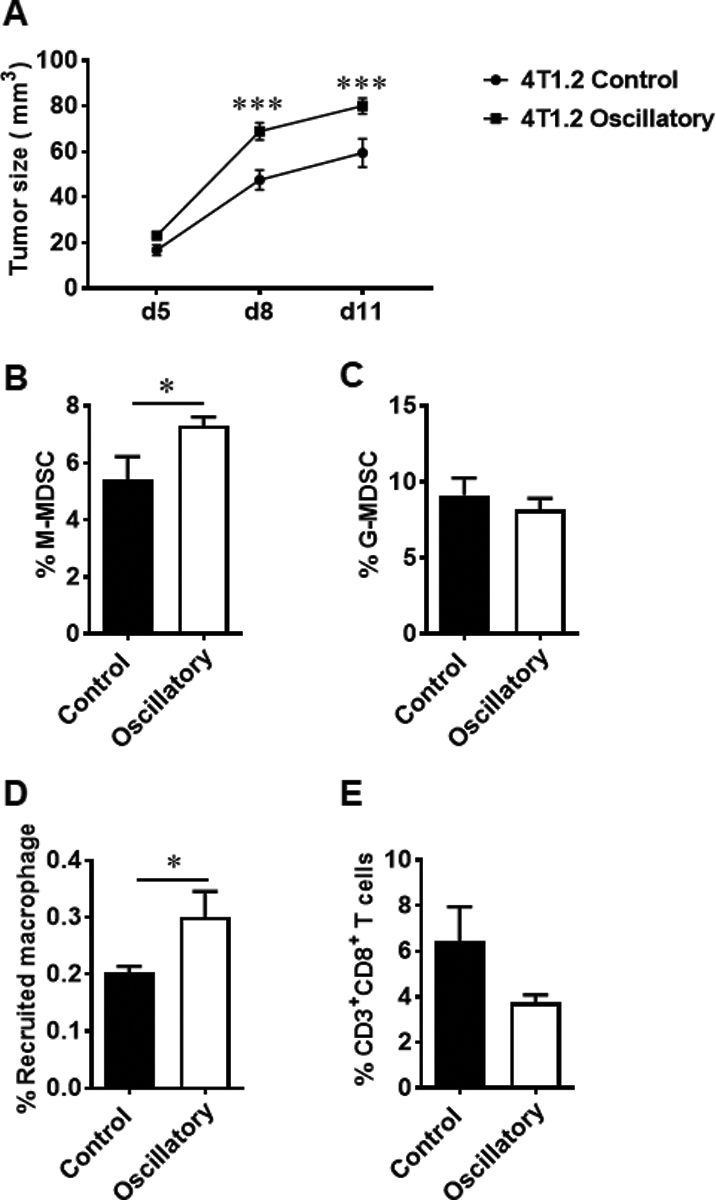
Oscillatory forces promote tumor growth and immunosuppression in the TME in vivo. Six- to eight-week-old female BALB/c mice were injected in the fourth mammary fat pad with 5 × 105 4T1.2 cells preconditioned with oscillatory strain or unstrained cells. a Tumor volumes were measured at the indicated time points after mammary fat pad injection of control or oscillatory strained 4T1.2 cells. “d” on x-axis indicates days after mammary fat pad injection of tumor cells. The infiltration of immune cells in tumor tissue was determined by FACS analysis of cells harvested from tumor tissue on day 14 post tumor implantation. b The percentage of CD11b+Ly6G−Ly6Chigh monocytic MDSCs was determined by FACS analysis of cells harvested from tumor tissue on day 14 post tumor implantation (n = 5 mice/group). c The percentage of CD11b+Ly6G+Ly6Clow granulocytic MDSCs in tumor tissue was evaluated by FACS analysis (n = 5 mice/group). d The percentage of recruited macrophages in tumor tissues was determined by FACS analysis (n = 5 mice/group). e The percentage of tumor infiltrated CD3+CD8+ T cells was evaluated by FACS analysis in the TME of mice implanted with oscillatory strained 4T1.2 cells compared to control group (n = 5 mice/group). Statistical significance was evaluated via a two-way ANOVA with Sidak’s multiple comparisons test or was determined using unpaired t tests.* P < 0.05, *** P < 0.005.
Oscillatory strain modulates exosome internalization by immunosuppressive cells in the TME
Tumor-derived exosomes, which function as mediators of intercellular communication, can deliver pro-tumorigenic signals to immune cells reprogramming their function in the TME26, 27. Mechanisms responsible for cellular reprogramming include cell surface signaling and/or internalization of TEXs by recipient cells promoting transcriptional/translational activities through miRNA, protein or other cargo transfer27. To further investigate whether tumor cell-derived exosomes are internalized by the immune cells to regulate the immune suppression in vivo, 4T1.2 cells labeled with a lipophilic dye, PKH67, with or without exposure to oscillatory strain were injected into the fourth mammary fat pad. Exosome internalization by the MDSCs and macrophages in the TME was then identified by PKH67 positive signal in these cell types. Exosome internalization by M-MDSCs and recruited macrophages in the breast TME was elevated in the TME of mice transplanted with 4T1.2-PKH cells preconditioned with oscillatory strain when compared to mice inoculated with control cells (Supplemental Fig. 3a and c). Exosome internalization by G-MDSC and M2 macrophages were not significantly different between oscillatory and control groups (Supplemental Fig. 3b and d). These results indicate that application of oscillatory strain on breast tumor cells not only enhances tumor cell-exosome secretion but also modulates immunosuppression in the TME by facilitating infiltration of MDSCs and macrophages via internalization of TEXs by MDSCs and recruited macrophages in the breast TME.
Exosome internalization by immune cells and tumor cells in the TME
To investigate whether TEXs are internalized by immune cells and tumor cells in the TME, 4T1.2 cells were injected into the fourth mammary fat pad. PKH67-labeled exosomes purified from conditioned media of 4T1.2 cells were injected directly into the tumor nodule on day 6 after tumor injection. Exosome internalization by the immune cells in the TME was identified as PKH67+CD45+ cells on days 2 (Fig. 6a and Supplemental Fig. S4a–b) and 8 (Fig. 6a and Supplemental Fig. S4c–d) after exosome injection whereas exosome internalization by tumor cells was identified as PKH67+CD45neg cells (Fig. 6b and Supplemental Fig. S5a–b and 5c–d). Further, exosome internalization by various immune cells were detected in TME on days 2 and 8 after exosome injection, including recruited macrophages (Fig. 6c and Supplemental Fig. S6a–b and Fig. S6c–d), M2 macrophages (Fig. 6d and Supplemental Fig. S7a–b and Fig. S7c–d), MDSCs (Fig. 6e–g and Supplemental Fig. S8a–b and Fig. S8c–d), CD4+ T cells (Fig. 6h and Supplemental Fig. S9a–b and Fig. S9c–d) and CD8+ T cells (Fig. 6i and Supplemental Fig. S9a–b and Fig. S9c–d). Additionally, the frequency of CD45+ cells in exosome+ cells was 69.14% while the frequency of CD45neg cells in exosome+ cells was 30.86% in the TME on day 2 after exosome injection (Supplemental Fig. S10a). The frequency of CD45+ cells in exosome+ cells was 84.14% while the frequency of CD45neg cells in exosome+ cells was 15.86% in the TME on day 8 after exosome injection (Supplemental Fig. S10b). Among exosomes+ cells, the frequency of MDSCs was higher than that of other studied immune cell population (Supplemental Fig. S10a–b). Moreover, confocal imaging of tumor tissue following intratumoral injection of exosomes identified PKH67+ cells in tumor tissue (Supplemental Fig. S11a–b). Together, these results indicate that TEXs can be internalized in vivo by immune cells and tumor cells in the TME.
Fig. 6.
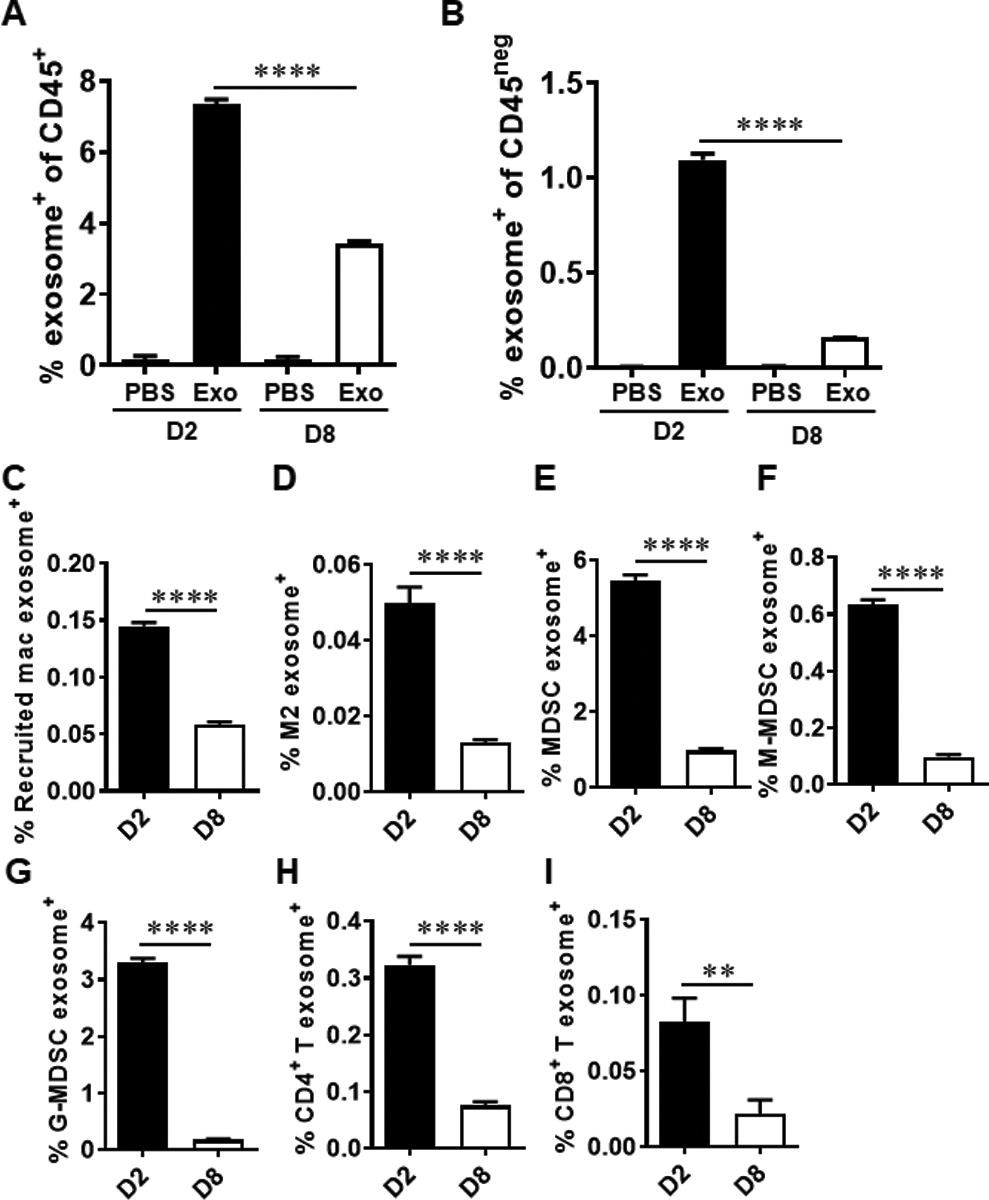
Internalization of exosomes by immune cells and tumor cells in TME. Six to eight-week-old female BALB/c mice were injected in the fourth mammary fat pad with 5 × 105 4T1.2 cells. At day 6, 7.5 × 108 PKH67-labeled 4T1.2 cell-derived exosomes or PBS were injected into the tumor nodule. On day 2 (D2) and day 8 (D8) after exosome injection, tumor tissues were harvested (n = 5 mice/group). The frequencies of exosome-positive cells in CD45+ cells (a) and CD45neg cells (b) was determined by FACS analyses. The frequencies of exosome-positive recruited macrophages (c), M2 macrophages (d), MDSCs (e), M-MDSCs (f), G-MDSCs (g), CD4+ T cells (h) and CD8+ T (i) cells in CD45+ cells were determined by FACS analyses. Statistical significance was evaluated using one-way ANOVA with Tukey’s multiple comparison testing (a and b). Statistical significance was determined using unpaired t tests (c to i). ** P < 0.01, **** P < 0.001.
Discussion
Mechanical forces generated during tumor growth contribute to the formation of an abnormal TME with elevated fluid and solid stresses23. These forces within the TME play a crucial role in tumor progression38. In this report, we show that mechanical strain induces phenotypic changes in BCa cells promoting invasive and pro-tumorigenic phenotypes that induce immunosuppression in the TME.
Mechanical forces within the breast TME are potent regulators of cancer progression. As breast tumors grow, forces accumulate in the TME, creating a complex scenario of elevated compression at the tumor interior, tension at the tumor periphery, and altered interstitial fluid flow. These forces within and around tumors impact cancer progression, invasion, and metastasis4, 39. Both constant and oscillatory forces would be present within the body, with oscillations likely due to tumor growth and changes in vasculature. Herein, constant strain, to mimic accumulated compressive forces within the tumor interior, and oscillating strain, to mimic the impact of altered intestinal fluid flow, have been specifically modulated to understand the impact of each on tumor growth and tumor-immune interactions.
There is increasing evidence for the involvement of interstitial pressure, stiffness, or hyperproliferative pressure acting predominantly as an enhancer of tumor progression22, 40. Numerous studies suggest that mechanical forces can regulate tumor growth and metastatic potential of cancer cells23, 41. Culturing mammary epithelial cells in matrices with high stiffness has been reported to increase the proliferation-specific gene signature in BCa42. Our observation that exposure to oscillatory forces results in a significant increase in the proliferation of BCa cells both in vitro and in vivo complements studies by Wozniak et al., who demonstrated that increased ECM rigidity can promote breast epithelial proliferation43. In our study, both ER+ and TNBC cells exposed to oscillatory strain showed increased proliferation and migration rates compared to control cells. However, only human TNBC cells, exposed to constant strain, had increased proliferation and migration rates compared to control cells. Regarding the distinct responses to constant strain by different subtypes of BCa cells, future studies to characterize transcriptional changes would be of interest to understand the molecular signaling pathways involved in tumor progression and metastasis in response to mechanical strain.
BCa invasion correlates with ECM stiffening and immune cell infiltration8. Our study identified that oscillatory strain promotes BCa cell migration in vitro. Further, in vivo studies showed increased immunosuppressive cell populations, such as M-MDSC and recruited macrophages, following implantation of BCa cells preconditioned with oscillatory strain, indicating that mechanical forces contribute to immune suppression in the TME. These phenotypic changes extended over 10 days when strained cells were utilized for in vivo studies, suggesting a lasting response to mechanical forces within the TME. While we did not specifically correlate our results with hormone receptor expression or proliferation rate, it is known that sensitivity of cancer cells to mechanical forces, such as stiffness, can be context-dependent22. Generally, it is thought that mechanoregulators (such as the FAK, ROCK/Rho, and Ras) regulate proliferation, but to our knowledge the reverse, in which proliferative status regulates mechano-responsiveness, has not be studied. Interestingly, ER+ breast cancers have a high incidence of bone metastasis, potentially indicating subtype-specific organ-tropism44 that may be driven, in part, by biomechanical microenvironmental effects, although there is no direct evidence of specific sensitivity to mechanoregulation in hormone driven breast cancers.
Tumor-derived exosomes regulate the TME by enhancing tumor cell growth, facilitating immune suppression and promoting tumor progression25–27, 45, 46. Conditioning mice with TEXs resulted in the accumulation of MDSCs and promotion of tumor growth46, 47. In this report, we show that TNBC cells exposed to oscillatory strain secreted more exosomes, which correlated with the increased cell proliferation, indicating that mechanical strain may enhance tumor cell growth by increasing the release of exosomes. These findings are consistent with numerous studies that TEXs can induce tumor cell proliferation through complex signaling networks involved in tumor cell-cell communication48, 49. Exosomes released from ER+ cells exposed to oscillatory strain showed increased percentage of CD63+ exosomes and decreased percentage of CD81+ exosomes compared to control cells, suggesting modulation of exosome populations by mechanical strain. Further studies are required to evaluate whether these populations play different functional roles within the TME.
EpCAM is a cell surface glycoprotein that is highly expressed in epithelial cancers50. Strong EpCAM overexpression was associated with enhanced invasion of breast cancer cell lines into extracellular matrix31. The expression of EpCAM on exosomes has been reported in breast cancer51, colon cancer52, ovarian cancer53, pancreatic cancer54. Although both CD81+EpCAM+ and CD63+EpCAM+ exosomes were detected in the conditional media from 4T1.2 cells, exposure to oscillatory strain did not affect the production of CD81+EpCAM+ or CD63+EpCAM+ exosomes from 4T1.2.
Tumor cells evade immune surveillance by upregulating the expression of immune checkpoint molecules, including PD-L1, which interacts with PD-1 receptor on T cells to elicit the immune checkpoint response55. Metastatic melanomas release PD-L1+ exosomes, which contribute to immunosuppression and serve as a predictor for anti-PD-1 therapy56. Recently it has been discovered that exosomal PD-L1 is a major regulator of tumor progression through its suppression of T cell activation in draining lymph nodes in prostate cancer30. PD-L1+ exosomes suppress T cell killing of breast cells and promote tumor growth in the TME57. Results of our study present evidence that mechanical strain may mediate immunosuppression through TEX PD-L1, as CD81+PD-L1+ and CD63+PD-L1+ exosome populations were increased after exposure to oscillatory strain. The observation that mechanical forces enhance exosomal PD-L1 provides insights to previous reports that exosomal PD-L1-mediates immunosuppression in BCa57, metastatic melanomas56, head and neck cancer58 and prostate cancer30.
Exosomes comprise a heterogeneous vesicle population in regards to tetraspanin expression. Therefore the biological meaning of the variation in frequency of CD63+ and CD81+ exosomes reported is unknown. While out of the scope of this work, it has been noted that tetraspanins may play a role in EV cargo selection59, therefore it is possible that the variation noted during exosome phenotyping may be indicative of changes in exosome cargo in response to mechanical strain, although this aspect was not evaluated in our study. This potential change in cargo is likely not responsible for the increases in migration and proliferation observed, as the variations in CD63+ and CD81+ exosomes were exclusive to one cell type.
Exosome internalization by the MDSCs and macrophages in the TME was identified by PKH67 positive signal from PKH67-labeled tumor cells. We cannot rule out the other uptake possibilities, including non-endosomal vesicles, such as microvesicles and apoptotic bodies, and fragments of dead tumor cells. ARF6 is a plasma membrane protein present on microvesicles and therefore can help differentiate microvesicles from exosomes which are endosomally-derived. Additionally, GRP94, a marker of endoplasmic reticulum was included to determine the contamination of cellular debris60. These are markers we routinely use to validate exosome preparations. To further confirm tumor-derived exosomes can be internalized by different cells in TME, we next performed experiments by intratumor injection of PKH67-labeled exosomes. The results provide direct evidence that TEXs can be internalized by MDSCs and M2 macrophages in vivo, further support our findings that exposure to oscillatory strain increases the infiltration of immunosuppressive myeloid-lineage cells that internalize TEXs in TME.
Exosomes derived from pancreatic cancer cells were shown to increase liver metastatic burden by transferring macrophage migration inhibitory factor to liver macrophages and by recruiting immune cells to initiate pre-metastatic niche formation in the liver9. Further, Chow and colleagues have demonstrated that macrophages internalize BCa-derived exosomes, resulting in macrophage immunomodulation through Toll-like receptor 2-mediated activation of NF-κB12 and MyD88 has been reported to play a pivotal role in tumor exosome-mediated expansion of MDSCs and tumor metastasis61. Interestingly, recent studies show that BCa-derived exosomes distributed predominantly to the lung, a frequent site of metastasis after systemic delivery, and were taken up by CD45+ cells including macrophages and CD11b+ myeloid cells47. Here, we report exosome internalization by M-MDSC and recruited macrophages was elevated in the TME of mice implanted with PKH labeled 4T1.2 cells exposed to oscillatory strain prior to inoculation when compared to unstrained control cells, showing that exposure to mechanical strain not only promotes the release of PD-L1+ exosomes, but also enhances their internalization by immunosuppressive cells, such as M-MDSCs and recruited macrophages. Future studies are required to characterize myeloid immunomodulation pathways induced by BCa-derived exosomes following exposure to oscillatory strain.
In summary, our data indicate that exposure to mechanical strain potentiates invasive and pro-tumorigenic phenotypes in BCa cells which promotes immunosuppression by altering proliferative and migratory potential of tumor cells and increasing the release of immunomodulatory exosomes which are internalized by immunosuppressive cells, thus facilitating enhanced immune-tumor cell crosstalk in the TME. Further efforts to investigate the potential mechanisms of mechanical stress involved in the tumor progression may provide new therapeutic approaches to target the TME.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge Marion Spell at the UAB Flow Cytometry Core Facilities for his technical assistance in acquiring sorted cell samples. Funding was provided by American Cancer Society - Institutional Research Grant Award IRG-60-001-53-IRG awarded to J.S.D, 1R01HL128502-01A1 awarded to J.S.D., Breast Cancer Research Foundation of Alabama Collaboration Award obtained by J.L.B., J.S.D. and A.F., and R01 CA184770. and BCRFA awarded to SP.
Footnotes
Conflict of Interest: There are no financial conflicts of interest
Reference
- 1.Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol. 2016;13:674–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stylianopoulos TMJ, Snuderl M, Mpekris F, Saloni R, Jain SR, and Jain RK. Coevolution of Solid Stress and Interstitial Fluid Pressure in Tumors During Progression: Implications for Vascular Collapse. Cancer Res. 2013;73:3833–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tse JM CG, Cheng G,Tyrrell JA, Wilcox-Adelman SA, Boucher Y, Jain RK, et al. Mechanical compression drives cancer cells toward invasive phenotype. PNAS. 2012;109:911–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Northey JJ, Przybyla L, Weaver VM. Tissue Force Programs Cell Fate and Tumor Aggression. Cancer Discov. 2017;7:1224–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sottnik JL, Dai J, Zhang H, Cambell B, Keller ET. Tumor-induced pressure in the bone microenvironment causes osteocytes to promote the growth of prostate cancer bone metastases. Cancer Res. 2015;75:2151–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seager RJ, Hajal C, Spill F, Kamm RD, Zaman MH. Dynamic interplay between tumour, stroma and immune system can drive or prevent tumour progression. Converg Sci Phys Oncol. 2017;3:034002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Acerbi I, Cassereau L, Dean I, Shi Q, Au A, Park C, et al. Human breast cancer invasion and aggression correlates with ECM stiffening and immune cell infiltration. Integr Biol (Camb). 2015;7:1120–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costa-Silva B, Aiello NM, Ocean AJ, Singh S, Zhang H, Thakur BK, et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol. 2015;17:816–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Becker A, Thakur BK, Weiss JM, Kim HS, Peinado H, Lyden D. Extracellular Vesicles in Cancer: Cell-to-Cell Mediators of Metastasis. Cancer Cell. 2016;30:836–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalluri R The biology and function of exosomes in cancer. J Clin Invest. 2016;126:1208–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chow A, Zhou W, Liu L, Fong MY, Champer J, Van Haute D, et al. Macrophage immunomodulation by breast cancer-derived exosomes requires Toll-like receptor 2-mediated activation of NF-kappaB. Sci Rep. 2014;4:5750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pironti G, Strachan RT, Abraham D, Mon-Wei Yu S, Chen M, Chen W, et al. Circulating Exosomes Induced by Cardiac Pressure Overload Contain Functional Angiotensin II Type 1 Receptors. Circulation. 2015;131:2120–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pisano M, Triacca V, Barbee KA, Swartz MA. An in vitro model of the tumor-lymphatic microenvironment with simultaneous transendothelial and luminal flows reveals mechanisms of flow enhanced invasion. Integr Biol (Camb). 2015;7:525–33. [DOI] [PubMed] [Google Scholar]
- 15.Jeon JS, Bersini S, Gilardi M, Dubini G, Charest JL, Moretti M, et al. Human 3D vascularized organotypic microfluidic assays to study breast cancer cell extravasation. Proc Natl Acad Sci U S A. 2015;112:214–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buchanan CF, Voigt EE, Szot CS, Freeman JW, Vlachos PP, Rylander MN. Three-dimensional microfluidic collagen hydrogels for investigating flow-mediated tumor-endothelial signaling and vascular organization. Tissue Eng Part C Methods. 2014;20:64–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Provenzano PP, Inman DR, Eliceiri KW, Keely PJ. Matrix density-induced mechanoregulation of breast cell phenotype, signaling and gene expression through a FAK-ERK linkage. Oncogene. 2009;28:4326–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goliwas KF, Richter JR, Pruitt HC, Araysi LM, Anderson NR, Samant RS, et al. Methods to Evaluate Cell Growth, Viability, and Response to Treatment in a Tissue Engineered Breast Cancer Model. Sci Rep. 2017;7:14167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tester AM, Ruangpanit N, Anderson RL, Thompson EW. MMP-9 secretion and MMP-2 activation distinguish invasive and metastatic sublines of a mouse mammary carcinoma system showing epithelial-mesenchymal transition traits. Clin Exp Metastasis. 2000;18:553–60. [DOI] [PubMed] [Google Scholar]
- 20.Hough KP, Wilson LS, Trevor JL, Strenkowski JG, Maina N, Kim YI, et al. Unique Lipid Signatures of Extracellular Vesicles from the Airways of Asthmatics. Sci Rep. 2018;8:10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thery C, Witwer KW, Aikawa E, Alcaraz MJ. Anderson JD, Andriantsitohaina R, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7:1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Broders-Bondon F, Nguyen Ho-Bouldoires TH, Fernandez-Sanchez ME, Farge E. Mechanotransduction in tumor progression: The dark side of the force. J Cell Biol. 2018;217:1571–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalli M, Stylianopoulos T. Defining the Role of Solid Stress and Matrix Stiffness in Cancer Cell Proliferation and Metastasis. Front Oncol. 2018;8:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Northcott JM, Dean IS, Mouw JK, Weaver VM. Feeling Stress: The Mechanics of Cancer Progression and Aggression. Front Cell Dev Biol. 2018;6:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barros FM, Carneiro F, Machado JC, Melo SA. Exosomes and Immune Response in Cancer: Friends or Foes? Front Immunol. 2018;9:730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greening DW, Gopal SK, Xu R, Simpson RJ, Chen W. Exosomes and their roles in immune regulation and cancer. Semin Cell Dev Biol. 2015;40:72–81. [DOI] [PubMed] [Google Scholar]
- 27.Whiteside TL. Exosomes and tumor-mediated immune suppression. J Clin Invest. 2016;126:1216–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo Y, Ji X, Liu J, Fan D, Zhou Q, Chen C, et al. Effects of exosomes on pre-metastatic niche formation in tumors. Mol Cancer. 2019;18:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hough KP, Chanda D, Duncan SR, Thannickal VJ, Deshane JS. Exosomes in immunoregulation of chronic lung diseases. Allergy. 2017;72:534–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poggio M, Hu T, Pai CC, Chu B, Belair CD, Chang A, et al. Suppression of Exosomal PD-L1 Induces Systemic Anti-tumor Immunity and Memory. Cell. 2019;177:414–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osta WA, Chen Y, Mikhitarian K, Mitas M, Salem M, Hannun YA, et al. EpCAM is overexpressed in breast cancer and is a potential target for breast cancer gene therapy. Cancer Res. 2004;64:5818–24. [DOI] [PubMed] [Google Scholar]
- 32.Guo P, Huang J, Wang L, Jia D, Yang J, Dillon DA, et al. ICAM-1 as a molecular target for triple negative breast cancer. Proc Natl Acad Sci U S A. 2014;111:14710–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9:162–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khaled YS, Ammori BJ, Elkord E. Myeloid-derived suppressor cells in cancer: recent progress and prospects. Immunol Cell Biol. 2013;91:493–502. [DOI] [PubMed] [Google Scholar]
- 35.Youn JI, Nagaraj S, Collazo M, Gabrilovich DI. Subsets of myeloid-derived suppressor cells in tumor-bearing mice. J Immunol. 2008;181:5791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141:39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murdoch C, Giannoudis A, Lewis CE. Mechanisms regulating the recruitment of macrophages into hypoxic areas of tumors and other ischemic tissues. Blood. 2004;104:2224–34. [DOI] [PubMed] [Google Scholar]
- 38.Provenzano PP, Keely PJ. Mechanical signaling through the cytoskeleton regulates cell proliferation by coordinated focal adhesion and Rho GTPase signaling. J Cell Sci. 2011;124:1195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jain RK, Martin JD, Stylianopoulos T. The role of mechanical forces in tumor growth and therapy. Annu Rev Biomed Eng. 2014;16:321–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stylianopoulos T, Martin JD, Snuderl M, Mpekris F, Jain SR, Jain RK. Coevolution of solid stress and interstitial fluid pressure in tumors during progression: implications for vascular collapse. Cancer Res. 2013;73:3833–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar S, Weaver VM. Mechanics, malignancy, and metastasis: the force journey of a tumor cell. Cancer Metastasis Rev. 2009;28:113–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whitfield ML, George LK, Grant GD, Perou CM. Common markers of proliferation. Nat Rev Cancer. 2006;6:99–106. [DOI] [PubMed] [Google Scholar]
- 43.Wozniak MA, Desai R, Solski PA, Der CJ, Keely PJ. ROCK-generated contractility regulates breast epithelial cell differentiation in response to the physical properties of a three-dimensional collagen matrix. J Cell Biol. 2003;163:583–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thaler JD, Achari Y, Lu T, Shrive NG, Hart DA. Estrogen receptor beta and truncated variants enhance the expression of transfected MMP-1 promoter constructs in response to specific mechanical loading. Biol Sex Differ. 2014;5:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alipoor SD, Mortaz E, Varahram M, Movassaghi M, Kraneveld AD, Garssen J, et al. The Potential Biomarkers and Immunological Effects of Tumor-Derived Exosomes in Lung Cancer. Front Immunol. 2018;9:819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiang X, Poliakov A, Liu C, Liu Y, Deng ZB, Wang J, et al. Induction of myeloid-derived suppressor cells by tumor exosomes. Int J Cancer. 2009;124:2621–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wen SW, Sceneay J, Lima LG, Wong CS, Becker M, Krumeich S, et al. The Biodistribution and Immune Suppressive Effects of Breast Cancer-Derived Exosomes. Cancer Res. 2016;76:6816–27. [DOI] [PubMed] [Google Scholar]
- 48.Maia J, Caja S, Strano Moraes MC, Couto N, Costa-Silva B. Exosome-Based Cell-Cell Communication in the Tumor Microenvironment. Front Cell Dev Biol. 2018;6:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Herlyn M, Steplewski Z, Herlyn D, Koprowski H. Colorectal carcinoma-specific antigen: detection by means of monoclonal antibodies. Proc Natl Acad Sci U S A. 1979;76:1438–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rupp AK, Rupp C, Keller S, Brase JC, Ehehalt R, Fogel M, et al. Loss of EpCAM expression in breast cancer derived serum exosomes: role of proteolytic cleavage. Gynecol Oncol. 2011;122:437–46. [DOI] [PubMed] [Google Scholar]
- 52.Tauro BJ, Greening DW, Mathias RA, Mathivanan S, Ji H, Simpson RJ. Two distinct populations of exosomes are released from LIM1863 colon carcinoma cell-derived organoids. Mol Cell Proteomics. 2013;12:587–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Im H, Shao H, Park YI, Peterson VM, Castro CM, Weissleder R, et al. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat Biotechnol. 2014;32:490–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Castillo J, Bernard V, San Lucas FA, Allenson K, Capello M, Kim DU, et al. Surfaceome profiling enables isolation of cancer-specific exosomal cargo in liquid biopsies from pancreatic cancer patients. Ann Oncol. 2018;29:223–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dong H, Strome SE, Salomao DR, Tamura H, Hirano F, Flies DB, et al. Tumor-associated B7–H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med. 2002;8:793–800. [DOI] [PubMed] [Google Scholar]
- 56.Chen G, Huang AC, Zhang W, Zhang G, Wu M, Xu W, et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. 2018;560:382–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang Y, Li CW, Chan LC, Wei Y, Hsu JM, Xia W, et al. Exosomal PD-L1 harbors active defense function to suppress T cell killing of breast cancer cells and promote tumor growth. Cell Res. 2018;28:862–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Theodoraki MN, Yerneni SS, Hoffmann TK, Gooding WE, Whiteside TL. Clinical Significance of PD-L1(+) Exosomes in Plasma of Head and Neck Cancer Patients. Clin Cancer Res. 2018;24:896–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Andreu Z, Yanez-Mo M. Tetraspanins in extracellular vesicle formation and function. Front Immunol. 2014;5:442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hough KP, Trevor JL, Strenkowski JG, Wang Y, Chacko BK, Tousif S, et al. Exosomal transfer of mitochondria from airway myeloid-derived regulatory cells to T cells. Redox Biol. 2018;18:54–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu Y, Xiang X, Zhuang X, Zhang S, Liu C, Cheng Z, et al. Contribution of MyD88 to the tumor exosome-mediated induction of myeloid derived suppressor cells. Am J Pathol. 2010;176:2490–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


