Abstract
Introduction:
The microtubule-associated protein tau (MAPT) gene is considered a strong genetic risk factor for Parkinson’s disease (PD) in Caucasians. MAPT is located within an inversion region of high linkage disequilibrium designated as H1 and H2 haplotype, and contains eight other genes which have been implicated in neurodegeneration. The aim of the current study was to identify common coding variants in strong linkage disequilibrium (LD) within the associated loci on chr17q21 harboring MAPT.
Methods:
Sanger sequencing of coding exons in 90 Caucasian late-onset PD (LOPD) patients was performed. Specific gene sequencing for LRRC37A, LRRC37A2, ARL17A and ARL17B was not possible given the high homology, presence of pseudogenes and copy number variants that are in the region, and therefore four genes (NSF, KANSL1, SPPL2C, and CRHR1) were included in the analysis. Coding variants from these four genes that did not perfectly tag (r2=1) the MAPT H1/H2 haplotype were genotyped in an independent replication series of Caucasian PD cases (N=851) and controls (N=730).
Results:
In the 90 LOPD cases we identified 30 coding variants. Eleven non-synonymous variants tagged the MAPT H1/H2 haplotype, including two SPPL2C variants (rs12185233 and rs12373123) that had high pathogenic combined annotation dependent depletion (CADD) scores of >20. In the replication series, the non-synonymous KANSL1 rs17585974 variant was in very strong LD with MAPT H1/H2 and had a high CADD score of 24.7.
Conclusion:
We have identified several non-synonymous variants across neighboring genes of MAPT that may warrant further genetic and functional investigation within the biological etiology of PD.
Keywords: MAPT, KANSL1, NSF, CRHR1, SPPL2C, H1 Haplotype, Parkinson’s disease
Introduction
Parkinson’s disease (PD) is a progressive, age-associated neurodegenerative movement disorder. PD is considered a multifactorial disease, whereby environment and genetics both contribute to disease risk. Although a handful of genes have been identified that harbor highly penetrant mutations, disease risk is primarily suspected to be influenced by multiple low penetrant population-based susceptibility variants. Genome-wide association approaches have now nominated over 90 susceptibility loci in Caucasians with one of the most highly replicated being on Chr17q21, containing the MAPT gene [1].
MAPT encodes for microtubule associated protein tau, a protein which is thought to stabilize the formation or facilitate flexibility of axonal microtubules [2, 3]. MAPT is located in a region of high disequilibrium (LD) spanning approximately 1.8 megabases and containing a 900 kilobase inversion polymorphism which defines two extended haplotypes, known as H1 and H2 [4]. H1 is the most common haplotype, with a frequency ~20% for the H2 allele in European populations; the H2 haplotype is completely absent or very rare in other populations [5]. Mutations in MAPT have been associated with many neurodegenerative diseases, and specifically the H1 haplotype is associated with increased risk of developing progressive supranuclear palsy (PSP) [6, 7], corticobasal degeneration (CBD) [8], and Alzheimer’s disease (AD) [9], as well as PD [10]. The MAPT H1 association linked to an increased risk of PD led to functional studies that determined the tau protein could influence α-synuclein aggregation and fibrillization [11–12].
However, the functional variant/s responsible for the MAPT H1 association signal with PD are still unknown. While continued research has focused on identifying variants within the MAPT gene that drive the PD association signal, few studies have investigated the other genes (LRRC37A, LRRC37A2, NSF, ARL17A, ARL17B, KANSL1, SPPL2C, and CRHR1), which are located within the inversion on Chr17q2-H1 haplotype and therefore could plausibly be driving the Caucasian MAPT PD association signal. In the current study, exons of non- MAPT genes on Chr17q2-H1 were Sanger sequenced in a cohort of patients with late-onset PD (LOPD). Identified variants were subsequently genotyped in a larger, independent, PD case-control cohort to assess association with disease in the context of the MAPT H1/H2 signal. Specifically, our aim was to identify common coding variants, in genes other than MAPT, in strong LD within the disease-associated haplotype on chr17q21.
Methods
Subjects
A total of 90 individuals, clinically diagnosed with sporadic, late-onset PD (age >50 at diagnosis) (LOPD) were initially included for exon sequencing as a discovery cohort (stage one). An independent cohort of 851 clinically diagnosed PD patients and 730 healthy controls were further recruited for a replication study (stage two). Cohort demographics are summarized in Table 1. All subjects were unrelated. PD diagnosis was determined using the Queens Square Brain bank criteria [13], and all patients were Caucasian, non-Hispanic and of European descent. Known carriers of pathogenic mutations in the leucine-rich repeat kinase 2 (LRRK2), α-synuclein (SNCA), vacuolar protein sorting-associated protein 35 (VPS35), PTEN-induced kinase 1 (PINK1), Parkin E3 ubiquitin protein ligase (PARKIN) and PARK7 (DJ1) genes were excluded. This study was approved by Mayo Clinic Institutional Review Board and written informed consent was obtained prior to commencement. All blood samples were collected at Mayo Clinic Jacksonville (FL, USA).
Table 1:
Cohort demographics.
| Series | N | Age of PD onset (years) | Early onset PD (<50 years) | Age at Study (years) | Number of Males | |
|---|---|---|---|---|---|---|
| Stage One Discovery |
PD cases | 90 | 69 ± 8 (51–90) | 0 (0.0%) | 71 ± 7 (40–90) | 60 (67%) |
| Stage Two Replication |
PD cases | 851 | 65 ± 12 (25–97) | 101 (12%) | 68 ± 11 (28–97) | 540 (63%) |
| Controls | 730 | N/A | N/A | 65 ± 13 (18–88) | 305 (42%) |
Age is given as the sample mean ± SD (minimum-maximum).
Sanger Sequencing
Genomic DNA was extracted from whole blood using Autogen FlexStar standard protocol methods (Autogen, Holliston, MA). A total of eight genes (LRRC37A, LRRC37A2, NSF, ARL17A, ARL17B, KANSL1, SPPL2C, and CRHR1) located within the chr17q21 H1/H2 inversion region (chr17:45,761,253–46,765,892; hg38) were initially considered for Sanger sequencing of coding exons (primers available upon request). However, specific gene sequencing for LRRC37A, LRRC37A2, ARL17A and ARL17B was not possible given the high homology, presence of pseudogenes and CNVs that are present in the region, and therefore four genes (NSF, KANSL1, SPPL2C, and CRHR1) were included in the analysis. Bidirectional Sanger sequencing was performed using established protocols on the Applied Biosystems 3730xl DNA analyzer (Thermo Fisher Scientific, Waltham, MA). Sequence data was analyzed using Applied Biosystems SeqScape software (version 2.5) (Thermo Fisher Scientific, Waltham, MA). Variant annotations were made using human build GRCh37. Variants were defined as common and rare if their minor allele frequency (MAF) was ≥1% or <1% respectively, and were labeled as H1/H2-defining if they were in perfect linkage disequilibrium (LD) (r2=1) with the MAPT H1/H2-tagging variant rs8070723. A variant was selected for further genotyping in the replication case-control series if it was (a) a non-synonymous variant or frameshift mutation, (b) not an H1/H2 tagging variant (r2≠1), and (c) not in complete LD with a different variant in the same gene that has already been selected for further genotyping in the replication series.
Genotyping
The H1/H2 tagging variant rs8070723 and H1c tagging variant rs242557 were genotyped using an ABI TaqMan™ allelic discrimination assay on Applied Biosystems 7900HT Real-time PCR System (Thermo Fisher Scientific, Waltham, MA) and were analyzed using SDS (version 2.2.2) software (Thermo Fisher Scientific, Waltham, MA).The genotype data for rs8070723 was then used to determine variants that defined the MAPT H1/H2 haplotype (i.e. perfect LD with rs8070723 with an r2=1). Additionally, Selected variants were genotyped using Agena Bioscience iPlex Gold chemistry (Agena Bioscience, San Diego, CA) and ABI Taqman™ SNP genotyping custom-designed assays (Thermo Fisher Scientific, Waltham, MA). Genotyping data was analyzed using Typer 4.0 (Agena Bioscience, San Diego, CA) and ABI QuantStudio Real-Time PCR (version 1.1) (Thermo Fisher Scientific, Waltham, MA) software respectively. The genotyping call rate was >95% for all variants
Statistical analysis
In the replication series, associations of common variants (MAF≥1%) with risk of PD were evaluated using logistic regression models that were adjusted for gender and age at blood draw. Odds ratios (ORs) and 95% confidence intervals were estimated, and each variant was examined under a dominant model (i.e. presence vs. absence of the minor allele). For rare variants (MAF<1%), the proportion of subjects with a copy of the minor allele was compared between PD patients and controls using Fisher’s exact test. Associations of common variants with age of PD onset were examined using linear regression models that were adjusted for gender; regression coefficients and 95% CIs were estimated and are interpreted as the change in mean age of PD onset corresponding to presence of the minor allele of the given variant. All statistical analyses were performed using R Statistical Software (version 3.6.1). P-values <0.05 were considered as statistically significant, and all statistical tests were two-sided. Combined annotation dependent depletion (CADD) scores were determined using the online CADD single nucleotide variant lookup tool (https://cadd.gs.washington.edu/snv); a CADD score >20 indicates a variant is among the 1% most deleterious for the gene.
Single-cell RNA-sequencing
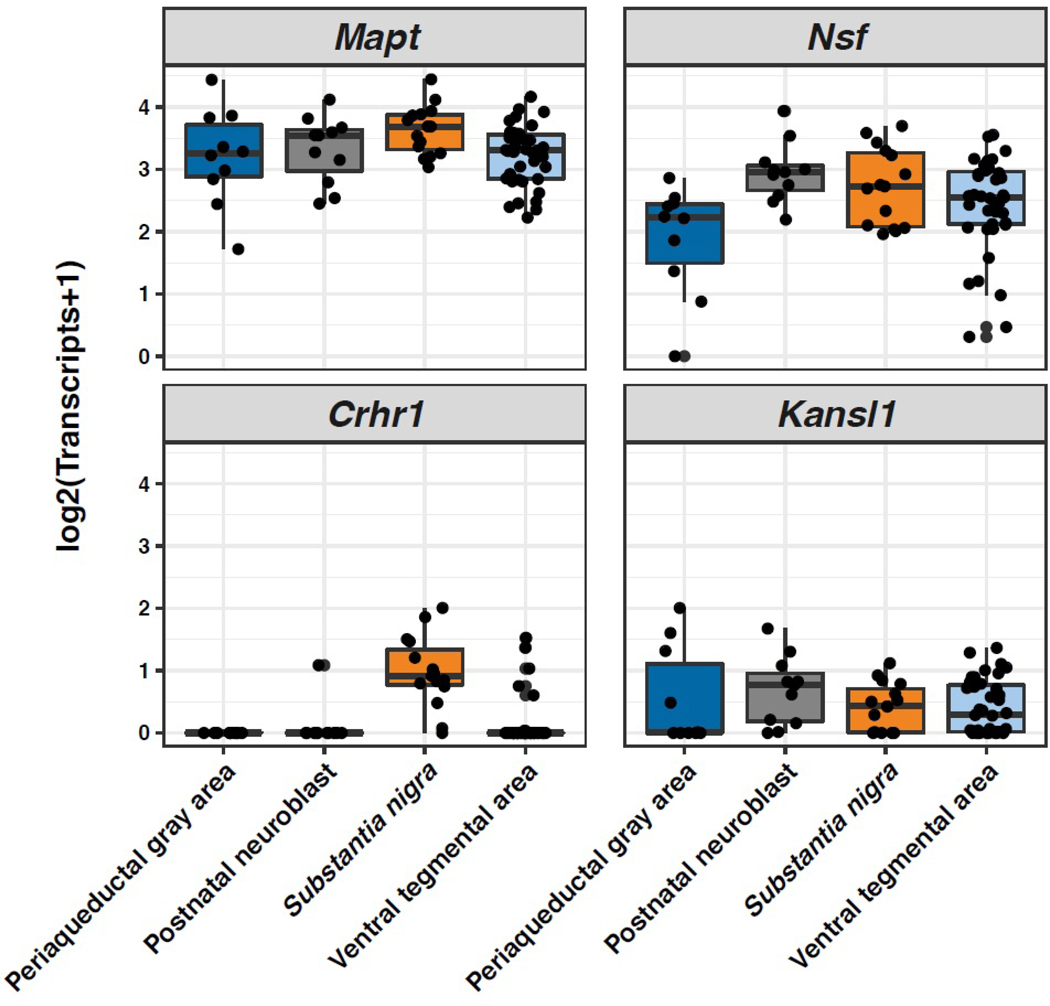
For exploration of the expression of genes in substantia nigra dopaminergic (DA) neurons, single-cell RNA-seq data from mouse postnatal day 7 (P7) midbrain neurons was downloaded (https://github.com/pwh124/sc-da-parkinsons) [14]. These populations included DA neurons from the periaqueductal grey area, substantia nigra, and ventral tegmental area as well as a postnatal neuroblast population [14]. Expression was visualized using ggplot2 [15] and custom scripts in the R statistical environment (https://www.r-project.org/).
Results
Discovery cohort
After Sanger sequencing of coding exons in four neighboring genes on the MAPT inversion (NSF, KANSL1, SPPL2C, and CRHR1) in 90 individuals with sporadic LOPD, a total of 51 variants were identified. Of these 51 variants, 40 were common (MAF ≥1%), 11 were rare (MAF <1%), and also among the 51 variants, 20 were in perfect LD with the H1/H2 haplotype tagging variant rs8070723 (r2=1). One NSF variant (rs748314870) was a frameshift mutation. We observed 29 variants that were non-synonymous, and 11 of those non-synonymous variants (located in KANSL1, SPPL2C and NSF) tagged the H1 haplotype (r2=1) (Table 2). Importantly, of these 11 non-synonymous H1/H2 tagging variants, two located in SPPL2C have a CADD score >20 (rs12373123 and rs12185233) (Table 3) estimating these variants be among the top 1% of those having a deleterious effect; also of note, NSF rs1238228075 had a CADD score of 19.2.
Table 2: Stage one discovery phase results.
Sanger sequencing of four neighboring genes (NSF, KANSL1, SPPL2C, and CRHR1) located 900kb around MAPT on chromosome 17 identified 51 variants in 90 individuals clinically diagnosed with sporadic, late-onset Parkinson’s disease.
| Gene | Exon | Genotypes | rsID | Amino Acid | Mutation type | Frequency | r2 | Included in Stage 2 Genotyping |
|---|---|---|---|---|---|---|---|---|
| CRHR1 | 2 | C/T | rs12936511 | P20P | Synonymous | Common | <0.01 | N |
| CRHR1 | 6 | G/A | rs75638861 | V161M | Nonsynonymous | Common | <0.01 | Y |
| CRHR1 | 8 | T/C | rs16940665 | T252T | Synonymous | Common | 1 | N |
| CRHR1 | 10 | C/T | rs141817026 | C287C | Synonymous | Common | 1 | N |
| KANSL1 | 1 | C/T | rs200649587 | D10D | Synonymous | Rare | <0.01 | N |
| KANSL1 | 1 | A/C | rs17585974 | K104T | Nonsynonymous | Common | 0.83 | Y |
| KANSL1 | 1 | T/G | rs17662889 | L138L | Synonymous | Common | 0.86 | N |
| KANSL1 | 1 | G/T | rs149566146 | G191C | Nonsynonymous | Rare | <0.01 | Y |
| KANSL1 | 1 | A/G | rs144882998 | N207S | Nonsynonymous | Rare | 0.02 | Y |
| KANSL1 | 1 | A/G | rs141110759 | H212R | Nonsynonymous | Rare | <0.01 | Y |
| KANSL1 | 1 | C/T | rs17662853 | T221I | Nonsynonymous | Common | <0.01 | Y |
| KANSL1 | 1 | A/G | rs35643216 | N225D | Nonsynonymous | Common | 0.86 | Y |
| KANSL1 | 1 | C/T | rs1881194 | S232S | Synonymous | Common | 0.90 | N |
| KANSL1 | 1 | C/G | rs2240758 | S337S | Synonymous | Common | 0.01 | N |
| KANSL1 | 1 | A/G | rs1881193 | R247R | Synonymous | Common | 1 | N |
| KANSL1 | 3 | A/G | rs17576165 | P497P | Synonymous | Common | 1 | N |
| KANSL1 | 6 | G/C | rs191986791 | R619R | Synonymous | Rare | <0.01 | N |
| KANSL1 | 7 | G/A | rs2277613 | P712P | Synonymous | Common | <0.01 | N |
| KANSL1 | 7 | T/C | rs34043286 | S718P | Nonsynonymous | Common | 1 | N |
| KANSL1 | 10 | T/C | rs17574604 | F860F | Synonymous | Common | 1 | N |
| KANSL1 | 12 | C/T | rs35833914 | D914E | Nonsynonymous | Common | 1 | N |
| KANSL1 | 12 | C/T | rs36076725 | F917F | Synonymous | Common | 0.96 | N |
| KANSL1 | 13 | C/T | rs7220988 | P1010L | Nonsynonymous | Common | 0.22 | Y |
| KANSL1 | 14 | A/G | rs201083879 | Q1057R | Nonsynonymous | Rare | 0.02 | Y |
| KANSL1 | 14 | T/C | rs34579536 | I1085T | Nonsynonymous | Common | 1 | N |
| NSF | 3 | T/- | rs748314870 | F63S fs 77Stop | Frameshift | Common | <0.01 | Y |
| NSF | 5 | C/A | rs1238328075 | N126K | Nonsynonymous | Common | 0.01 | N |
| NSF | 10 | G/A | rs2074406 | V361M | Nonsynonymous | Common | 0.07 | N* |
| NSF | 12 | G/A | rs373218599 | V431M | Nonsynonymous | Rare | 0.02 | Y |
| NSF | 13 | C/T | rs757532604 | T476M | Nonsynonymous | Common | 0.01 | Y |
| NSF | 18 | C/T | -- | S662S | Synonymous | Rare | <0.01 | N |
| NSF | 19 | G/A | rs199533 | K702K | Synonymous | Common | 0.96 | N |
| SPPL2C | 1 | C/T | rs117261590 | P68S | Nonsynonymous | Rare | <0.01 | Y |
| SPPL2C | 1 | G/A | rs17763658 | R123Q | Nonsynonymous | Common | 0.05 | Y |
| SPPL2C | 1 | G/T | rs142955406 | G167W | Nonsynonymous | Rare | 0.02 | Y |
| SPPL2C | 1 | G/A | rs929223 | E209K | Nonsynonymous | Common | 0.03 | Y |
| SPPL2C | 1 | T/C | rs62621252 | S224P | Nonsynonymous | Common | 1 | N |
| SPPL2C | 1 | G/A | rs242944 | R303H | Nonsynonymous | Common | 0.30 | Y |
| SPPL2C | 1 | G/A | rs148362814 | R307Q | Nonsynonymous | Common | <0.01 | Y |
| SPPL2C | 1 | G/A | rs62054815 | A332T | Nonsynonymous | Common | 1 | N |
| SPPL2C | 1 | C/T | rs150431364 | L380L | Synonymous | Rare | 0.02 | N |
| SPPL2C | 1 | G/C | rs12185233 | R461P | Nonsynonymous | Common | 1 | N |
| SPPL2C | 1 | A/G | rs12185268 | I471V | Nonsynonymous | Common | 1 | N |
| SPPL2C | 1 | C/T | rs12185235 | T477T | Synonymous | Common | 1 | N |
| SPPL2C | 1 | G/A | rs171443 | S542S | Synonymous | Common | 0.04 | N |
| SPPL2C | 1 | T/C | rs11079725 | D554D | Synonymous | Common | 1 | N |
| SPPL2C | 1 | T/C | rs12373123 | S601P | Nonsynonymous | Common | 1 | N |
| SPPL2C | 1 | G/A | rs12373139 | G620R | Nonsynonymous | Common | 1 | N |
| SPPL2C | 1 | C/G | rs12373142 | P643R | Nonsynonymous | Common | 1 | N |
| SPPL2C | 1 | T/C | rs12373124 | H649H | Synonymous | Common | 1 | N |
| SPPL2C | 1 | G/A | rs12373140 | Q653Q | Synonymous | Common | 1 | N |
Common: MAF≥0.01; Rare: MAF<0.01; H1/H2-tagging: SNPs are in complete LD (r2=1.00) with MAPT H1/H2-defining SNP rs8070723.
indicates SNP excluded from genotyping in stage 2 because it was in complete LD with rs757532604 which was genotyped.
Table 3:
Non-synonymous SNPs in complete linkage-disequilibrium to MAPT H1/H2-tagging SNP rs8070723 (r2=1.00).
| Gene | Base Position | rsID | AA | Minor allele | CADD Score * | MAF (%) east Asian | MAF (%) South Asian | MAF (%) non-Finnish |
|---|---|---|---|---|---|---|---|---|
| SPPL2C | 45845576 | rs62621252 | S224P | C | 0.004 | 0.07 | 7.63 | 21.20 |
| SPPL2C | 45845900 | rs62054815 | A332T | A | 0.001 | 0.07 | 7.62 | 21.15 |
| SPPL2C | 45846288 | rs12185233 | R461P | C | 25.6 | 0.07 | 7.62 | 21.17 |
| SPPL2C | 45846317 | rs12185268 | I471V | G | 0.001 | 0.07 | 7.62 | 21.17 |
| SPPL2C | 45846707 | rs12373123 | S601P | C | 22.7 | 0.08 | 7.62 | 21.17 |
| SPPL2C | 45846764 | rs12373139 | G620R | A | 0.526 | 0.07 | 7.62 | 21.16 |
| SPPL2C | 45846834 | rs12373142 | P643R | G | 0.117 | 0.07 | 7.62 | 21.13 |
| KANSL1 | 46039753 | rs34043286 | S718P | C | 15.71 | 0.07 | 7.60 | 21.15 |
| KANSL1 | 46033175 | rs35833914 | D914E | T | 4.236 | 0.07 | 7.48 | 20.87 |
| KANSL1 | 46031540 | rs34579536 | I1085T | C | 8.024 | 0.07 | 7.59 | 21.05 |
| NSF | 46637515 | rs1238328075 | N126K | A | 19.19 | N/A† | N/A† | N/A† |
Base positions are aligned to homo sapiens chromosome build GRCh38, allele frequencies are obtained from the publicly available gnomAD database. AA= amino acid. CADD= Combined Annotation Dependent Depletion.
A CADD greater than 20 indicates a variant that is in the top 1% of deleterious variants.
NSF rs1238328075 has sequence coverage below 10 and therefore the MAF is not available (N/A) from the gnomAD database.
Replication cohort
After exclusion of NSF rs2074406 from further analysis due to its complete LD with a different NSF variant (rs757532604), 17 non-synonymous variants and the frameshift mutation were selected for genotyping in the replication series of 786 PD patients and 751 controls in order to evaluate whether variants in genes neighboring MAPT may be driving the PD association signal that is observed for the MAPT H1/H2 haplotype. When evaluating associations between these variants and PD risk (Table 4), no variants were more strongly associated with risk of PD than the H1/H2 defining rs8070723 variant (OR=0.57, P<0.001) in terms of association ORs. However, similar but slightly weaker associations were noted for both KANSL1 rs35643216 (OR=0.62, P<0.001) and KANSL1 rs17585974 (OR=0.61, P<0.001); both of these KANSL1 variants were in strong LD with rs8070723 (r2≥0.83) and the second of these rs17585974, has a CADD score of 24.7. We attempted to look in single-cell RNA-seq data from mouse midbrain neurons to assess expression of these PD candidate genes in disease relevant tissue. This revealed the expression of Crhr1, Mapt, Nsf and Kansl1 (Figure 1) in this population of cells critical to PD pathogenesis (https://pwh124.shinyapps.io/expressionshiny/), consistent with their posited roles in modulating risk, individually or in combination [14]. Expression levels of the other genes (Lrrc37a, Arl17a/b and Sppl2c) were too low to accurately measure or not detectible.
Table 4: Stage two replication phase results.
Associations of genotyped common and rare, non-synonymous, and non-H1/H2 tagging variants with risk of late-onset Parkinson’s disease in independent cohorts of N=786 unrelated, clinically diagnosed PD patients and N=751 unrelated, healthy controls. Non-H1/H2 tagging variants had an r2 < 1.00 linkage disequilibrium score relative to MAPT H1/H2-defining SNP rs8070723. Base positions are aligned to Homo sapiens chromosome build GRCh38. For common variants (MAF≥1%), ORs, 95% CIs, and p-values result from logistic regression models that were adjusted for age and sex. ORs correspond to presence of the minor allele. For rare variants (MAF<1%), the proportion of subjects with a copy of the minor allele was compared between PD patients and controls using Fisher’s exact test.
| Gene | Base Position | rsID | AA | Minor allele | MAF (cases) | MAF (controls) | OR (95% CI) | P value | CADD * |
|---|---|---|---|---|---|---|---|---|---|
| CRHR1 | 45829607 | rs75638861 | V161M | A | 1.50% | 1.60% | 0.92 (0.51, 1.68) | 0.79 | 10.5 |
| SPPL2C | 45845108 | rs117261590 | P68S | T | 0.36% | 0.21% | - | 0.52 | 0 |
| SPPL2C | 45845274 | rs17763658 | R123Q | A | 7.39% | 6.96% | 1.02 (0.76, 1.37) | 0.9 | 7.33 |
| SPPL2C | 45845405 | rs142955406 | G167W | T | 0.12% | 0.14% | - | 1 | 21.8 |
| SPPL2C | 45845531 | rs929223 | E209K | A | 2.27% | 3.04% | 0.84 (0.50, 1.39) | 0.49 | 11.98 |
| SPPL2C | 45845814 | rs242944 | R303H | G | 41.30% | 46.76% | 0.80 (0.64, 0.99) | 0.045 | 0 |
| SPPL2C | 45845826 | rs148362814 | R307Q | A | 0.65% | 0.21% | - | 0.1 | 0.01 |
| MAPT | 45942346 | rs242557 | -- | A | 39.92% | 35.07% | 1.15 (0.93, 1.41) | 0.19 | 0 |
| MAPT | 46003698 | rs8070723 | -- | G | 17.84% | 25.79% | 0.57 (0.46, 0.70) | <0.001 | 0 |
| KANSL1 | 46171833 | rs17585974 | K104T | G | 15.17% | 20.60% | 0.61 (0.49, 0.76) | <0.001 | 24.7 |
| KANSL1 | 46171573 | rs149566146 | G191C | A | 0.18% | 0.00% | - | 0.25 | 22.9 |
| KANSL1 | 46171524 | rs144882998 | N207S | C | 0.06% | 0.07% | - | 1 | 10.7 |
| KANSL1 | 46171509 | rs141110759 | H212R | C | 0.06% | 0.00% | - | 1 | 1.15 |
| KANSL1 | 46171482 | rs17662853 | T221I | T | 15.24% | 14.62% | 1.04 (0.83, 1.31) | 0.72 | 23.2 |
| KANSL1 | 46171471 | rs35643216 | N225D | C | 15.25% | 20.43% | 0.62 (0.50, 0.77) | <0.001 | 10.71 |
| KANSL1 | 46032108 | rs7220988 | P1010L | T | 44.76% | 40.06% | 1.30 (1.05, 1.61) | 0.018 | 18.02 |
| KANSL1 | 46031624 | rs201083879 | Q1057R | C | 0.12% | 0.00% | - | 0.5 | 21.6 |
| NSF | 46626698 | rs748314870 | F63S fs 77Stop | T/- | 1.85% | 2.12% | 0.82 (0.39, 1.74) | 0.61 | 27.6 |
| NSF | 46694579 | rs373218599 | V431M | A | 0.06% | 0.00% | - | 1 | 14.89 |
| NSF | 46704811 | rs757532604 | T476M | T | 22.13% | 20.87% | 1.06 (0.87, 1.31) | 0.55 | 24.7 |
AA= Amino Acid. MAF= Minor Allele Frequency. OR=odds ratio. CI=confidence interval. CADD= Combined Annotation Dependent Depletion.
A CADD greater than 20 indicates a variant that is in the top 1% of deleterious variants.
Figure 1.
Boxplots displaying the RNA expression of Mapt, Nsf, Crhr1, and Kansl1 in mouse midbrain dopaminergic cell populations identified in postnatal day 7 (P7) mice [14]. The levels of other genes (Lrrc37a, Arl17a/b and Sppl2c) are not detectible. The box represents the interquartile range and whiskers on the boxplots represent +/−1.5 interquartile range. Each dot represents expression measured in a single cell as log2-transformed transcript counts.
Associations with age of PD onset
We next examined associations with age of PD onset for the MAPT rs8070723 H1/H2 tagging variant, the MAPT H1c haplotype partial tagging variant rs242557, and the two aforementioned KANSL1 non-H1/H2 tagging variants (rs35643216 and rs17585974). Consistent with the findings of a recent GWAS [16], there was not a significant association with of age of PD onset for MAPT rs8070723 (β: 0.76, 95% CI: −0.98 to 2.49, P=0.39), MAPT rs242557 (β: −0.16, 95% CI: −1.82 to 1.49, P=0.85), KANSL1 rs35643216 (β: 1.12, 95% CI: −0.67 to 2.91, P=0.22), or KANSL1 rs17585974 (β: 1.19, 95% CI: −0.60 to 2.97, P=0.19).
Discussion
This study set out to investigate if variation outside of the MAPT gene could account for the PD GWAS signal at Chr17q21 in Caucasians. We identified a number of H1/H2 tagging non-synonymous variants in genes other than MAPT, including two SPPL2C variants (rs12185233 and rs12373123) with CADD scores >20 indicating high likelihood of a deleterious effect. We did not observe any stronger associations with PD risk for common non-synonymous non-H1/H2 tagging variants relative to the H1/H2 signal. However, slightly weaker associations were noted for two KANSL1 variants (rs35643216 and rs17585974; ORs 0.62 and 0.61); these two variants were in strong LD with MAPT H1/H2, and interestingly KANSL1 rs17585974 has a high CADD score of 24.7. Also of note, while not quite reaching a CADD >20, NSF rs1238328075 and KANSL1 rs34043286 have relatively high CADD scores 19.19 and 15.71 respectively and should not be excluded from being functionally important. Though future functional studies are clearly needed in order to establish whether any of the aforementioned variants may truly be the causal variant that drives the MAPT H1/H2 association in PD, the results provided herein represent an important and necessary first step toward the identification of potential causal variants and the subsequent conduction of such functional studies.
A closer look at the functionality of the other genes is warranted. KANSL1 codes for KAT8 Regulatory NSL Complex Subunit 1, which encodes a nuclear protein that is involved with histone acetylation with the MLL1 and NSL1 complexes [17]. Notably a recent study has implicated KANSL1 (and KAT8 another potential PD GWAS hit) in regulating PINK1-directed mitophagy nominating variation within the gene as driving the Chr17q21 association signal [18]. Differences in KANSL1 expression levels have also been observed in brains of individuals with PD, AD, and frontotemporal dementia [19, 20], which suggests a potential role in neurodegeneration although this may be simply driven by the broader H1/H2 association and not be specific. Additionally, we have reported single cell transcriptional analyses of mouse midbrain dopamine neurons in which we reported the expression of Crhr1, Mapt, Kansl1 and Nsf within nigral dopamine neurons [14]. The expression of Sppl2C was very low or not detectible however, SPPL2C codes for Signal Peptide Peptidase Like 2C, which is a member of the GxGD‐type intramembrane aspartyl proteases family, which have emerged as key components of driving pathologies in AD and viral infections [21]. Even though not being well-characterized, recently SPPL2C candidate substrates have been demonstrated to cluster and impair vesicular trafficking which accelerates retention of cargo proteins in the endoplasmic reticulum, and disrupts subcellular compartmentation [21]; dysfunction in synaptic vesicle trafficking is well characterized in PD pathogenesis [22].
Although links can be made for KANSL1 and SPPL2C in PD pathogenesis, a direct link to the well-established PD gene LRRK2 can be made for NSF. NSF codes for a N-Ethylmaleimide Sensitive Factor, Vesicle Fusing ATPase. The N-terminal of NSF is required for SNAP-SNARE complex binding [22], and the D1 domain is essential for ATP binding and hydrolysis, which controls NSF activity managing synaptic vesicle endocytosis [23, 24]. Belluzzi et al., demonstrated that NSF is directly phosphorylated by LRRK2 at T645 resulting in enhanced ATPase activity and disrupted synaptic vesicle trafficking [24, 25]. Synaptic vesicle release and recycling is one of the major pathways implicated in PD etiology and is also linked to α-synuclein dysfunction at the synapse [26, 27].
A number of limitations within the study design did not let us fully resolve the causative gene at this locus. For example, specific gene sequencing for LRRC37A, LRRC37A2, ARL17A and ARL17B was not possible given the high homology, presence of pseudogenes and CNVs that are present in the region. LRRC37A codes for Leucine Rich Repeat Containing-37, Member A, and ARL17A and ARL17B encode ADP-Ribosylation Factor-Like 17A and −17B respectively. LRR motifs are important for intermolecular or intercellular interactions with exogenous factors in the immune system and/or with different cell types in the developing nervous system [28]. These genes could not be excluded as potentially influencing susceptibility to PD phenotype.
Although the sample population chosen was sporadic to address the GWAS signal specifically we did observe two variants in KANSL1 (both CADD >20) and one variant (CADD >14) in NSF that were detected in PD cases but not in controls. The function of these variants have not been characterized, yet both variants in KANSL1 are in the pathogenic range, CADD <20.38 is benign and CADD > 33.25 is pathogenic [29]. Further sequencing of patients with familial PD may identify pathogenic mutations that would help discriminate the disease-related gene or genes on the chr17q21 haplotype. It is important to note that next generation sequencing data currently available does not have adequate coverage of all coding exons and therefore should be used with caution for identifying variants and their significance.
Another approach would be to exploit ethnic-diversity to narrow down the region of association and pinpoint the relevant gene. Interestingly, in association studies in Asian populations there is no evidence of a signal at Chr17q21 for PD and the H2 haplotype is absent in this population [30]. We have included population frequencies from publicly available dataset (gnomAD) in Table 4 to highlight the ethnic-specific nature of the alleles. If the variants are frequent in the Asian population it would potentially rule those out as driving the signal in Caucasian populations. Using populations with different haplotype structure and genomic architecture may be a viable way to fine-map GWAS signals and nominate functional genes/variants.
In conclusion, our work explored the association of common coding variation around the Chr17q21 PD GWAS signal that do not map to the MAPT gene. Although no association was observed that was stronger than the established H1/H2 association, a number of non-synonymous variants were identified that also tag H1/H2 across at least three genes (KANSL1, SPPL2C and CRHR1) and may represent other functional variants that influence disease risk. Further biological testing of variants in cell and animal models for modulation of PD relevant phenotypes such as alpha-synuclein aggregation/toxicity or PINK1-PRKN mitophagy will be needed to establish the disease risk at the Chr17q21 locus.
MAPT H1 is a consistent Parkinson’s disease association locus on chr17q21
Chr17q21 H1 extended haplotype is a complex genetic region of inversion
Other genes/variants in complete linkage disequilibrium may be responsible
Potentially damaging variants in KANSL1, NSF and SPPL2C
Acknowledgments
The authors would like to thank all those who have contributed to our research, particularly the patients and families who donated DNA samples. The Mayo Clinic is an American Parkinson Disease Association (APDA) Information and Referral Center, an APDA Center for Advanced Research, a Lewy Body Dementia Association Research Center of Excellence and was a Morris K. Udall Parkinson’s Disease Research Center of Excellence (NINDS P50 NS072187). CL was the recipient of a FRSQ postdoctoral fellowship and was a 2015 Younkin Scholar supported by the Mayo Clinic Alzheimer’s Disease and Related Dementias Genetics program. ASM and PWH were supported in part by awards from NIH (NS62972 and MH106522) and by HELIS, Helis Medical Research Foundation to ASM. PJM is supported by Mayo Foundation for Medical Research, NIH; R01 NS110085, U54 NS110435 Lewy Body Dementia Center without Walls, American Parkinson’s Disease Association Mayo Clinic Center for Advanced Research, and an Ed and Ethel Moore Award from Florida department of Public Health. WS is partially supported by NIH [R01/RF1 NS085070, R01 NS110085, U54 NS110435 and R56 AG062556], the Department of Defense Congressionally Directed Medical Research Programs (CDMRP) [W81XWH-17-1-0248], the Florida Department of Health - Ed and Ethel Moore Alzheimer’s Disease Research Program [9AZ10], the Michael J. Fox Foundation for Parkinson’s Research (MJFF), the American Parkinson Disease Association (APDA), and the Mayo Clinic Center for Biomedical Discovery (CBD).OAR is supported by the National Institutes of Health (NIH; R01 NS78086; U54 NS100693), the US Department of Defense (W81XWH-17-1-0249), Lewy Body Dementia Center WithOut Walls U54 NS110435, The Michael J. Fox Foundation, the Mayo Clinic LBD Functional Genomics Program and The Little Family Foundation.
Footnotes
Financial Disclosures of all authors
Alexandra I. Soto-Beasley reports no relevant financial disclosures
Ronald L. Walton reports no relevant financial disclosures
Rebecca R. Valentino reports no relevant financial disclosures
Paul W. Hook reports no relevant financial disclosures
Loyal A. Goff reports no relevant financial disclosures
Catherine Labbé reports no relevant financial disclosures
Michael G. Heckman reports no relevant financial disclosures
Patrick W. Johnson reports no relevant financial disclosures
Ryan J. Uitti reports no relevant financial disclosures
Pamela J. McLean no relevant financial disclosures
Wolfdieter Springer no relevant financial disclosures
Andrew S. McCallion no relevant financial disclosures
Zbigniew K. Wszolek is partially supported by the Mayo Clinic Center for Regenerative Medicine, the gifts from The Sol Goldman Charitable Trust, Albertson Parkinson’s Research Foundation and the Donald G. and Jodi P. Heeringa Family, and by the Haworth Family Professorship in Neurodegenerative Diseases fund. He serves as PI or Co-PI on Abbvie, Inc. (M15–562, M15–563, and laboratory based grant), and Biogen, Inc. (228PD201) grants. He serves as PI of the Mayo Clinic American Parkinson Disease Association (APDA) Information and Referral Center, and as Co-PI of the Mayo Clinic APDA Center for Advanced Research.
Owen A. Ross received support from R01-NS078086, P50-NS072187, U54 NS100693, U54 NS110435, the US Department of Defense (W81XWH-17–1-0249), the Mayo Clinic LBD Functional Genomics Program, The Little Family Foundation, and the Michael J. Fox Foundation.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Nalls MA, Blauwendraat C, Vallerga CL, Heilbron K, Bandres-Ciga S, Chang D, Tan M, Kia DA, Noyce AJ, Xue A, Bras J, Young E, von Coelln R, Simón-Sánchez J, Schulte C, Sharma M, Krohn L, Pihlstrom L, Siitonen A, Iwaki H, Leonard H, Faghri F, Raphael Gibbs J, Hernandez DG, Scholz SW, Botia JA, Martinez M, Corvol J-C, Lesage S, Jankovic J, Shulman LM, Tienari P, Majamaa K, Toft M, Brice A, Yang J, Gan-Or Z, Gasser T, Heutink P, Shulman JM, Wood N, Hinds DA, Hardy J, Morris HR, Gratten J, Visscher PM, Graham RR, Singleton AB, Parkinson’s disease genetics: identifying novel risk loci, providing causal insights and improving estimates of heritable risk, bioRxiv (2019) 388165. [Google Scholar]
- [2].Kadavath H, Hofele RV, Biernat J, Kumar S, Tepper K, Urlaub H, Mandelkow E, Zweckstetter M, Tau stabilizes microtubules by binding at the interface between tubulin heterodimers, Proc Natl Acad Sci U S A 112(24) (2015) 7501–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Qiang L, Sun X, Austin TO, Muralidharan H, Jean DC, Liu M, Yu W, Baas PW, Tau Does Not Stabilize Axonal Microtubules but Rather Enables Them to Have Long Labile Domains, Curr Biol 28(13) (2018) 2181–2189 e4. [DOI] [PubMed] [Google Scholar]
- [4].Zody MC, Jiang Z, Fung HC, Antonacci F, Hillier LW, Cardone MF, Graves TA, Kidd JM, Cheng Z, Abouelleil A, Chen L, Wallis J, Glasscock J, Wilson RK, Reily AD, Duckworth J, Ventura M, Hardy J, Warren WC, Eichler EE, Evolutionary toggling of the MAPT 17q21.31 inversion region, Nat Genet 40(9) (2008) 1076–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Evans W, Fung HC, Steele J, Eerola J, Tienari P, Pittman A, Silva R.d., Myers A, Vrieze FW-D, Singleton A, Hardy J, The tau H2 haplotype is almost exclusively Caucasian in origin, Neurosci Lett 369(3) (2004) 183–185. [DOI] [PubMed] [Google Scholar]
- [6].Hoglinger GU, Melhem NM, Dickson DW, Sleiman PM, Wang LS, Klei L, Rademakers R, de Silva R, Litvan I, Riley DE, van Swieten JC, Heutink P, Wszolek ZK, Uitti RJ, Vandrovcova J, Hurtig HI, Gross RG, Maetzler W, Goldwurm S, Tolosa E, Borroni B, Pastor P, Cantwell LB, Han MR, Dillman A, van der Brug MP, Gibbs JR, Cookson MR, Hernandez DG, Singleton AB, Farrer MJ, Yu CE, Golbe LI, Revesz T, Hardy J, Lees AJ, Devlin B, Hakonarson H, Muller U, Schellenberg GD, Identification of common variants influencing risk of the tauopathy progressive supranuclear palsy, Nature genetics 43(7) (2011) 699–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Heckman MG, Brennan RR, Labbé C, Soto AI, Koga S, DeTure MA, Murray ME, Petersen RC, Boeve BF, van Gerpen JA, Uitti RJ, Wszolek ZK, Rademakers R, Dickson DW, Ross OA, Association of MAPT Subhaplotypes With Risk of Progressive Supranuclear Palsy and Severity of Tau Pathology, JAMA Neurology (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kouri N, Ross OA, Dombroski B, Younkin CS, Serie DJ, Soto-Ortolaza A, Baker M, Finch NC, Yoon H, Kim J, Fujioka S, McLean CA, Ghetti B, Spina S, Cantwell LB, Farlow MR, Grafman J, Huey ED, Ryung Han M, Beecher S, Geller ET, Kretzschmar HA, Roeber S, Gearing M, Juncos JL, Vonsattel JP, Van Deerlin VM, Grossman M, Hurtig HI, Gross RG, Arnold SE, Trojanowski JQ, Lee VM, Wenning GK, White CL, Hoglinger GU, Muller U, Devlin B, Golbe LI, Crook J, Parisi JE, Boeve BF, Josephs KA, Wszolek ZK, Uitti RJ, Graff-Radford NR, Litvan I, Younkin SG, Wang LS, Ertekin-Taner N, Rademakers R, Hakonarsen H, Schellenberg GD, Dickson DW, Genome-wide association study of corticobasal degeneration identifies risk variants shared with progressive supranuclear palsy, Nat Commun 6 (2015) 7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Allen M, Kachadoorian M, Quicksall Z, Zou F, Chai HS, Younkin C, Crook JE, Pankratz VS, Carrasquillo MM, Krishnan S, Nguyen T, Ma L, Malphrus K, Lincoln S, Bisceglio G, Kolbert CP, Jen J, Mukherjee S, Kauwe JK, Crane PK, Haines JL, Mayeux R, Pericak-Vance MA, Farrer LA, Schellenberg GD, Parisi JE, Petersen RC, Graff-Radford NR, Dickson DW, Younkin SG, Ertekin-Taner N, Association of MAPT haplotypes with Alzheimer’s disease risk and MAPT brain gene expression levels, Alzheimer’s Research & Therapy 6(4) (2014) 39–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Skipper L, Wilkes K, Toft M, Baker M, Lincoln S, Hulihan M, Ross OA, Hutton M, Aasly J, Farrer M, Linkage disequilibrium and association of MAPT H1 in Parkinson disease, Am J Hum Genet 75(4) (2004) 669–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Giasson BI, Forman MS, Higuchi M, Golbe LI, Graves CL, Kotzbauer PT, Trojanowski JQ, Lee VM, Initiation and synergistic fibrillization of tau and alpha-synuclein, Science 300(5619) (2003) 636–40. [DOI] [PubMed] [Google Scholar]
- [12].Guo JL, Covell DJ, Daniels JP, Iba M, Stieber A, Zhang B, Riddle DM, Kwong LK, Xu Y, Trojanowski JQ, Lee VM, Distinct alpha-synuclein strains differentially promote tau inclusions in neurons, Cell 154(1) (2013) 103–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hughes AJ, Daniel SE, Kilford L, Lees AJ, Accuracy of clinical diagnosis of idiopathic Parkinson’s disease: a clinico-pathological study of 100 cases, J Neurol Neurosurg Psychiatry 55(3) (1992) 181–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hook Paul W., McClymont Sarah A., Cannon Gabrielle H., Law William D., Jennifer Morton A, Goff Loyal A., and McCallion Andrew S.. 2018. “Single-Cell RNA-Seq of Mouse Dopaminergic Neurons Informs Candidate Gene Selection for Sporadic Parkinson Disease.” American Journal of Human Genetics 102 (3): 427–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wickham H 2016. ggplot2: Elegant Graphics for Data Analysis. Springer Publ; Ed.1: 1–213. [Google Scholar]
- [16].Blauwendraat C, Heilbron K, Vallerga CL, Bandres-Ciga S, von Coelln R, Pihlstrom L, Simon-Sanchez J, Schulte C, Sharma M, Krohn L, Siitonen A, Iwaki H, Leonard H, Noyce AJ, Tan M, Gibbs JR, Hernandez DG, Scholz SW, Jankovic J, Shulman LM, Lesage S, Corvol JC, Brice A, van Hilten JJ, Marinus J, andMe Research T, Eerola-Rautio J, Tienari P, Majamaa K, Toft M, Grosset DG, Gasser T, Heutink P, Shulman JM, Wood N, Hardy J, Morris HR, Hinds DA, Gratten J, Visscher PM, Gan-Or Z, Nalls MA, Singleton AB, International C Parkinson’s Disease Genomics, Parkinson’s disease age at onset genome-wide association study: Defining heritability, genetic loci, and alpha-synuclein mechanisms, Mov Disord 34(6) (2019) 866–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sheikh BN, Guhathakurta S, Akhtar A. The non-specific lethal (NSL) complex at the crossroads of transcriptional control and cellular homeostasis. EMBO Rep. 2019;20(7):e47630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Soutar MPM, Melandri D, Annuario E, Monaghan AE, Welsh NJ, D’Sa K, Guelfi S, Zhang D, Pittman A, Trabzuni D, Pan KS, Kia DA, Bictash M, Gandhi S, Houlden H, Cookson MR, Wood NW, Singleton AB, Hardy J, Whiting PJ, Blauwendraat C, Whitworth AJ, Manzoni C, Ryten M, Lewis PA, Plun-Favreau H, Regulation of mitophagy by the NSL Complex underlies genetic risk for Parkinson’s disease at Chr16q11.2 and on the MAPT H1 allele, bioRxiv 2020January06896241; doi: 10.1101/2020.01.06.896241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ferrari R, Wang Y, Vandrovcova J, Guelfi S, Witeolar A, Karch CM, Schork AJ, Fan CC, Brewer JB, International FTDGC, International C Parkinson’s Disease Genomics, P. International Genomics of Alzheimer’s, Momeni P, Schellenberg GD, Dillon WP, Sugrue LP, Hess CP, Yokoyama JS, Bonham LW, Rabinovici GD, Miller BL, Andreassen OA, Dale AM, Hardy J, Desikan RS, Genetic architecture of sporadic frontotemporal dementia and overlap with Alzheimer’s and Parkinson’s diseases, Journal of neurology, neurosurgery, and psychiatry 88(2) (2017) 152–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Witoelar A, Jansen IE, Wang Y, Desikan RS, Gibbs JR, Blauwendraat C, Thompson WK, Hernandez DG, Djurovic S, Schork AJ, Bettella F, Ellinghaus D, Franke A, Lie BA, McEvoy LK, Karlsen TH, Lesage S, Morris HR, Brice A, Wood NW, Heutink P, Hardy J, Singleton AB, Dale AM, Gasser T, Andreassen OA, Sharma M, International NABEC Parkinson’s Disease Genomics Consortium, I. United Kingdom Brain Expression Consortium, Genome-wide Pleiotropy Between Parkinson Disease and Autoimmune Diseases, JAMA neurology 74(7) (2017) 780–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Papadopoulou AA, Müller SA, Mentrup T, Shmueli MD, Niemeyer J, Haug‐Kröper M, von Blume J, Mayerhofer A, Feederle R, Schröder B, Lichtenthaler SF, Fluhrer R, Signal peptide peptidase‐like 2c impairs vesicular transport and cleaves SNARE proteins, EMBO reports 20(3) (2019) e46451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lotharius J, Brundin P, Pathogenesis of parkinson’s disease: dopamine, vesicles and α-synuclein, Nature Reviews Neuroscience 3(12) (2002) 932–942. [DOI] [PubMed] [Google Scholar]
- [23].Matveeva EA, Whiteheart SW, Vanaman TC, Slevin JT, Phosphorylation of the N-ethylmaleimide-sensitive factor is associated with depolarization-dependent neurotransmitter release from synaptosomes, J Biol Chem 276(15) (2001) 12174–81. [DOI] [PubMed] [Google Scholar]
- [24].Belluzzi E, Gonnelli A, Cirnaru MD, Marte A, Plotegher N, Russo I, Civiero L, Cogo S, Carrion MP, Franchin C, Arrigoni G, Beltramini M, Bubacco L, Onofri F, Piccoli G, Greggio E, LRRK2 phosphorylates pre-synaptic N-ethylmaleimide sensitive fusion (NSF) protein enhancing its ATPase activity and SNARE complex disassembling rate, Mol Neurodegener 11 (2016) 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Pan P-Y, Li X, Wang J, Powell J, Wang Q, Zhang Y, Chen Z, Wicinski B, Hof P, Ryan TA, Yue Z, Parkinson’s Disease-Associated LRRK2 Hyperactive Kinase Mutant Disrupts Synaptic Vesicle Trafficking in Ventral Midbrain Neurons, J Neurosci 37(47) (2017) 11366–11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Burre J, Sharma M, Tsetsenis T, Buchman V, Etherton MR, Sudhof TC, Alpha-synuclein promotes SNARE-complex assembly in vivo and in vitro, Science 329(5999) (2010) 1663–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sun J, Wang L, Bao H, Premi S, Das U, Chapman ER, Roy S, Functional cooperation of alpha-synuclein and VAMP2 in synaptic vesicle recycling, Proc Natl Acad Sci U S A 116(23) (2019) 11113–11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Giannuzzi G, Siswara P, Malig M, Marques-Bonet T, Program NCS, Mullikin JC, Ventura M, Eichler EE, Evolutionary dynamism of the primate LRRC37 gene family, Genome Res 23(1) (2013) 46–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].van der Velde KJ, de Boer EN, van Diemen CC, Sikkema-Raddatz B, Abbott KM, Knopperts A, Franke L, Sijmons RH, de Koning TJ, Wijmenga C, Sinke RJ, Swertz MA, GAVIN: Gene-Aware Variant INterpretation for medical sequencing, Genome Biol 18(1) (2017) 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Foo JN, Chew EGY, Chung SJ, Peng R, Blauwendraat C, Nalls MA, Mok KY, Satake W, Toda T, Chao Y, Tan LCS, Tandiono M, Lian MM, Ng EY, Prakash KM, Au WL, Meah WY, Mok SQ, Annuar AA, Chan AYY, Chen L, Chen Y, Jeon BS, Jiang L, Lim JL, Lin JJ, Liu C, Mao C, Mok V, Pei Z, Shang HF, Shi CH, Song K, Tan AH, Wu YR, Xu YM, Xu R, Yan Y, Yang J, Zhang B, Koh WP, Lim SY, Khor CC, Liu J, Tan EK, Identification of Risk Loci for Parkinson Disease in Asians and Comparison of Risk Between Asians and Europeans: A Genome-Wide Association Study, JAMA Neurol (2020); April 20:e200428. doi: 10.1001/jamaneurol.2020.0428. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]