Abstract
A successful acute inflammatory response results in the elimination of infectious agents by neutrophils and monocytes, followed by resolution and repair through tissue-resident and recruited macrophages. Resolvins (D-series and E-series) are pro-resolving lipid mediators involved in resolution and tissue repair, who’s intracellular signaling remains of interest. Here, we report that D-series resolvins (RvD1- RvD5) activate phospholipase D (PLD), a ubiquitously expressed membrane lipase enzyme activity in modulating phagocyte functions. The mechanism for PLD-mediated actions of Resolvin-D5 (RvD5) in polarizing macrophages (M1-like towards M2-like) was found to be two-pronged: (a) RvD5 inhibits post-transcriptional modifications, by miRs and 3’exonucleases that process PLD2 mRNA, thus increasing PLD2 expression and activity; and (b) RvD5 enhances PLD2-S6Kinase signaling required for membrane expansion and efferocytosis. In an in vivo model of second organ reflow injury, we found that RvD5 did not reduce lung neutrophil myeloperoxidase levels in PLD2−/− mice compared to WT and PLD1−/− mice, confirming a novel role of PLD2 as the isoform in RvD5-mediated resolution processes. These results demonstrate that RvD5-PLD2 are attractive targets for therapeutic interventions in vascular inflammation such as ischemia-reperfusion injury and cardiovascular diseases.
Keywords: Macrophages, Inflammation, Resolution, Phospholipase D, Resolvins D-series, Polarization
INTRODUCTION
Acute inflammation is a response initiated by harmful stimuli, such as infections or injury, which activate endogenous mediators (1). These chemical mediators can generate a local inflammatory exudate. They also signal the activation of the first line of defense mechanism, infiltrating plasma proteins and innate inflammatory cells such as neutrophils, monocytes and macrophages by extravasation to the site of inflammatory insult (2). Macrophages are key players in chronic inflammatory diseases and are phagocytic cells involved in the clearance of apoptotic neutrophils a process that is central to the resolution phase (3).
Macrophages play a critical role in maintaining tissue homeostasis (4–6) in pathologies including but not limited to ischemia/reperfusion injury, tumor microenvironment, obesity, and conditions of chronic inflammation (7–11). Their diverse functions, including cytokine production and phagocytosis, place macrophages at the balance of pro-inflammation and resolution, both necessary components of the healing process (5, 6). Macrophages can polarize into M1 or M2 phenotypes (4, 5). M1 cells are deemed as pro-inflammatory and have a high microbicidal capacity and secrete pro-inflammatory cytokines, such as interleukin-1β (IL-1β), interleukin-12 (IL-12) and tumor necrosis factor α (TNF-α) (7). In contrast, M2 cells mediate resolution of inflammation by secreting interleukin-10 (IL-10) and transforming growth factor β (TGF-β) (7). Pro-inflammatory signals, such as toll-like receptor (TLR) ligands and interferon γ (IFNγ), induce polarization to the M1 phenotype, while anti-inflammatory signals, such as interleukin-4 (IL-4) and IL-10, induce polarization to the M2 phenotype (9). Macrophage phenotype switch is critical to proper macrophage function and tissue homeostasis. However, disruption of the M1/M2 balance results in a number of different pathologies (5, 7, 12).
The process of inflammation has two classic phases, with an inflammatory insult the body initiates an acute-inflammatory response, which eliminates foreign agents, followed by resolution and repair, where tissue-resident macrophages have important roles. Resolution, the process of returning to baseline conditions occurs via a well-regulated endogenous anti-inflammation program aimed to restore homeostasis (13). Resolvins belong to a new superfamily of specialized proresolving lipid mediators that have specific roles in the resolution of inflammation and enhance tissue regeneration and repair (13–16). Resolvins stimulate innate killing mechanisms to manage bacterial loads and stimulate clearance of bacteria (3). For example, Resolvin E1 exhibits a time-dependent response to inflammation resolution. During the short or acute inflammatory response period (0–4h), resolvins promote phagocytosis via activation of S6Kinase that phosphorylates ribosomal S6 protein resulting in phagosome formation and hence phagocytosis (14).
Phospholipase D (PLD) is a ubiquitously expressed membrane-associated lipase that hydrolyzes phosphatidylcholine (PC) into free choline and phosphatidic acid (PA) (15–17). PLD is upregulated in response to various cell stressors, such as hypoxia and nutrient starvation (18, 19). Moreover, it has been shown that PLD2−/− mice produce less pro-inflammatory cytokines in a sepsis model (20). The product of the PLD catalytic reaction, PA, is itself a mitogen and a critical secondary messenger that activates many downstream pathways leading to cell growth and proliferation, vesicle trafficking and cell migration (15–17). Additionally, PA is conical in shape and carries a negative charge, meaning its accumulation in the membrane results in membrane curvature needed for cell migration (19, 21–23). Although PLD/PA play a role in macrophage adhesion (21, 24), no role of PLD in macrophage polarization has been described as of yet.
Phospholipase D (PLD, has two major mammalian isoforms, PLD1 and PLD2) has a pivotal role in cellular activities such as chemotaxis and phagocytosis (17, 25–30). Aberrant PLD expression and activity is implicated in chronic inflammation (19, 31–34) and other cellular functions (17, 35). PLD is also actively involved in pro-inflammatory cytokine recruitment, reactive oxygen species (ROS) generation, chemotaxis, and cell invasion (36–39). In this report, we set out to investigate the resolvins and the potential role of PLD as an intracellular signaling molecules in their actions in acute inflammation and resolution.
MATERIALS AND METHODS
Materials.
Human macrophages were derived from peripheral blood mononuclear cells (PBMCs) purchased from Boston Children’s Hospital Blood Bank. RAW264.7 murine macrophages (cat. # TIB-71) and DMEM (cat. # 30–2002) were obtained from ATCC (Manassas, VA, USA). RPMI 1640 with L-glutamine and 25 mM HEPES (cat. # SH30255.01) and ECL reagent (cat. # RPN2106) were from GE Healthcare Life Sciences (Logan, UT, USA). Fetal bovine serum (heat inactivated) (cat. # 900–108) and Penicillin/Streptomycin (10,000 units penicillin/10,000 mg/ml streptomycin) (cat. # 400–109) were from Gemini Bio-Products (West Sacramento, CA, USA). 5×10−3 M EDTA, pH 8.0 (cat. # 15575–038) was from Life Technologies (Carlsbad, CA, USA). Recombinant mouse M-CSF (cat. # 315–02) was from PeproTech (Rocky Hill, NK, USA). Sterile-filtered Histopaque 1119 (cat. #11191) was from Sigma-Aldrich (St. Louis, MO, USA). Mouse isotope control antibody (cat. # 553476) was obtained from BD Biosciences (San Diego, CA). shRNAs were purchased from Santa Cruz biotechnology (cat. # sh-44000-SH (shPLD1) and sh-44001-SH (shPLD2)). miR-138 (cat. # 002284) and miR-887 (cat.# 002374) were purchased from Taqman (cat.# 4427975). Resolvins were obtained from Dr. Charles Serhan’s lab and Cayman Chemicals (cat. # 10007279 CAS# 810668–37-2 (RvD2) and cat. # 10007280 CAS# 578008–43-2 (RvD5)).
Animals.
Mice used in the present study were 6–8 weeks old male or female wild-type C57BL/6J (Charles River Laboratories, Charleston, SC, USA), PLD1−/− or PLD2−/− mice (weighing 20–25 g). PLD1−/− were obtained from Dr. Yasunori Kanaho’s laboratory, University of Tsukuba, Tennodai, Japan with exon 13 removed (40). PLD2−/− were from Dr. Gilbert Di Paolo’s laboratory, Columbia University with exons 13–15 removed (41). The PLD1−/− and PLD2−/− mice were backcrossed with C57BL/6J mice for >7 generations (40, 41). The mice were maintained in a temperature- and light-controlled environment with unrestricted access to food (laboratory standard rodent diet 5001; PMI Nutrition International, St. Louis, MO, USA) containing 4.5% total fat with 0.3% ω−3 fatty acids and <0.02% C20:4 and were provided ad libitum. The mice received veterinary care every day, and experiments were performed in accordance with the Harvard Medical School Standing Committee on Animals guidelines for animal care (Protocol# 02570). Experiments for this manuscript have also followed the National Institutes of Health Guide for the Care and use of Laboratory Animals (NIH Publications No. 8023, revised 1978).
Ischemia-reperfusion-induced second-organ injury.
Mice were anesthetized by i.p. injection of ketamine/xylazine mixture (80mg/kg;10mg/kg). Hind limb ischemia was induced using rubber band tourniquets tied on each hind limb as previously described (42). Hind limb ischemia was induced for 1 h, after which the tourniquets were removed to begin reperfusion. Resolvin D5 (RvD5) was administered at 0.5 μg/mouse in vehicle (PBS + 0.1% ethanol) and compared to vehicle alone or no ischemia reperfusion control. RvD5 was administered intravenously ~5 min prior to the start of reperfusion. At the end of this reperfusion period (2 h), these mice were euthanized with an overdose of anesthetic (isoflurane) and cervical dislocation. The lungs were quickly harvested and either flash frozen in liquid nitrogen and stored at −80°C or fixed and stored in 4% paraformaldehyde for H&E and immunohistochemistry. The right lungs (flash frozen) from individual mice were homogenized and centrifuged, and the tissue levels of myeloperoxidase (MPO) in the supernatants were determined using a mouse MPO ELISA (R&D Systems, Minneapolis, MN, USA). The fixed tissues (left lungs) were sectioned and stained for hematoxylin and eosin (H&E) (AML laboratories Inc., FL, USA) to study its histology and degree of tissue damage.
Myeloperoxidase (MPO) assay.
MPO is an enzyme that is mostly present in neutrophils and correlates with the extent of neutrophil infiltration into tissues. The MPO assay was performed using the MPO Mouse Myeloperoxidase DuoSet ELISA (R&D Systems, Minneapolis, MN) (cat# DY3667). Lungs were harvested from mice 2 hours after reperfusion. Samples were washed in cold phosphate-buffered saline (PBS, pH 7.4), immediately frozen on dry ice and stored at −80°C. For MPO assay, the lung samples were thawed, weighed, and homogenized in 1x PBS (pH 7.4) and centrifuged (1000 x g, 5 min, 4°C). The resulting supernatant was used for the MPO assay. Assay plates were prepared by coating the plate with capture antibody overnight at room temperature. The plates were then washed and blocked with blocking solution for 2h. After blocking, samples were added with respective control and standards. After thorough washing, detection antibody was added and incubated at room temperature for 2 h. Then the wells were washed thoroughly and streptavidin-HRP at 1:100 dilution was added for 20 minutes, followed by substrate for 20 min at room temperature. The reaction was stopped using a stop solution (2N H2SO4). The samples were read in a micro-plate reader at 450 nm with wavelength correction set to 540 nm or 570 nm. Results were expressed as units per microgram (wet weight) of protein in tissue.
Histopathology of lung tissue.
Lung tissue was fixed in 4% paraformaldehyde. The formalin fixed-tissue samples were used for sectioning for unstained sections or tissue histology H&E staining (AML Laboratory Inc., FL). EVOS™ XL Core Imaging System was used for microscopic analysis.
Macrophage culture and polarization to M1 and M2 phenotypes.
Human monocytes were isolated from de-identified peripheral blood Leukopaks obtained from Boston Children’s Hospital Blood Bank (Boston, MA). Blood was obtained from healthy human volunteers giving informed consent under protocol #1999-P-001297 approved by the Partners Human Research Committee. All procedures were conducted in accordance with the relevant guidelines and regulations. The leukocytes-rich pack was used to isolate peripheral blood mononuclear cells (PBMCs) by Ficoll-Histopaque 1077–1 (Sigma-Aldrich, St. Louis, MO) density gradient. PBMCs were differentiated into macrophages (M0) using GM-CSF (20 ng/mL) or M-CSF (20 ng/mL) in RPMI culture media (Lonza, Walkersville, MD, USA) containing 10% FBS (Invitrogen, Carlsbad, CA, USA), 5 mM L-Glutamine (Lonza), and 1% penicillin and streptomycin (Lonza) for 7 days with media changes on days 3 and 6 at 37° C in an incubator with a humidified atmosphere of 5% CO2. For selected experiments (indicated as such in the figures’ legend) RAW264.7 murine macrophages were used instead of human macrophages. RAW264.7 cells were purchased from ATCC and maintained in DMEM (cat. # 30–2002) with 10% FBS and 1% penicillin and streptomycin at 37° C in an incubator with a humidified atmosphere of 5% CO2. For polarization of differentiated M0 macrophages towards M1 and M2 phenotypes, published methods (43) were followed. Briefly, M ‘zero’ denoted M0 macrophages were polarized to M1 macrophages by the addition of 100 ng/mL LPS + 20 ng/mL IFN-γ to cultures and maintained for 1 day. M ‘zero’/M0 macrophages were polarized to M2 macrophages by the addition of 20 ng/mL IL-4 to cultures and maintained for 2 days. At the end of these time periods, M0, M1 or M2 macrophages were plated in a 6-well plate at 1×106 cells/well or in 8-well chamber slides at 5×104 cells/well overnight before experiments. For experiments, D-series Rv solution in ethanol was evaporated under a steam of nitrogen gas, followed by resuspension in PBS pH 7.2 and water-bath sonication. D-series Rv solution in PBS was added to culture media to attain a final concentration of 10 nM (44–47).
Macrophage phagocytosis measurements.
For phagocytosis, macrophages were incubated with fluorescent E. coli. and fluorescence was measured by real-time microscopy, unless otherwise indicated. Human macrophages were pre-incubated with vehicle (DPBSCa+/Mg+) or D-series Rv (10 nM) in chamber slides kept in a Stage Top Incubation system for microscopes equipped with a built-in digital gas mixer and temperature regulator (TOKAI HIT model INUF-K14). Incubations were maintained for 15 minutes at 37 °C before addition of 1:50 green fluorescence protein (BacLightGFP)-labeled E. coli (5×108 CFU/mL) (7 μg/mL, BacLight, Molecular Probes, Eugene, OR, USA). Fluorescence was assessed after 60 min (37 °C) using a BZ9000 real–time (BIOREVO) inverted fluorescence phase-contrast microscope (×20 objective) equipped with a monochrome/color-switching camera using BZ-II Viewer software (Keyence, Itasca, IL, USA). Incucyte™ Basic software computes green fluorescence intensity of phagocytosed E. Coli by macrophages from confocal images. In selected experiments (to directly compare phagocytosis with efferocytosis) phagocytosis of E. Coli was also measured by flow cytometry with 3 × 105 cells treated with BacLight labeled E. Coli in the ratio of 1:50 cells:bacteria.
Macrophage efferocytosis.
For efferocytosis, macrophages were incubated with apoptotic human polymorphonuclear neutrophils (PMN). PMN were isolated by density-gradient Ficoll-Histopaque from human peripheral blood. Peripheral blood was obtained from healthy human volunteers giving informed consent under protocol # 1999-P-001297 approved by the Partners Human Research Committee. Isolated neutrophils were allowed to undergo apoptosis by plating 1 × 107 cells/mL in 5 mL DPBSCa+/Mg+ for 24 h in 100 mm x 20 mm Petri dishes and then stained with 10 μM, CellTrace™ CFDA (Molecular Probes, Eugene, OR, USA) for 1h. Human macrophages were pre-incubated with vehicle (DPBSCa+/Mg+ + 0.1% ethanol) or D-series Rv (10 nM) for 15 minutes at 37 °C before addition of CFDA-labeled apoptotic PMNs (48, 49). Apoptotic PMNs were added to 3×105 macrophages in a ratio of 5:1. Apoptotic PMNs were added into 6-well plates with macrophages for 1 hour, after the cells were washed thoroughly and kept on ice, to stop any additional efferocytosis. This 10nM concentration of RvDs was selected based on results of earlier studies (44–47). These cells were harvested and subject to flow cytometry staining to measure mean fluorescence intensity as a read of efferocytosis with appropriate controls. A dilution 1:50 of Trypan blue was added to quench extracellular fluorescence.
Cell culture and transfections.
Human macrophages were transfected with plasmid DNA, HA-PLD1 or myc-PLD2 for overexpression of PLD1 or PLD2 for 48 h or shRNA, shPLD1 (cat. # sc-44000-SH) or shPLD2 (cat. # sc-44001-SH) (Santa Cruz Biotechnology, Dallas, TX) for silencing PLD1 or PLD2 for 72 h using JetPEI macrophage transfection reagent (cat. # 103–05N, Polyplus transfection, NY). RAW 264.7 macrophages were transfected with 1–2 μg plasmid DNA for 18–24 h, HA-PLD1 or myc-PLD2 using Amaxa® Cell Line Nucleofector® Kit V reagent (cat. # VCA-1003, Lonza, Switzerland).
Flow cytometry.
106 cells in 100 μL FACS buffer were stained with fluorochrome-labelled antibodies for macrophage type-specific cell surface markers: anti-mouse CD23 (cat. # 101618, clone B3B4, Per.CP/Cy5.5, 7 μl, Biolegend, CA), anti-mouse CD38 (cat. # 102705, clone 90, FITC, 3 μl, Biolegend, San Diego, CA), anti-mouse CD80 (cat. # 104707, clone 16–10A1, PE, 3 μl, Biolegend, CA), anti-mouse CD206 (cat. # 141708, clone C068C2, APC, 3 μl, Biolegend, CA), anti-human CD80 (cat. # 305208, clone 2D10, PE, 10 μl, BD Pharmingen, CA), anti-human CD86 (cat. # 555660, clone 2331, APC, 10 μl, BD Pharmingen, CA), anti-human CD163 (cat. # 326512, clone RM3/1, PerCP-Cy5.5, 15 μl, BD Pharmingen, CA), anti-human CD206 (cat. # 551135, clone 19.2, FITC, 10 μl, BD Pharmingen, CA). Cell counts were obtained before flow staining by trypan blue exclusion and normalized to single stained and isotype controls on a FACSCanto II (BD Biosciences) or BD Accuri c6 (BD Biosciences) and FlowJo Ver. 10 software (Tree Star, Ashland, OR, USA) or BD FACS Xpress software were used for analyses.
PLD activity.
Total PLD activity was measured by an enzyme assay using [3H]-butanol in an in vitro transphosphatylation reaction to yield [3H]-phosphobutanol. Briefly, lysed macrophages (50 μg) were processed for PLD2 activity in PC8 liposomes and [3H]n-butanol beginning with the addition of the following reagents (final concentrations): 3.5 mM PC8 phospholipid, 1 μM PIP2, 45 mM HEPES (pH 7.8), and 1.0 μCi [3H]n-butanol in a liposome form, as described (27, 50, 51). Samples were incubated for 20 min at 30oC with continuous shaking. Addition of 0.3 ml ice-cold chloroform/methanol (1:2) stopped the reactions. Lipids were isolated, dried (under N2) and suspended in chloroform:methanol (9:1) and then spotted on thin-layer chromatography plates along with 1,2-dipalmitoyl-sn-glycero-3-phosphobutanol (PBut) (Avanti Polar Lipids, Inc., AL) authentic standards. The amount of [3H]-phosphatidylbutanol ([3H]-PBut) that co-migrated with PBut standards (Rf~0.45+0.36) was measured by scintillation spectrometry and background subtracted. Results were expressed as total PLD enzymatic activity as dpm/mg protein/min.
Quantitative Real-time Reverse Transcription Polymerase Chain Reaction (qRT-PCR) for mRNA measurements.
Reverse transcription, coupled to qPCR, was performed following published technical details (52). Total RNA was isolated from macrophages with the RNeasy mini-kit following the manufacturer’s protocol (Qiagen, Valencia, CA, USA). RNA concentrations were determined using a Nano-Drop spectrophotometer, and samples were normalized to 29 ng RNA/μl. Reverse transcription was performed with 2 μg of RNA, 210 ng of random hexamers/primers, 500 μM dNTPs, 84 units of RNase OUT and 210 units of Moloney murine leukemia virus reverse transcriptase and incubated at 42°C for 55 min. Quantitative PCR reactions were run with 100 ng of total input cDNA (3.45 μl), 10 μl of the gene expression assay (RT2 SYBR Green ROX qPCR Master Mix) (cat. # 330520, Qiagen, Valencia, CA, USA) and 1 μl of the relevant mouse RT2 qPCR Primer Assay. The following mouse primer sets were used from Qiagen (Valencia, CA, USA): TBP (PPM03560F) (used as housekeeping gene), PLD1 (PPH02835A), PLD2 (PPH02787A), S6K (PPH00791F), PLD3 (PPH02828A), PLD4 (PPH19319A) and PLD6 (PPH08933A), Actin (PPM02945B), PPARg (PPM05108C). Quantitative PCR conditions for the Stratagene Cycler were: 95°C for 10 min and then 50 cycles of the next 2 steps: 30 s 95°C and then 1 min 55°C, followed by 1 cycle of 1 min 95°C, 30 s 55°C and 30 s 95°C to establish the dissociation curves. The “cycle threshold” Ct values were chosen from the linear part of the PCR amplification curve where an increase in fluorescence can be detected at >10 S.E. above the background signal. DCT of sample was calculated as: ΔCTsample= CTGOI – CTGAPDH. Then, arithmetic mean of “control” ΔCTGOI from individual experiments (i.e., arithmetic mean of “control” ΔCTsample from one experiment with technical duplicates) was calculated for computing ΔΔCT= ΔCTsample – Arithmetic mean of “control” ΔCTsample. Relative gene expression was computed as 2−ΔΔCT (53).
qRT-PCR for micro-RNA quantification.
To measure miR-138 and miR-887 expression levels, cells that were polarized and treated with vehicle or Resolvins were used for RNA lysates using the Taqman micro-RNA Cells-to-CT kit according to the manufacturer’s protocol (Life Technologies; Cat. # 4391848). RNA concentrations were determined using a NanoDrop, and samples were normalized to ~66 ng/μl RNA. Reverse transcription was performed in a 15 μl reaction volume with 1 μg of RNA, 1.5 μl 10T Buffer, 1 mM dNTPs, 3.8 units of RNase Inhibitor and 1 μl of Multiscribe Reverse Transcriptase and incubated in one-cycle at 16 °C for 30 min, 42 °C for 30 min and then 85 °C for 5 min. Quantitative PCR reactions were run in a 20 μl reaction volume using 10 μl Taqman Master Mix, ~88 ng of total input cDNA, 1 μl of the relevant micro-RNA gene expression assay (6-carboxyfluorescein (FAM)-labeled) multiplexed with U6 housekeeping gene. Taqman miR primers and fluorescent probes were from Life Technologies. Quantitative PCR conditions for the Stratagene Cycler were: 95 °C for 10 min and then 40 cycles of the next 3 steps: 15 s 95 °C and then 1 min 60 °C. Cycle threshold Ct values were obtained as indicated for mRNA qRT-PCR (above paragraph).
SDS-PAGE and Western blot analyses.
To confirm the presence of endogenous PLD1, PLD2 and S6K proteins in RAW264.7 macrophages, we performed SDS-PAGE and western blot analyses specific for each of these three proteins, as well as using TBP as the equal protein loading control. Approximately 150 μg of protein lysate (lysis buffer composition was: 50 mM HEPES, pH 7.2, 100 μM Na3VO4, 0.1% Triton X-100 and 1 mg/ml each of protease inhibitors (aprotinin and leupeptin) was loaded per each lane of the SDS-gels that were then used for Western blot analyses. For western-blot analyses, rabbit PLD1 (F-12) IgG (Santa Cruz Biotechnology, Santa Cruz, CA, USA) (cat. # sc-28314), rabbit PLD2 (N-term) IgG (Abgent, San Diego, CA, USA) (cat. # AP14669a), rabbit S6K IgG (49D7), S6 (cat. # 2217) and pS6 (cat. # 4858). (Cell Signaling Technology, Danvers, MA, USA) (cat. # 2708) and rabbit TBP IgG (Cell Signaling Technology) (cat. # 8515) were utilized as primary antibodies according to the manufacturers’ recommendations. Anti-rabbit or -mouse IgG HRP antibodies were from Cell Signaling Technology (cat. # 7074 and 7076, respectively). Immunoreactivities were detected using enhanced chemiluminescence (ECL) reagents from GE Healthcare (Pittsburgh, PA, USA) (cat. # RPN2106) and autoradiograph film.
Statistical Analysis.
The sample size for experiments was chosen empirically based on previous studies to ensure adequate statistical power. Data presented in the figures as bars are mean ± Standard Error of the Mean (SEM) (standard deviation/n1/2, where n is the sample size). Experiments were performed in technical triplicates (for qPCR measurements or for functional assays) or technical duplicates (for PLD activity assays or for flow cytometry) for n=3–5 independent experiments. Statistical significance (*p < 0.05, **p < 0.01, and ***p < 0.001) between means was assessed by two-tailed t test in case of experiments with only two groups (control and test). For multiple comparisons, one-way analysis of variance (ANOVA) with Tukeyʼs multiple comparison (or comparisons to controls) post hoc tests were used. Analyses of data were conducted using GraphPad Prism 7 software (San Diego, CA).
RESULTS
PLD expression and activity in macrophages.
Since well-appreciated mechanisms separating self-limited acute inflammation from delayed resolution exists (3, 13), we questioned whether PLD would play a role in the resolution phase of acute inflammation in macrophages that are critical in resolution. In order to study the role of PLD in inflammation and resolution, we measure the expression levels of PLD in the different macrophage subpopulations that occur in the inflammation-resolution process. Specifically, we measured basal gene expression for Pld1 and Pld2 in M1 and M2 macrophages (Fig. S1A) relative to M0; basal levels of protein (Fig. S1B–C) and endogenous lipase activity from M0, M1 and M2 macrophages (Fig. S1D). M1 macrophages had a level of basal activity higher than the M0 used as control. Pld1 gene and PLD1 protein were highly expressed in M1-like macrophages, whereas Pld2 gene and PLD2 protein were highly expressed in M2 like macrophages. Our interest was to investigate the basis for the two isoforms, PLD1 and PLD2, that were differentially expressed in the inflammatory vs pro-resolving (M2) macrophages and whether PLD2 that is highly expressed in M2-like macrophages has a role in the resolution of inflammation.
D series Resolvins impact PLD’s gene expression and enzyme activity in macrophages.
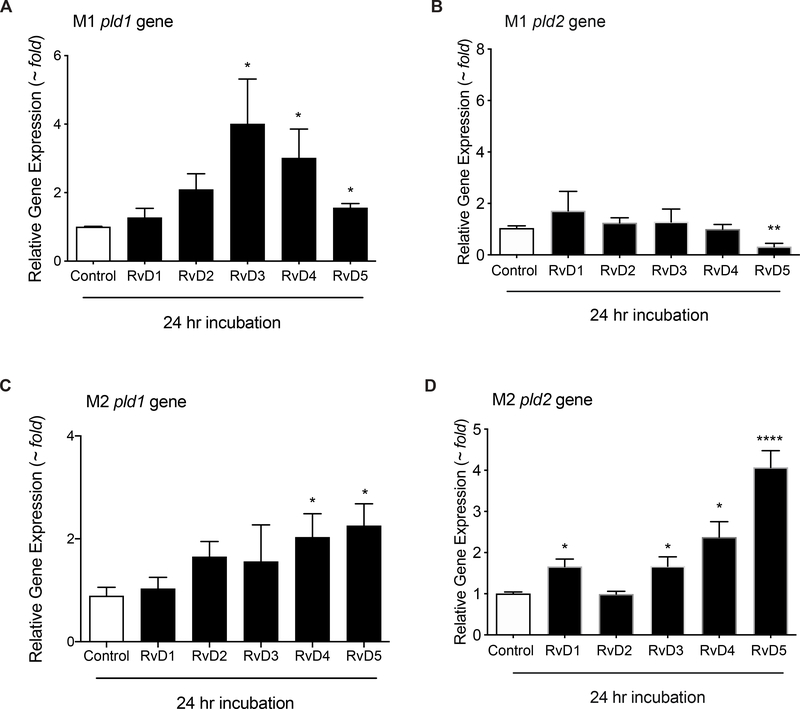
Having observed the basal expression of PLD1 and PLD2 in M1 and M2 macrophages, we next determined if Resolvins, a class of Specialized Pro-resolving Mediators (SPMs), had any effect on PLD mRNA levels in macrophages. We focused our interest on Resolvins of the D-series (RvDs), including RvD1, RvD2, RvD3, RvD4 and RvD5, and their effect on PLD activity of differentiated macrophages. We treated M0, M1 and M2 macrophages with vehicle (0.1% ethanol) or 10 nM D-series Resolvins (RvD1–5). In Supplemental Figs. S2, S3 report the impact of different periods of incubation of Resolvins-D1–5 (at 10 nM) on Pld1 (Fig. S2) and Pld2 (Fig. S3) gene expression in M0, M1 or M2 macrophages. These time points were chosen since resolvins are involved in resolution of acute inflammation and start their actions early on in inflammation soon after macrophages arrive to the site of inflammation. Here we observed that both RvD4 and RvD5 increased Pld1 gene expression in unpolarized M0 macrophages at 6 and 18 h post-treatment (Figs. S2). RvD4 alone increased Pld2 gene expression, while RvD1–4 increased Pld2 gene expression in the non-polarized M0 macrophages at 18h (Fig S3). Since M0 macrophages are functionally naïve, we used them for comparison as controls for polarization and macrophage phenotype functionality. We observed that even though RvD1 or RvD3 had some punctual effect on M2’s at 24h, both RvD4 and RvD5 consistently gave the largest increase on altering PLD gene expression. Since 6 h of incubations of Resolvins with macrophages was not enough to elicit statistically significant changes in PLD gene expression, we concentrate in Fig. 1 on 24 h of incubation with macrophages. We observed a statistically significant increase in Pld1 gene expression in M1 (Fig. 1A) and M2 macrophages (Fig. 1A). Interestingly, RvD5 decreased Pld2 gene expression in M1 macrophages (Fig. 1C), whereas RvD5 induced a robust increase in Pld2 gene expression in M2 macrophages (Fig. 1D). In fact, in M2 macrophages, other resolvins (RvD1, RvD3 and RvD4) were also able to induce an increase in Pld2 gene expression, albeit at lower levels than RvD5.
Figure 1. D-series Resolvins actions on PLD1 and PLD2 gene expression in M1 and M2 macrophages.
Effect of D-series Resolvins (each at 10 nM) on endogenous PLD1 (A,B) or PLD2 (C,D) gene expression in M1 or M2 macrophages. For each condition, 1×106 cells were treated with 10 nM of each of the indicated Resolvins-D series for 24 hours in continuous culture. Gene expression was measured by qRT-PCR (n=3 with technical duplicates). Data presented are means ± SEM; statistical significance (*p < 0.05, **p < 0.01, and ***p < 0.001) was evaluated with one-way ANOVA and Tukey’s post hoc comparing samples with controls (vehicle only) for each panel. See supplemental figures S2 and S3 for a full range of Pld1 and Pld2 gene changes at different time lengths of macrophages incubation with RvDs.
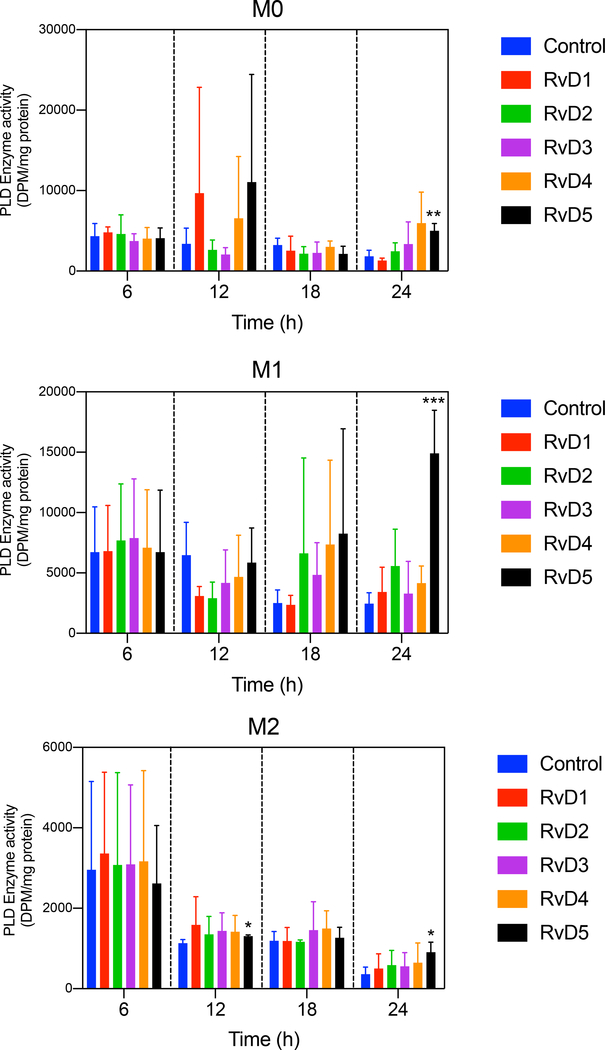
We next measured PLD enzyme activity at 6, 12, 18 or 24 h in M0, M1 and M2 macrophages (Fig. 2) following treatment with D-series resolvins. Both RvD4 and RvD5 regulated PLD activity in M0, M1 and M2 macrophages in a time-dependent fashion (significantly increased after 24 h incubation with RvD5). As observed earlier for PLD gene expression, RvD4 and RvD5 each maximally stimulate PLD activity. It should also be noted that the in vitro PLD assay monitors the combined activity of both PLD1 and PLD2 and PLD enzyme activation is regulated by factors such as substrate presentation and membrane translocation (50). Since activity is upregulated by RvD4 and RvD5 and so is PLD1 gene expression (and PLD2 is downregulated) it seems plausible that the effects of RvD4 and RvD5 on M1 are mediated via PLD1. Conversely, since the main PLD isoform expressed in M2 is PLD2, it is plausible that the actions of RvD4 and RvD5 with M2 are mediated via PLD2. However, since PLD enzyme activity in the cellular milieu namely within intact cells was not directly monitored it is thus a limitation of the present experiments.
Figure 2. Actions of D-series Resolvins on PLD enzymatic activity in M0, M1 and M2 macrophages.
Effect of D-series Resolvins (each at 10 nM) on total PLD activity in M0, M1, or M2 macrophages. For each condition, 1×106 cells were treated with 10 nM of each of the indicated Resolvins-D series at 6 (n=6),12 (n=3),18 (n=3) or 24 (n=3) hours in continuous culture. Total PLD activity was measured by an enzyme assay using [3H]-butanol in an in vitro transphosphatidylation reaction to yield [3H]-phosphobutanol. Data presented are means ± SEM; statistical significance (*p < 0.05, **p < 0.01, and ***p < 0.001) was evaluated with one-way ANOVA and Tukey’s post hoc comparing samples with controls (vehicle only) at a given time point in each panel.
We were also interested in investigating other Pld genes in these cells. There are PLD enzyme isoforms other than PLD1 or PLD2, namely PLD3, PLD 4 and PLD 6. The scheme (Supplemental Fig. S4A) shows the “classical” (PLD1 and PLD2) and “non-classical” (PLD3, PLD4 and PLD6), (note: PLD5 has not been fully described), where mammalian PLD architecture depends on the protein architecture regulatory domains. All PLD’s have one or two HKD “lipase signature” domains, but only PLD1 and PLD2 have important regulatory domains (PX and PH) that allow the protein to anchor strongly to cellular or the intracellular membranes. In macrophages, we observed that not all Pld (i.e. Pld1–6) genes are equally expressed. Supplemental Fig. S4B compares the basal gene expression levels of Pld1, Pld2, Pld3, Pld4, and Pld6 in M1 and M2 macrophages, indicating that the levels of basal expression of Pld1 and Pld2 are the highest. We measured the gene expression levels of non-classical Plds in M1, and M2 macrophages treated with 10 nM D-series Resolvins for 24 h. We observed that after 24 h of D-series resolvins treatment, RvD5 reduced the non-classical Pld genes expression in M1 macrophages and activation in M2 macrophages, except pld6 gene expression which was inhibited by RvD2 in M1 macrophages (Supplemental Fig. S4C–E). Overall, Pld3, Pld4 and Pld6 follow the same trend of expression in response to these D-series Resolvins as Pld2 (Figs. 1B,1D). Taken together, these results show that RvD5 consistently increased expression of PLD in pro-resolving M2 macrophages, while reducing PLD expression in pro-inflammatory-M1 macrophages, with the exception of PLD1.
Resolvin D5 increases macrophage PLD function.
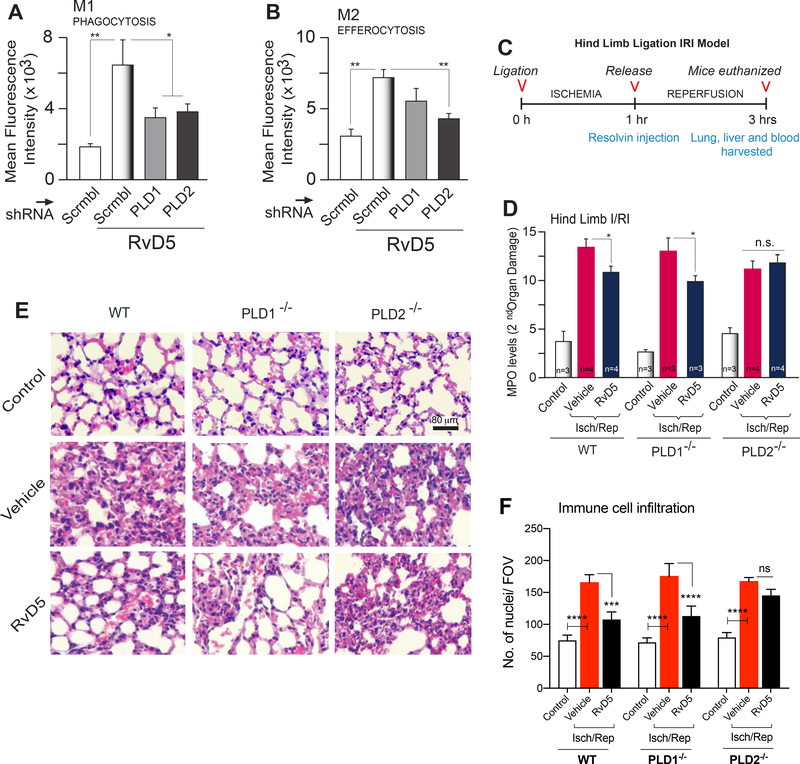
To investigate the direct actions of RvD5 on regulating PLD, we performed macrophage functional assays by silencing PLD1 or PLD2 in human macrophages (silencing was confirmed by measuring PLD enzyme activity), followed by RvD5 treatment for the duration of the assays. We found that silencing PLD1 or PLD2 significantly inhibited RvD5’s action on phagocytosis of E. coli by M1 macrophages (Fig. 3A) whereas, only PLD2 silencing affected RvD5’s action on efferocytosis of apoptotic neutrophils by M2 macrophages (Fig. 3B). Thus, both PLD1 and PLD2 are essential for RvD5-mediated functions on macrophage phagocytosis (function of M1 macrophages), while only PLD2 isoform is important for efferocytosis (function of M2 macrophages).
Figure 3. RvD5 protects mice from second organ injury in a PLD dependent manner.
(A) Phagocytosis. Fluorescence intensity of phagocytosis of BacLightGFP-labeled E. coli by real-time microscopy of M1 macrophages upon silencing PLD1 or PLD2 with RNAi for 72 hr, followed by incubation with 10 nM RvD5 for 1 hr. E. coli was added in a ratio of 1:50 to 3×105 macrophages (n=3). (B) Efferocytosis. Fluorescence intensity of fluorescent-(CellTraceTM-CFDA)-labeled apoptotic neutrophils (PMNs) was measured by flow cytometry, using M2 macrophages upon silencing PLD1 or PLD2 with RNAi for 72 hr, followed by incubation for 1 hr with 10 nM RvD5. A macrophage to CFDA-labeled apoptotic PMN ratio was added at 1:5 with 3×105 cells/well (n=3). (C) Schematic timeline of the used hind limb ischemia/reperfusion injury (HLIR) procedure. (D) Myeloperoxidase (MPO) levels (units/μg (wet weight) of protein in tissue) were measured using ELISA from lung lysates of WT, PLD1−/− or PLD2−/− mice treated with vehicle or 10 nM of RvD5. (E-F) Representative photomicrographs (from a total of 10 fields/per condition observed) showing pathology of lung sections by H&E staining and quantification of cell infiltration from control, vehicle or RvD5 treated mice. Scale bar=80 μm. Results are expressed as mean ± SEM; statistical significance (*p < 0.05, **p < 0.01, and ***p < 0.001; n.s.=not significant) was evaluated with one-way ANOVA and Tukey’s post hoc comparing samples with controls (Scrbl=scrambled RNA in (A,B) or control in WT mice in (D)).
RvD5 activates PLD-mediated inflammation resolution in vivo.
Furthermore, to validate the in vivo relevance of the effects of RvD5-PLD signaling in sterile injury and inflammation, we performed ischemia reperfusion injury and assessed second-organ injury to the lungs. It has been previously shown that RvD2 prevents reflow injury in the lungs by impeding neutrophil infiltration and functions (49). To this date a role of RvD5 in second organ injury has not been studied. Our lab has demonstrated PLD’s role in phagocyte functions, cell migration in many cell types, including neutrophils and macrophages (23, 31–33), in pathological conditions such as cancer and atherosclerosis (23, 30). Uncontrolled inflammation is linked to defective generation of pro-resolving mediators, such as in asthma, which suggests that resolution of acute inflammation is critical (54). As the damaged, ischemic tissue initiates an intense innate immune response (inflammation), we next determined if RvD5 could decrease second organ injury mediated by PLD and leukocyte-mediated tissue injury. RvD5 or vehicle control were administered to PLD1−/−, PLD2−/− and WT mice in a second organ ischemia reperfusion injury after hind limb ischemia.
Hind limb ischemia was performed for 1 h at which time vehicle or 500 ng of RvD5 were administered by i.v. injection and then reperfusion was allowed for 2 h. The outline used for the hind limb ischemia/reperfusion injury (HLIR) is given in Fig. 3C. Following completion of reperfusion, second organ reperfusion lung injury was assessed by myeloperoxidase (MPO) to determine PMN infiltration (Fig. 3D) of lung lysates from WT, PLD1−/− or PLD2−/− treated with vehicle or 500 ng RvD5 at the beginning of reperfusion. Albeit the levels of MPO in the vehicle treated group of PLD2−/− mice did not increase as much as WT or PLD1−/− mice, the difference in MPO levels was not significant. Of interest, we found that RvD5 reduced MPO levels in both WT and PLD1−/− mice, indicating a reduction in PMN infiltration. In contrast, RvD5 did not affect the lung levels of MPO in the PLD2−/− mice. Based on these results, we conclude that RvD5 functions via the PLD2 isoform and not the PLD1 isoform.
We also performed H&E staining on the lung tissue sections to observe lung pathology (second-organ injury) after hind-limb ischemia reperfusion injury (Fig. 3E). We observed increased neutrophil infiltration and loss of honeycomb lattice structure in the WT, PLD1−/− and PLD2−/− mice compared to non-ischemia controls. We also quantified total immune cell infiltration in the lungs by counting nuclei per field of view in H&E images (Fig. 3F) that is in accordance with the MPO data. Lung injury, neutrophil and other immune cell infiltrates were reduced in RvD5 treated mice in WT and PLD1−/−, but not in PLD2−/− confirming that RvD5 functions via PLD2 during HLIR injury. Based on these results, we conclude that PLD2 is the important isoform for RvD5-mediated effects on neutrophil infiltration in vivo.
M1 to M2 macrophage polarization: a role for PLD.
Resolvins are well known to have a role in promoting macrophage “phenotype switch” toward M2-like phenotype (55, 56), which results in increased efferocytosis by M2 macrophages and promotes resolution of inflammation (43, 49). Studying the role of PLD altering M1 and M2 specific cell surface markers’ expression in the non-polarized M0 macrophages as indicated in the previous figures, led us to further investigate the role of PLD in the process of inflammation and resolution. After an inflammatory insult, macrophages polarize to M1 phenotype and later undergo transient polarization to M2 macrophages to promote anti-inflammation and resolution via efferocytosis (43, 49). To study if PLD had a role in macrophage polarization during inflammation and later in resolution, we undertook two different approaches: (a) PLD silencing and transfection of non-polarized M0 macrophages; and (b) PLD silencing and transfection of M1 macrophages. Figs. 4A–B indicate that non-differentiated macrophages (M0) can be induced to express an M1-like phenotype with PLD ectopic expression; that is, overexpressed PLD2, significantly increased M1 marker expression CD80 and decreased M2 surface marker expression CD206 (Fig. 4A). As controls for this and the next series of experiments, PLD activity upon either silencing by RNAi or ectopic overexpression of PLD1 or PLD2 expression plasmids in M1 macrophages is shown in Fig. 4C. Thus, we found that an increase in PLD expression induces macrophage polarization, from M0-M1 confirming a role of PLD early in inflammation. Interestingly, silencing PLD2 gene in M0 macrophages increased M2 marker, CD206 expression suggesting a potential role for PLD2 in M2 macrophage polarization and anti-inflammation. However, since PLD is well-known to be upregulated upon an inflammatory insult, it is likely that a phase and stimuli-dependent role of PLD exists, first in inflammation and later in resolution of inflammation.
Figure 4. M1 to M2 macrophage reprogramming induced by PLD.
(A,B) Flow cytometry analysis of surface expression of polarization markers for CD80 and CD86-positive cells (M1 markers) or CD163 and CD206-positive cells (M2 markers). Experiments were performed in M’zero’ (M0) macrophages overexpressing either PLD1 or PLD2 (1.5 μg DNA plasmid per condition) after 24 hr transfection (n=3) (A) or silenced with shPLD1 or shPLD2 (100 nM per condition) after 72 hr transfection with shRNAs (“Scrbl”, scrambled RNA control) (n=4) (B). Shown in (C) are the transfection controls to show efficiency of either overexpression or silencing of PLD1 and PLD2 plasmids or shRNA, respectively (n=3). (D) Flow cytometry analysis of surface expression of polarization markers for CD38/CD86-positive cells (M1 markers) or CD23/CD206-positive cells (M2 markers) in M1 macrophages upon ectopic overexpression of PLD1 or PLD2 plasmids (n=3). See Supplementary Figure S9 for representative flow cytometry plots. (E) Gene expression of CD206 using qRT-PCR with or without RvD5 (10 nM) treatment in culture for 24 hrs (n=3). (F) Phagocytosis of M1 macrophages silenced with PLD1 or PLD2 shRNA. Fluorescence intensity of phagocytosis of BacLightGFP-labeled E. coli was measured by real-time microscopy of M1 macrophages upon silencing PLD1 or PLD2 with 100 nM shRNA (“Scrbl”, scrambled RNA control) for 72 hr, followed by incubation with 10 nM RvD5 for 1 hr. A cell to E. coli ratio was 1:50 with 3×105 cells. (inset=example of fluorescent-field view from the microscope) (n=4 independent experiments with a sample size of ~60 cells per condition/FOV). (G) Efferocytosis of M1 macrophages after transfection of PLD1 or PLD2 expression plasmids. Fluorescence intensity of green fluorescence intensity from efferocytosis of green fluorescence (CellTraceTM-CFDA)-labeled apoptotic neutrophils (PMNs) was measured by flow cytometry, upon transfection of PLD1 or PLD2 expression plasmids for 48 hr, followed by incubation for 1 hr with 10 nM RvD5. A macrophage to CFDA-labeled apoptotic PMN was added to a 1:5 ratio with 3×105 cells/well (n=3). (H) Effect of ResolvinsD1–5 on surface expression of polarization markers on M1 and M2 macrophages, measured by flow cytometry (n=3). Data in bar graphs are mean ± SEM; statistical significance (*p < 0.05, **p < 0.01, and ***p < 0.001) was evaluated with one-way ANOVA and Tukey’s post hoc multiple comparisons.
To investigate a phase contingent role of PLD in resolution, we began the experiments with M1 macrophages instead of M0. PLD is upregulated during inflammation, although to date, a phase and time specific role of PLD in late inflammation and resolution have not been studied. M1 macrophages were transfected with plasmids to overexpress PLD1 or PLD2 or with shRNA to silence PLD1 or PLD2. Flow cytometric analysis of CD38/CD86-positive cells (M1 markers) or CD23/CD206-positive cells (M2 markers) was then conducted (Fig. 4D). See gaiting strategy on Supplemental Fig. S5. Since CD206 is an M2 phenotype marker, we investigated CD206 gene expression in M1 macrophages in the presence of RvD5, wherein we found that RvD5 by itself induces the expression of CD206 (Fig. 4E).
Given that PLD plays an important role in macrophage phagocytosis (34, 57), we next assessed whether PLD would alter the macrophage phenotype-specific functions. To investigate this, we silenced PLD1 or PLD2 and measured phagocytosis of E. coli by M1 macrophages (Fig. 4F). We observed that silencing PLD1 or PLD2 significantly decreased E. coli phagocytosis by M1 macrophages. The inset in Fig. 4F presents a visual control of cells silenced with either PLD1 or PLD2 shRNA. The red bar indicates the level of phagocytosis of E. coli by M2 macrophages treated and measured in similar conditions as M1. When PLD2 was silenced in M1 macrophages, E. coli phagocytosis decreased. Thus, silencing PLD2 in M1 macrophages led to decreased phagocytosis confirming PLD’s role in early inflammation.
We also measured efferocytosis, a known function of resolving macrophages, and found that overexpressing PLD2 in M1 macrophages significantly augmented efferocytosis (Fig. 4G). The blue bar indicates the level of efferocytosis of M2 macrophages treated and measured in similar conditions as M1. Thus, when PLD2 was overexpressed in M1 macrophages (inducing an “M2-like” phenotype), apoptotic PMN efferocytosis increased. Interestingly, PLD1 did not affect efferocytosis which can be attributed to its cellular localization within organelle and vesicle membranes essential for phagolysosome formation essential for phagocytosis, whereas PLD2 localizes in the plasma membrane important for scavenger receptor detection of apoptotic cells. These results are is accordance with our previous study showing the crucial role of PLD2 signaling in scavenger receptor signaling in phagocytes (30). All together, we found that PLD had a role in phenotype-switch from M1 to M2 with the expected functions of those cell subpopulations. Thus, we identified a course-dependent role of PLD in macrophage polarization from M0-M1 and M1-M2.
Lastly, incubation of macrophages with RvD5 resulted in elevated CD163 M2 surface protein expression but no changes on CD80 M1 surface protein expression when compared to control (Fig. 4H). RvD1 is recognized as an inducer of macrophage polarization (55, 56, 58) but such similar role for RvD5 has not been previously described. Thus, findings in Fig. 4H and Fig. 1D confirm that RvD5 induces polarization to M2-like phenotype, along with increasing PLD2 expression. In summary, after finding out differential expression of PLD1 and PLD2 in the different polarized macrophage subpopulations, we observed that overexpressing PLD2 decreased M1 markers and increased M2 marker expression in M1 macrophages and showed an increase in apoptotic PMN efferocytosis by these macrophages. This is also supported by data showing RvD5 increasing M2 surface marker expression. Hence, these results confirmed our earlier findings, indicating a role of PLD in macrophage polarization and thus function.
Mechanism for PLD-mediated effect of RvD5 in macrophages: Involvement of miRs and PARN.
Because PLD2 mediates both phagocytosis and efferocytosis, while PLD1 mediates only phagocytosis, we decided to understand the possible mechanism(s) behind this newly uncovered role of RvD5-PLD2 signaling. For this, we first analyzed two post-transcriptional mechanisms that could target PLD2 transcripts and could be altered by RvD5: miRs and PARN mRNA deadenylase.
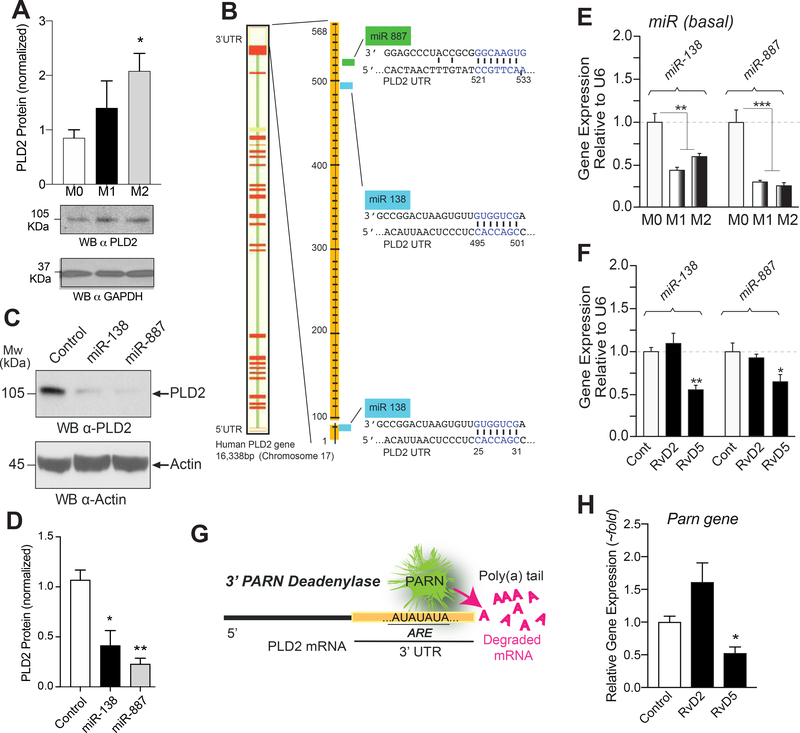
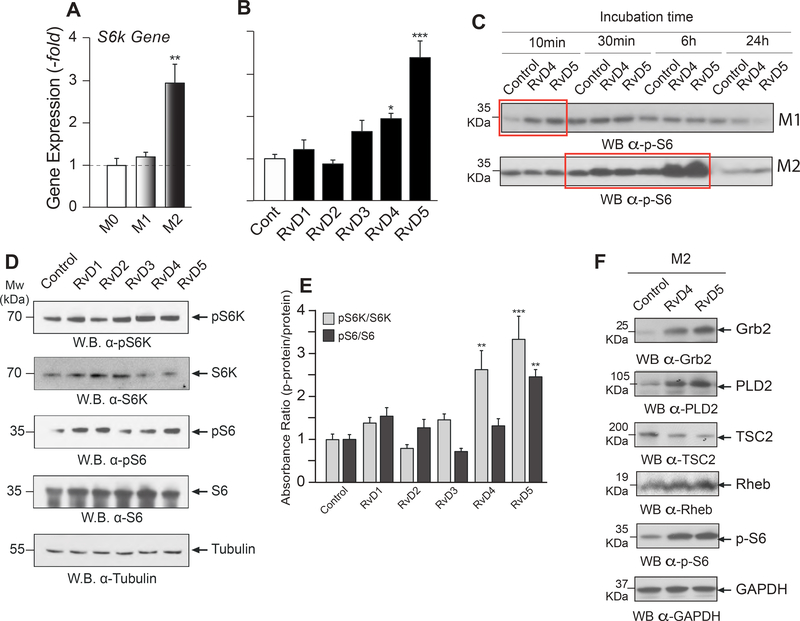
Protein expression of PLD2 (normalized to GAPDH) derived from Western blot analysis in M0, M1 and M2 resting macrophages (Fig. 5A) shows that PLD2 protein is highly expressed in M2 macrophages with respect to M0 and M1. There could be several reasons for this. We have studied previously post-translational modification of PLD2 transcripts and reported how that is under regulation of certain miRs (18, 22). For macrophages, we selected miR-138 and miR-887 that bind to the 3’UTR of PLD2 (Fig. 5B). The demonstration that transfected miR-138 and miR-887 decrease PLD2 expression in M0 macrophages is shown in Figs. 5C and 5D. The levels of endogenous miR expression are higher in resting M0 macrophages than in M1 or M2 macrophages (Fig. 5E), which could partially explain the results of protein expression. Furthermore, levels of miR expression in response to 10 nM RvD5 are further reduced in M2 macrophages (Fig. 5F). Our miRNA results are in agreement with previous findings on the role of Resolvins in miR biology (59–63). In the present study, we found that RvD5 inhibits PLD2-miRs (miR-138 and miR-887).
Figure 5. Mechanism for PLD-mediated actions of RvD5 in macrophages involving miRs and PARN.
(A) Protein expression of PLD2 (normalized to GAPDH) derived from Western blot analysis in M0, M1 and M2 macrophages (n=3). (B) Scheme depicting the 3’UTR of PLD2 with predicted target sites for miR-138 and miR-887 (n=3). (C-D) Demonstration that transfected miR-138 and miR-887 decrease PLD2 expression in macrophages (n=3). (E) Levels of endogenous PLD-miR expression (by qRT-PCR) in M0, M1 or M2 macrophages (n=3). (F) Levels of PLD-miR expression from macrophages that had been incubated with or without 10 nM RvD2 or RvD5 for 6 hrs (n=3). (G) Scheme depicting the 3’ exonuclease activity of Poly(A) Ribonuclease (PARN) as cleaving the poly-A tail of PLD mRNA transcripts in the cell cytoplasm, leading to mRNA decay (n=3). (H) Effect of RvD2 or RvD5 on PARN gene expression (n=3). Data from RAW264.7 macrophages are represented as means ± SEM; statistical significance (*p < 0.05, **p < 0.01, and ***p < 0.001) was evaluated with one-way ANOVA and Tukey’s post hoc comparing samples with controls (vehicle only) for each panel.
There are other possible post-transcriptional ways to regulate PLD2 expression, for example 3’exonucleases that can process mRNA transcripts in the cytoplasm, thus elevating the rate of mRNA decay (64). The scheme presented in Fig. 5G depicts the 3’ exonuclease activity of Poly(A) Ribonuclease (PARN) as cleaving the poly-A tail of mRNA transcripts leading to the ultimate destruction of mRNA by exosomes and P-bodies. Incubation of macrophages with RvD5 decreases the basal level of Parn gene expression (Fig. 5H). PARN silencing increases PLD2 expression (64), an effect that is also observed upon treatment with RvD5. Therefore, inhibition of PARN by RvD5 explains increased PLD2 expression. These findings confirm that RvD5 inhibits post-transcriptional regulators of PLD2 such as PLD2-miRs and PARN in M2 macrophages.
Potential intracellular mechanism for RvD5 signaling in macrophages
To understand the signaling cascade underlying the newly uncovered role of RvD5 signaling via PLD2, we analyzed two intracellular pathways in which PLD effectors are implicated, S6K and Actin polymerization (65) in phagocytes, and asked if they could be altered by RvD5. Earlier studies have shown some D-series Resolvins regulate phosphorylation of S6 in phagocytes to promote phagocyte functions and cellular processes indispensable for the resolution of inflammation (66). S6K is a morphogenic protein (67) that acts through Actin activation and is involved in cell shape change. Basal S6K gene expression was highest in M2 macrophages (Fig. 6A). We observed a significantly positive increase on S6K mRNA levels following RvD4 and especially RvD5 treatment in M2 macrophages (Fig. 6B). This was similar to PLD2 upregulation by RvD5 in M2’s described earlier (Fig. 1D).
Figure 6. Intracellular mechanism for RvD5 signaling involving S6K.
(A-E) S6K analysis. (A) Basal S6k gene expression by qRT-PCR in M0, M1, and M2 macrophages (n=3). (B) S6k gene expression changes upon 10 nM RvD1–5 treatment of M2 macrophages (n=3). (C) Effects of vehicle (Control), RvD4 or RvD5 (at 10 nM) on protein S6 phosphorylation [p(T232)-S6] in M1 or M2 macrophages (n=4). (D) Effects of RvD1–5 (at 10 nM) on S6K and phospho(T381)-S6K; S6 protein and phospho (T232)-S6 (with tubulin being loading control) (n=3). (E) Densitometry of protein bands such as those shown in panel D, with the calculated ratio ‘phospho-protein’/’protein’ for S6K and for S6, in response to Resolvins-D1–5 relative to control. (F) Analysis of proteins in the Grb2-PLD2 and PLD2-S6K signaling pathways (Grb2, PLD2, TSC2, Rheb and pS6) in M2 macrophages (n=4). Bottom row: Loading control GAPDH. Experiments from RAW264.7 macrophages are represented as mean ± SEM; statistical significance (*p < 0.05, **p < 0.01, and ***p < 0.001) was evaluated with one-way ANOVA and Tukey’s post hoc multiple comparisons.
We also observed a very large effect of RvD5 on S6 phosphorylation (Fig. 6C and Supplemental Figs. S6A–B), a ribosomal protein that is a natural substrate of S6K. S6K and phospho (T381)-S6K; S6 protein and phospho (T232)-S6 are shown in Western blots (Fig. 6D). Phosphorylation of S6K is essential for its kinase activity on its substrate, i.e. phosphorylation of protein S6. Densitometry of protein bands such as those shown in panel 6E, with the calculated ratio ‘phospho-protein’/’protein’ for S6K and for S6, in response to Resolvins i.e. RvD1-RvD5 (Fig. 6E) indicate the large effect of RvD4 and, especially RvD5.
Important signaling mediators in PLD-S6K signaling include Grb2, TSC2, Rheb and pS6 crucial for phagocyte phenotype and thus functions (65). Fig. 6F shows RvD4 and RvD5 increase Grb2 and PLD2 and decrease PLD2’s downstream signaling molecule TSC2. This downregulation of TSC2 is accompanied by phosphorylation of protein S6 as observed in Fig. 6F and Supplemental Fig. S6C. Based on these findings we hypothesized a potential role of PLD2 in RvD5-S6K signaling in macrophages, which is in accordance with our in vivo data (Fig. 3). However, this does not eliminate the possibility that RvD5 may modulate PLD2 and S6K independently. In order to address this, we studied the relationship between PLD and S6K levels in macrophages. We overexpressed PLD1 or PLD2 in macrophages and observed that an increase in PLD2, and not PLD1, also showed an increase in S6K gene expression (Supplemental Fig. S6D). Thus, we speculate that RvD5 actions in macrophages could potentially be via PLD2-S6K signaling cascade.
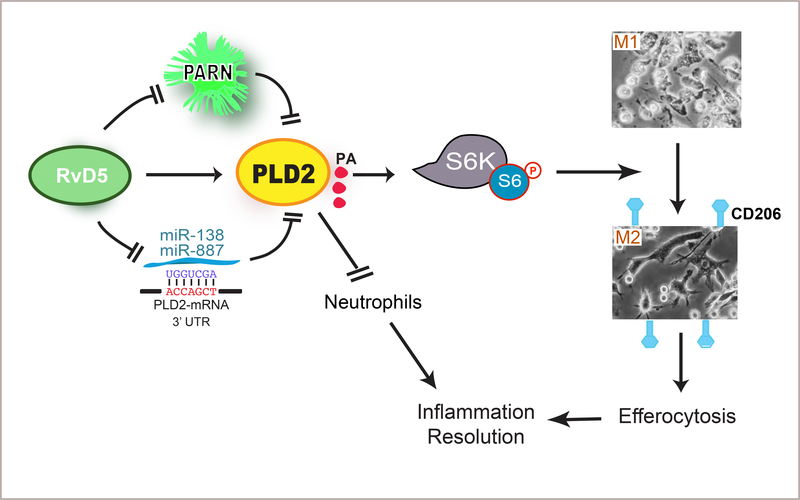
Model summarizing the main findings.
Schematic showing a proposed model of Resolvins D-series acting on macrophage M1-M2 conversion and neutrophil activity mediated by PLD2 signaling (Fig. 7). PLD2 is required for RvD5’s actions on neutrophil actions in promoting resolution of inflammation. RvD5 reverses the inhibition of PLD2 by either PARN or by miR-138 or miR-887 thus increasing PLD2 expression and promoting PLD2 activity. This could potentially initiate PLD2 activation of S6K. Specifically, we found that D-series resolvins activate PLD2 in M2 macrophages resulting in increased phospho-S6 and hence phagocytosis and efferocytosis as well as CD206 cell surface expression. This contributes to the M1 to M2 polarization in that we found that PLD induces macrophage polarization by increasing M2 markers and decreasing M1 markers. The M2 macrophages can then carry out the key function of efferocytosis central to intact resolution mechanisms in the inflammatory response.
Figure 7. Summary model of RvD5-PLD2 signaling in macrophages and neutrophils for resolution of acute inflammation.
A new model providing insight into the intracellular signaling mechanisms of RvD5’s action in macrophages and neutrophils as well as the RvD5-PLD2 cooperation in eliciting a phenotype switch from pro-inflammatory M1 to pro-resolving M2 macrophages. Herein, RvD5 reverses inhibition of PLD2 by either PARN or PLD2-miRs, thus increasing PLD2 expression and promoting PLD2 activity. This increase in PLD2 potentially initiates activation of S6K leading to CD206 cell surface expression confirming M1-M2 phenotype switch. The M2 macrophages then carry out the key function of efferocytosis needed for resolution of inflammation. Likewise, RvD5 acts via PLD2 in preventing neutrophil exacerbated inflammation and reflow injury, thus facilitating resolution of inflammation.
DISCUSSION
The acute inflammatory response is a complex host defense process that involves recruitment of leukocytes which undergo both phenotypic and functional changes involving several signaling mediators including specialized pro-resolving lipid mediators (SPMs) that culminate in resolution. Unresolved inflammation that fails to attain homeostasis will lead to chronic inflammation (2,14). Depending on the site of inflammation, specific macrophage phenotypes are known to be critical for the initiation and in the resolution phase of inflammation. The findings reported in this study, focus on the first line of defense mechanism involving macrophages and neutrophils during inflammation and resolution. Phospholipase D (PLD) is a major signaling molecule in leukocytes, essential for actin cytoskeletal rearrangement (68), chemotaxis (69) and phagocytosis (33, 34, 57). PLD is well-studied for its role in early inflammation where it is required for pseudopodia formation and phagocytosis (34, 70) in macrophages. To this date, the role of PLD in macrophage polarization and more importantly in resolution of acute inflammation has not been reported. Herein, we demonstrated for the first time that PLD mediates macrophage polarization activated by specific D-series resolvins.
Evidence accumulated in the last two decades demonstrate that microenvironmental cues such as infection or tissue injury can polarize macrophages to two well-characterized heterogeneous populations (71). Subject to the microenvironmental signals, macrophages can be polarized to either classically activated M1 macrophages or alternatively activated M2 macrophages. These M1 macrophages are considered pro-inflammatory composed of ROS- and proteolytic enzymes-rich granules. M1 macrophages secrete a number of pro-inflammatory mediators and their main function is to kill and/or phagocytose pathogens. The M2 phenotype macrophages are considered anti-inflammatory and pro-resolving based on their ability to carry out efferocytosis of apoptotic cells (5, 7, 12) as well as production of the lion’s share of resolvins and other SPMs (43, 47). We demonstrated that both PLD protein and activity are higher in M1 and M2 macrophages compared to amounts in M ‘zero’ M0 macrophages (Fig. S1). Also, the D-series resolvins regulate PLD gene expression (Fig. 1) as well as lipase activity (Fig. 2) in both the M1 and M2 cells compared to M0 macrophages.
Resolvin-mediated signaling is different in pro-inflammatory (M1) compared to their signaling in (M2)-like pro-resolving macrophages exhibiting differential impact on PLD signaling (Fig. 4). PLD is well-known to be upregulated in activated neutrophils and macrophages during inflammation (65) and yet to date there are no reports of PLDs role in the resolution phase of inflammation. Herein, we elucidated a previously unreported role of PLD, specifically PLD2, in resolution of inflammation which promotes macrophage polarization to M2 phenotype and their function, efferocytosis (Fig. 4). Increases in resolvins promote timely resolution of inflammation (57), restoring homeostasis. Resolvin D5 (RvD5) has recently been shown to be produced by M2 macrophages in response to E. coli (47), protect against intestinal inflammation and injury (72). Hence, the intracellular signaling of RvD5’s actions are of interest. In the present study, we demonstrated that RvD5-mediated macrophage polarization to M2 by means of PLD2, as the intracellular signaling molecule. Results presented in Figures 5 and 6, with M1 and M2 macrophages, RvD5 signals via PLD2, phosphorylation of S6 protein (Fig. 6), and increases CD206 surface expression (M2 marker), with increased efferocytosis of apoptotic neutrophils (Fig. 4). These findings provide evidence for a novel signaling axis for RvD5 via PLD2, S6 and CD206 in promoting M2 macrophage polarization and their function in resolution of inflammation. Moreover, we found that in macrophages, the mechanism of RvD5 action on PLD expression involves post-transcriptional modification by means of PLD-specific miRs and a deadenylase, called PARN (Fig. 5). Taken together, these findings indicate that RvD5 via PLD2 modulates resolution of acute inflammation. 15-LOX, a crucial enzyme in Rv biosynthesis is known to be induced along with CD163 and CD206 in pro-resolving M2 macrophages (47, 73). Our findings can be a basis for exploring in further detail the mechanism of action of RvD5 on PLD2 and whether PLD2 has a feedback mechanism to regulate RvD5 biosynthesis by inducing 15-LOX in pro-resolving macrophages. Also, future studies may determine whether PLD2 activation is downstream of the recently identified target receptor of RvD5, GPR101 (74).
In light of our findings that resolvins regulate PLD in inflammation resolution, we decided to further investigate using an in vivo model of rapidly progressing acute inflammatory response as in the setting of hind-limb ischemia/reperfusion injury (49). Using this model of leukocyte mediated second organ injury, we evaluated how PLD and RvD5 regulate neutrophils, the first line of host defense, that rapidly and abundantly infiltrate sites of inflammation. In humans, blunt physical trauma such as in a car accident or injury or myocardial infarction are very common insults causing blood flow obstruction, and local inflammation followed by restoration, known as ischemia reperfusion injury (75). Ischemia/reperfusion injury results in second-organ damage wherein leukocytes, predominantly neutrophils accumulate in organs such as the lungs exacerbating inflammation along with releasing reactive oxygen species (ROS) and lysosomal enzymes. Earlier studies demonstrated that resolvins limit neutrophils infiltration (49) whereas PLD promotes neutrophil chemotaxis during inflammation known to be acting in the initiation phase (76). In the present study, we found that RvD5 in the absence of PLD2 is unable to reduce neutrophil infiltration and thus exacerbates inflammation and PMN numbers in the lungs. These results demonstrate PLD2 is important in RvD5’s signaling pro-resolving activities (Fig. 3). This new information on the connection between PLD2-RvD5 provides specific new targets that may have potential implications for improving human health.
The findings reported here are a substantial departure from the status quo of investigating PLD2’s role in inflammation and cancer (77, 78), thus warranting further research in resolution of inflammation involving PLD2. Resolvins have been shown to reduce cancer burden in mice (79) and recent studies have shown that E-series resolvins act via PLD in goblet cell functions in rat conjunctivitis (80). Additional studies are necessary to confirm the actions of PLD specific inhibitors on resolvins in the resolution of inflammation and alternatively studying other potential roles of PLD in this context. Understanding and addressing the role and functions of PLD isoforms in both acute inflammation and its natural self-limited resolution should help in treating chronic inflammatory diseases and improving patient outcomes clinically. PLD possesses both lipase as well as a GEF (Guanine nucleotide exchange factor) activities which are crucial for cell migration and cell proliferation (27–29) during chronic inflammation. Given PLD is implicated in cardiovascular diseases (30, 81) and cancer (23, 40), in future studies it will be of interest to assess the role of resolvins in regulating both PLD2-GEF and lipase activities in chronic inflammation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Yasunori Kanaho and Dr. Gilbert Di Paolo for providing us the PLD2 and PLD1 knockout mice, respectively. We thank Dr. Kristen Fite for helpful suggestions and comments on this study. The following grants to Dr. Cambronero (J.G.C) have supported this work: HL056653-14 from the National Institutes of Health (NIH) and 13GRNT17230097 from the American Heart Association. Experiments in the CNS lab Boston,MA were supported by National Institutes of Health grant P01GM095467 (CNS). This work is dedicated in loving memory of our dearest colleague, mentor and friend, Prof. Julian Gomez-Cambronero. The experiments reported herein were initiated while Dr. Gomez-Cambronero was on sabbatical in the CNS lab in Boston before his untimely passing.
ABBREVIATIONS
- Arg1
Arginase I
- CD
Cluster of differentiation
- DHA
Docosahexanoic acid
- EPA
Eicosapentanoic acid
- GPR32
G-protein coupled receptor 32
- HLIR
Hind-limb ischemia reperfusion
- IFNγ
Interferon gamma
- IL-12
Interleukin 12
- IL-4
Interleukin 4
- iNOS2
Inducible Nitric oxide synthase 2
- LPS
Lipopolysaccharide (Bacterial)
- miR
micro RNA
- MPO
Myeloperoxidase
- PA
Phosphatidic acid
- PARN
Poly(A)-tail ribonuclease
- PBDM
Peripheral blood-derived macrophages
- PC
Phosphatidyl choline
- PLD
Phopsholipase D
- PLD1−/−-
Phospholipase D1 knockout
- PLD2−/−
Phospholipase D2 knockout
- PMN
Polymorphonuclear neutrophils
- PPARγ
Peroxisome-proliferation antagonist receptor gamma
- ROS
Reactive oxygen species
- RvD1
7S,8R,17S-trihydroxy-4Z,9E,11E,13Z,15E,19Z-docosahexaenoic acid, Resolvin D1
- RvD2
7S,16R,17S-trihydroxy-4Z,8E,10Z,12E,14E,19Z-docosahexaenoic acid, Resolvin D2
- RvD3
4S,11R,17S-trihydroxy-5Z,7E,9E,13Z,15E,19Z-docosahexaenoic acid, Resolvin D3
- RvD4
4S,5,17S-trihydroxy-6E,8E,10E,13E,15Z,19Z-docosahexaenoic acid, Resolvin D4
- RvD5
7S,17S-dihydroxy-4Z,8E,10Z,13Z,15E,19Z-docosahexaenoic acid, Resolvin D5
- SEM
Standard Error of the Mean
- S6
Ribosomal protein S6
- phospho S6
Phosphorylated ribosomal protein S6
- S6K
p70-S6 Kinase
- SPMs
Specialized pro-resolving lipid mediators
- TGF-β
Transforming growth factor beta
- TNFα
Tumor necrosis factor alpha
- WT
Wild-type
Footnotes
Conflict of interest disclosure: The authors declare no competing financial interests.
This is the peer reviewed version of the following article: The FASEB Journal 2020, which has been published in final form at https://doi.org/10.1096/fj.201903025RR. This article may be used for non-commercial purposes in accordance with Wiley Terms and Conditions for Use of Self-Archived Versions.
REFERENCES
- 1.Medzhitov R (2008) Origin and physiological roles of inflammation. Nature 454, 428–435 [DOI] [PubMed] [Google Scholar]
- 2.Perkins GD, Nathani N, McAuley DF, Gao F, and Thickett DR (2007) In vitro and in vivo effects of salbutamol on neutrophil function in acute lung injury. Thorax 62, 36–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Serhan CN, Chiang N, and Van Dyke TE (2008) Resolving inflammation: dual anti-inflammatory and pro-resolution lipid mediators. Nat Rev Immunol 8, 349–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gordon S, and Taylor PR (2005) Monocyte and macrophage heterogeneity. Nature reviews. Immunology 5, 953–964 [DOI] [PubMed] [Google Scholar]
- 5.Wynn TA, and Barron L (2010) Macrophages: master regulators of inflammation and fibrosis. Seminars in liver disease 30, 245–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bashir S, Sharma Y, Elahi A, and Khan F (2016) Macrophage polarization: the link between inflammation and related diseases. Inflammation research : official journal of the European Histamine Research Society … [et al.] 65, 1–11 [DOI] [PubMed] [Google Scholar]
- 7.Tan HY, Wang N, Li S, Hong M, Wang X, and Feng Y (2016) The Reactive Oxygen Species in Macrophage Polarization: Reflecting Its Dual Role in Progression and Treatment of Human Diseases. Oxid Med Cell Longev 2016, 2795090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dandekar RC, Kingaonkar AV, and Dhabekar GS (2011) Role of macrophages in malignancy. Annals of maxillofacial surgery 1, 150–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou D, Huang C, Lin Z, Zhan S, Kong L, Fang C, and Li J (2014) Macrophage polarization and function with emphasis on the evolving roles of coordinated regulation of cellular signaling pathways. Cellular signalling 26, 192–197 [DOI] [PubMed] [Google Scholar]
- 10.Lecca D, Trincavelli ML, Gelosa P, Sironi L, Ciana P, Fumagalli M, Villa G, Verderio C, Grumelli C, Guerrini U, Tremoli E, Rosa P, Cuboni S, Martini C, Buffo A, Cimino M, and Abbracchio MP (2008) The recently identified P2Y-like receptor GPR17 is a sensor of brain damage and a new target for brain repair. PloS one 3, e3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rausch ME, Weisberg S, Vardhana P, and Tortoriello DV (2008) Obesity in C57BL/6J mice is characterized by adipose tissue hypoxia and cytotoxic T-cell infiltration. International journal of obesity 32, 451–463 [DOI] [PubMed] [Google Scholar]
- 12.Braga TT, Agudelo JS, and Camara NO (2015) Macrophages During the Fibrotic Process: M2 as Friend and Foe. Frontiers in immunology 6, 602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Serhan CN, and Savill J (2005) Resolution of inflammation: the beginning programs the end. Nat Immunol 6, 1191–1197 [DOI] [PubMed] [Google Scholar]
- 14.Ohira T, Arita M, Omori K, Recchiuti A, Van Dyke TE, and Serhan CN (2010) Resolvin E1 receptor activation signals phosphorylation and phagocytosis. J Biol Chem 285, 3451–3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nelson RK, and Frohman MA (2015) Physiological and pathophysiological roles for phospholipase D. J Lipid Res 56, 2229–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foster DA, Salloum D, Menon D, and Frias MA (2014) Phospholipase D and the maintenance of phosphatidic acid levels for regulation of mammalian target of rapamycin (mTOR). J Biol Chem 289, 22583–22588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gomez-Cambronero J (2014) Phospholipase D in cell signaling: from a myriad of cell functions to cancer growth and metastasis. J Biol Chem 289, 22557–22566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fite K, Elkhadragy L, and Gomez-Cambronero J (2016) A Repertoire of MicroRNAs Regulates Cancer Cell Starvation by Targeting Phospholipase D in a Feedback Loop That Operates Maximally in Cancer Cells. Mol Cell Biol 36, 1078–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schonberger T, Jurgens T, Muller J, Armbruster N, Niermann C, Gorressen S, Sommer J, Tian H, di Paolo G, Scheller J, Fischer JW, Gawaz M, and Elvers M (2014) Pivotal role of phospholipase D1 in tumor necrosis factor-alpha-mediated inflammation and scar formation after myocardial ischemia and reperfusion in mice. Am J Pathol 184, 2450–2464 [DOI] [PubMed] [Google Scholar]
- 20.Lee SK, Kim SD, Kook M, Lee HY, Ghim J, Choi Y, Zabel BA, Ryu SH, and Bae YS (2015) Phospholipase D2 drives mortality in sepsis by inhibiting neutrophil extracellular trap formation and down-regulating CXCR2. J Exp Med 212, 1381–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Speranza F, Mahankali M, Henkels KM, and Gomez-Cambronero J (2014) The molecular basis of leukocyte adhesion involving phosphatidic Acid and phospholipase d. J Biol Chem 289, 28885–28897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fite K, and Gomez-Cambronero J (2016) Down-regulation of MicroRNAs (MiRs) 203, 887, 3619 and 182 Prevents Vimentin-triggered, Phospholipase D (PLD)-mediated Cancer Cell Invasion. J Biol Chem 291, 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henkels KM, Boivin GP, Dudley ES, Berberich SJ, and Gomez-Cambronero J (2013) Phospholipase D (PLD) drives cell invasion, tumor growth and metastasis in a human breast cancer xenograph model. Oncogene 32, 5551–5562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gobel K, Schuhmann MK, Pankratz S, Stegner D, Herrmann AM, Braun A, Breuer J, Bittner S, Ruck T, Wiendl H, Kleinschnitz C, Nieswandt B, and Meuth SG (2014) Phospholipase D1 mediates lymphocyte adhesion and migration in experimental autoimmune encephalomyelitis. Eur J Immunol 44, 2295–2305 [DOI] [PubMed] [Google Scholar]
- 25.Gomez-Cambronero J, Morris AJ, Henkels KM (2016) PLD protein-protein interactions with signaling molecules and modulation by PA. Methods In Enzymology In press. [DOI] [PubMed] [Google Scholar]
- 26.Gomez-Cambronero J, and Kantonen S (2014) A river runs through it: how autophagy, senescence, and phagocytosis could be linked to phospholipase D by Wnt signaling. J Leukoc Biol 96, 779–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahankali M, Henkels KM, and Gomez-Cambronero J (2013) A GEF-to-phospholipase molecular switch caused by phosphatidic acid, Rac and JAK tyrosine kinase that explains leukocyte cell migration. J Cell Sci 126, 1416–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahankali M, Henkels KM, Alter G, and Gomez-Cambronero J (2012) Identification of the catalytic site of phospholipase D2 (PLD2) newly described guanine nucleotide exchange factor activity. J Biol Chem 287, 41417–41431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahankali M, Peng HJ, Henkels KM, Dinauer MC, and Gomez-Cambronero J (2011) Phospholipase D2 (PLD2) is a guanine nucleotide exchange factor (GEF) for the GTPase Rac2. Proc Natl Acad Sci U S A 108, 19617–19622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ganesan R, Henkels KM, Wrenshall LE, Kanaho Y, Di Paolo G, Frohman MA, and Gomez-Cambronero J (2018) Oxidized LDL phagocytosis during foam cell formation in atherosclerotic plaques relies on a PLD2-CD36 functional interdependence. J Leukoc Biol 103, 867–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park DW, Lee HK, Lyu JH, Chin H, Kang SW, Kim YJ, Bae YS, and Baek SH (2013) TLR2 stimulates ABCA1 expression via PKC-eta and PLD2 pathway. Biochem Biophys Res Commun 430, 933–937 [DOI] [PubMed] [Google Scholar]
- 32.Mancini F, and Ciervo A (2015) Enzymatic characterization of Chlamydophila pneumoniae phospholipase D. New Microbiol 38, 59–66 [PubMed] [Google Scholar]
- 33.Iyer SS, Agrawal RS, Thompson CR, Thompson S, Barton JA, and Kusner DJ (2006) Phospholipase D1 regulates phagocyte adhesion. J Immunol 176, 3686–3696 [DOI] [PubMed] [Google Scholar]
- 34.Iyer SS, Barton JA, Bourgoin S, and Kusner DJ (2004) Phospholipases D1 and D2 coordinately regulate macrophage phagocytosis. J Immunol 173, 2615–2623 [DOI] [PubMed] [Google Scholar]
- 35.Gomez-Cambronero J, Di Fulvio M, and Knapek K (2007) Understanding phospholipase D (PLD) using leukocytes: PLD involvement in cell adhesion and chemotaxis. J Leukoc Biol 82, 272–281 [DOI] [PubMed] [Google Scholar]
- 36.Usatyuk PV, Kotha SR, Parinandi NL, and Natarajan V (2013) Phospholipase D signaling mediates reactive oxygen species-induced lung endothelial barrier dysfunction. Pulm Circ 3, 108–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koch T, Seifert A, Wu DF, Rankovic M, Kraus J, Borner C, Brandenburg LO, Schroder H, and Hollt V (2009) mu-opioid receptor-stimulated synthesis of reactive oxygen species is mediated via phospholipase D2. J Neurochem 110, 1288–1296 [DOI] [PubMed] [Google Scholar]
- 38.Usatyuk PV, Gorshkova IA, He D, Zhao Y, Kalari SK, Garcia JG, and Natarajan V (2009) Phospholipase D-mediated activation of IQGAP1 through Rac1 regulates hyperoxia-induced p47phox translocation and reactive oxygen species generation in lung endothelial cells. J Biol Chem 284, 15339–15352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee HY, Kang HK, Jo EJ, Kim JI, Lee YN, Lee SH, Park YM, Ryu SH, Kwak JY, and Bae YS (2004) Trp-Lys-Tyr-Met-Val-Met stimulates phagocytosis via phospho-lipase D-dependent signaling in mouse dendritic cells. Exp Mol Med 36, 135–144 [DOI] [PubMed] [Google Scholar]
- 40.Chen Q, Hongu T, Sato T, Zhang Y, Ali W, Cavallo J-A, van der Velden A, Tian H, Di Paolo G, Nieswandt B, Kanaho Y, and Frohman MA (2012) Key roles for the lipid signaling enzyme phospholipase d1 in the tumor microenvironment during tumor angiogenesis and metastasis. Science signaling 5, ra79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oliveira TG, Chan RB, Tian H, Laredo M, Shui G, Staniszewski A, Zhang H, Wang L, Kim T-W, Duff KE, Wenk MR, Arancio O, and Di Paolo G (2010) Phospholipase d2 ablation ameliorates Alzheimer’s disease-linked synaptic dysfunction and cognitive deficits. The Journal of neuroscience : the official journal of the Society for Neuroscience 30, 16419–16428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chiang N, Gronert K, Clish CB, O’Brien JA, Freeman MW, and Serhan CN (1999) Leukotriene B4 receptor transgenic mice reveal novel protective roles for lipoxins and aspirin-triggered lipoxins in reperfusion. J Clin Invest 104, 309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dalli J, and Serhan CN (2012) Specific lipid mediator signatures of human phagocytes: microparticles stimulate macrophage efferocytosis and pro-resolving mediators. Blood 120, e60–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krishnamoorthy S, Recchiuti A, Chiang N, Yacoubian S, Lee CH, Yang R, Petasis NA, and Serhan CN (2010) Resolvin D1 binds human phagocytes with evidence for proresolving receptors. Proc Natl Acad Sci U S A 107, 1660–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dalli J, Winkler JW, Colas RA, Arnardottir H, Cheng CY, Chiang N, Petasis NA, and Serhan CN (2013) Resolvin D3 and aspirin-triggered resolvin D3 are potent immunoresolvents. Chem Biol 20, 188–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spite M, Norling LV, Summers L, Yang R, Cooper D, Petasis NA, Flower RJ, Perretti M, and Serhan CN (2009) Resolvin D2 is a potent regulator of leukocytes and controls microbial sepsis. Nature 461, 1287–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Werz O, Gerstmeier J, Libreros S, De la Rosa X, Werner M, Norris PC, Chiang N, and Serhan CN (2018) Human macrophages differentially produce specific resolvin or leukotriene signals that depend on bacterial pathogenicity. Nat Commun 9, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oh SF, Pillai PS, Recchiuti A, Yang R, and Serhan CN (2011) Pro-resolving actions and stereoselective biosynthesis of 18S E-series resolvins in human leukocytes and murine inflammation. J Clin Invest 121, 569–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chiang N, Dalli J, Colas RA, and Serhan CN (2015) Identification of resolvin D2 receptor mediating resolution of infections and organ protection. J Exp Med 212, 1203–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liscovitch M, Czarny M, Fiucci G, and Tang X (2000) Phospholipase D: molecular and cell biology of a novel gene family. Biochem J 345 Pt 3, 401–415 [PMC free article] [PubMed] [Google Scholar]
- 51.Hatton N, Lintz E, Mahankali M, Henkels KM, and Gomez-Cambronero J (2015) Phosphatidic Acid Increases Epidermal Growth Factor Receptor Expression by Stabilizing mRNA Decay and by Inhibiting Lysosomal and Proteasomal Degradation of the Internalized Receptor. Mol Cell Biol 35, 3131–3144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Henkels KM, Miller TE, Ganesan R, Wilkins BA, Fite K, and Gomez-Cambronero J (2016) A Phosphatidic Acid (PA) conveyor system of continuous intracellular transport from cell membrane to nucleus maintains EGF receptor homeostasis. Oncotarget 7, 47002–47017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Livak KJ, and Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 54.Levy BD, Bonnans C, Silverman ES, Palmer LJ, Marigowda G, Israel E, Severe Asthma Research Program, N. H. L., and Blood I. (2005) Diminished lipoxin biosynthesis in severe asthma. Am J Respir Crit Care Med 172, 824–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hsiao HM, Sapinoro RE, Thatcher TH, Croasdell A, Levy EP, Fulton RA, Olsen KC, Pollock SJ, Serhan CN, Phipps RP, and Sime PJ (2013) A novel anti-inflammatory and pro-resolving role for resolvin D1 in acute cigarette smoke-induced lung inflammation. PLoS One 8, e58258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Titos E, Rius B, Gonzalez-Periz A, Lopez-Vicario C, Moran-Salvador E, Martinez-Clemente M, Arroyo V, and Claria J (2011) Resolvin D1 and its precursor docosahexaenoic acid promote resolution of adipose tissue inflammation by eliciting macrophage polarization toward an M2-like phenotype. J Immunol 187, 5408–5418 [DOI] [PubMed] [Google Scholar]
- 57.Corrotte M, Chasserot-Golaz S, Huang P, Du G, Ktistakis NT, Frohman MA, Vitale N, Bader MF, and Grant NJ (2006) Dynamics and function of phospholipase D and phosphatidic acid during phagocytosis. Traffic 7, 365–377 [DOI] [PubMed] [Google Scholar]
- 58.Schmid M, Gemperle C, Rimann N, and Hersberger M (2016) Resolvin D1 Polarizes Primary Human Macrophages toward a Proresolution Phenotype through GPR32. J Immunol 196, 3429–3437 [DOI] [PubMed] [Google Scholar]
- 59.Fredman G, and Serhan CN (2011) Specialized proresolving mediator targets for RvE1 and RvD1 in peripheral blood and mechanisms of resolution. Biochem J 437, 185–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fredman G, Li Y, Dalli J, Chiang N, and Serhan CN (2012) Self-limited versus delayed resolution of acute inflammation: temporal regulation of pro-resolving mediators and microRNA. Sci Rep 2, 639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krishnamoorthy S, Recchiuti A, Chiang N, Fredman G, and Serhan CN (2012) Resolvin D1 receptor stereoselectivity and regulation of inflammation and proresolving microRNAs. Am J Pathol 180, 2018–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li Y, Dalli J, Chiang N, Baron RM, Quintana C, and Serhan CN (2013) Plasticity of leukocytic exudates in resolving acute inflammation is regulated by MicroRNA and proresolving mediators. Immunity 39, 885–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Recchiuti A, Krishnamoorthy S, Fredman G, Chiang N, and Serhan CN (2011) MicroRNAs in resolution of acute inflammation: identification of novel resolvin D1-miRNA circuits. FASEB J 25, 544–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miller TE, and Gomez-Cambronero J (2017) A feedback mechanism between PLD and deadenylase PARN for the shortening of eukaryotic poly(A) mRNA tails that is deregulated in cancer cells. Biol Open 6, 176–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gomez-Cambronero J (2011) The exquisite regulation of PLD2 by a wealth of interacting proteins: S6K, Grb2, Sos, WASp and Rac2 (and a surprise discovery: PLD2 is a GEF). Cell Signal 23, 1885–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chiang N, de la Rosa X, Libreros S, and Serhan CN (2017) Novel Resolvin D2 Receptor Axis in Infectious Inflammation. J Immunol 198, 842–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Henkels KM, Mallets ER, Dennis PB, and Gomez-Cambronero J (2015) S6K is a morphogenic protein with a mechanism involving Filamin-A phosphorylation and phosphatidic acid binding. FASEB J 29, 1299–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Colley WC, Sung TC, Roll R, Jenco J, Hammond SM, Altshuller Y, Bar-Sagi D, Morris AJ, and Frohman MA (1997) Phospholipase D2, a distinct phospholipase D isoform with novel regulatory properties that provokes cytoskeletal reorganization. Curr Biol 7, 191–201 [DOI] [PubMed] [Google Scholar]
- 69.Lehman N, Di Fulvio M, McCray N, Campos I, Tabatabaian F, and Gomez-Cambronero J (2006) Phagocyte cell migration is mediated by phospholipases PLD1 and PLD2. Blood 108, 3564–3572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kantonen S, Hatton N, Mahankali M, Henkels KM, Park H, Cox D, and Gomez-Cambronero J (2011) A novel phospholipase D2-Grb2-WASp heterotrimer regulates leukocyte phagocytosis in a two-step mechanism. Mol Cell Biol 31, 4524–4537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mantovani A, Sica A, and Locati M (2005) Macrophage polarization comes of age. Immunity 23, 344–346 [DOI] [PubMed] [Google Scholar]
- 72.Gobbetti T, Dalli J, Colas RA, Federici Canova D, Aursnes M, Bonnet D, Alric L, Vergnolle N, Deraison C, Hansen TV, Serhan CN, and Perretti M (2017) Protectin D1n-3 DPA and resolvin D5n-3 DPA are effectors of intestinal protection. Proc Natl Acad Sci U S A 114, 3963–3968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Newson J, Stables M, Karra E, Arce-Vargas F, Quezada S, Motwani M, Mack M, Yona S, Audzevich T, and Gilroy DW (2014) Resolution of acute inflammation bridges the gap between innate and adaptive immunity. Blood 124, 1748–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Flak MB, Koenis DS, Sobrino A, Smith J, Pistorius K, Palmas F, and Dalli J (2020) GPR101 mediates the pro-resolving actions of RvD5n-3 DPA in arthritis and infections. J Clin Invest 130, 359–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Collard CD, and Gelman S (2001) Pathophysiology, clinical manifestations, and prevention of ischemia-reperfusion injury. Anesthesiology 94, 1133–1138 [DOI] [PubMed] [Google Scholar]
- 76.Abdulnour RE, Howrylak JA, Tavares AH, Douda DN, Henkels KM, Miller TE, Fredenburgh LE, Baron RM, Gomez-Cambronero J, and Levy BD (2018) Phospholipase D isoforms differentially regulate leukocyte responses to acute lung injury. J Leukoc Biol 103, 919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gomez-Cambronero J (2010) New concepts in phospholipase D signaling in inflammation and cancer. ScientificWorldJournal 10, 1356–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kang DW, Choi KY, and Min do S (2014) Functional regulation of phospholipase D expression in cancer and inflammation. J Biol Chem 289, 22575–22582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sulciner ML, Serhan CN, Gilligan MM, Mudge DK, Chang J, Gartung A, Lehner KA, Bielenberg DR, Schmidt B, Dalli J, Greene ER, Gus-Brautbar Y, Piwowarski J, Mammoto T, Zurakowski D, Perretti M, Sukhatme VP, Kaipainen A, Kieran MW, Huang S, and Panigrahy D (2018) Resolvins suppress tumor growth and enhance cancer therapy. J Exp Med 215, 115–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lippestad M, Hodges RR, Utheim TP, Serhan CN, and Dartt DA (2018) Signaling pathways activated by resolvin E1 to stimulate mucin secretion and increase intracellular Ca(2+) in cultured rat conjunctival goblet cells. Exp Eye Res 173, 64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stegner D, Thielmann I, Kraft P, Frohman MA, Stoll G, and Nieswandt B (2013) Pharmacological inhibition of phospholipase D protects mice from occlusive thrombus formation and ischemic stroke--brief report. Arterioscler Thromb Vasc Biol 33, 2212–2217 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.