Abstract
Aims:
The present report aimed to investigate the underlying genes and pathways of high glucose driving cholangiocarcinoma (CCA) aggressiveness.
Main methods:
We screened and compared the gene expression profiles obtained by RNA sequencing, of CCA cells cultured in high and normal glucose. Results from the transcriptomic analysis were confirmed in additional cell lines using in vitro migration-invasion assay, Western blotting and immunocytofluorescence.
Key findings:
Data indicated that high glucose increased the expression of interleukin-1β (IL-1β), an upstream regulator of nuclear factor-κB (NF-κB) pathway, through the nuclear localization of NF-κB. High glucose-induced NF-κB increased the migration and invasion of CCA cells and the expression of downstream NF-κB targeted genes associated with aggressiveness, including interleukin-6, a potent triggering signal of the signal transducer and activator of transcription 3 (STAT3) pathway. Such effects were reversed by inhibiting NF-κB nuclear translocation which additionally reduced the phosphorylation of STAT3 at Y705.
Significance:
These results indicate that NF-κB is activated by high glucose and they suggest that NF-κB interaction with STAT3 enhances CCA aggressiveness. Therefore, targeting multiple pathways such as STAT3 and NF-κB might improve CCA treatment outcome especially in condition such as hyperglycemia.
Keywords: High glucose, diabetes mellitus, nuclear factor-kappa B, interleukin-1β, transcriptomics, cholangiocarcinoma, biliary tract cancer, STAT3
1. Introduction
The association of diabetes mellitus (DM) and cancers is well documented. The increased risk and progression of cancer due to DM was speculated as a result of several diabetogenic conditions, such as high insulin level, sustained low grade inflammation, and high glucose level [1]. In the past decade, high glucose was proven as the direct link between DM and cancers, possibly independent from other factors [2, 3]. To date, many reports demonstrated that high glucose plays a critical role in the promotion and progression of almost all cancer hallmarks [2, 4]. The promoting effects of high glucose on aggressiveness of cancer; namely proliferation, migration and invasion, were reported in breast [5, 6], pancreas [7, 8], colon [9], and liver cancers, including hepatocellular carcinoma [10] as well as cholangiocarcinoma (CCA) [11].
CCA is the second prevalent primary liver cancer and its incidence is globally increasing [12]. Considered a rare cancer in western countries, its prevalence is relatively high in Southeast Asia, especially in Northeast Thailand [13]. The major risk factor in this area is liver fluke, Opisthorchis viverrini, infection [14] but other risk factors have also been reported [15]. As the high prevalence of DM and CCA almost geographically overlaid [16, 17], the association between DM and liver fluke-associated CCA was recently reviewed [18]. The effect of diabetogenic conditions, especially high glucose, on the carcinogenesis and progression of CCA deserve further analysis. It is well documented that carcinogenesis and progression of cancer are multi-step processes, regulated by multiple pathways that interplay for cancer survival [19]. Understanding the signal pathways underlying these processes may offer an opportunity to develop a specific strategy for an effective treatment, especially in patients with diabetogenic conditions.
We previously reported that high glucose promoted the proliferation and metastatic potential of CCA cells by activating STAT3 pathways [11]. The promoting effects were investigated in vitro as well as in human tumor tissues. We recently showed that the inhibition of STAT3 nuclear translocation by specific inhibitor and metformin suppressed growth and metastatic related phenotypes in CCA cells [20]. Using phospho-kinases array, our previous study also showed that high glucose induces phosphorylation of many signaling molecules in CCA cells. We, hence, hypothesized that other signaling pathways may collaborate with STAT3 to promote high glucose-enhanced CCA cells aggressiveness.
In the present study, a set of differentially expressed genes related to high glucose and progressive phenotypes of CCA cells were identified in CCA cells cultured in high glucose versus normal glucose, and in highly metastatic cells by RNA sequencing (RNA-seq). The activation of the NF-κB signaling cascade was found to be related to aggressive phenotypes under high glucose condition. A mutual regulation of downstream targets of NF-κB and STAT3 related to aggressive phenotypes was observed. These findings suggested that NF-κB/STAT3 are co-activated and co-regulated under the high glucose induced aggressiveness of CCA.
2. Materials and Methods
2.1. Cell culture and treatment
The human CCA cell lines, KKU-213A and KKU-213B, were established from Thai O. viverrini-associated CCA patients [21] and obtained from the Japanese Collection of Research Bioresources (JCRB) Cell Bank, Osaka, Japan. Cells were cultured in normal glucose (NG: 5.6 mM) or high glucose (HG: 25 mM) Dulbecco Modified Eagle’s Medium (Invitrogen, Carlsbad, CA) for at least 5 passages allowing their adaptation in different glucose environments and labeled respectively as NG or HG cells [11]. The media were changed every two days and cells were subcultured at 80% confluence to maintain logarithmic growth. A highly metastatic subline of KKU-213A, named KKU-213A-L5 or L5 cells, was established as previously described [22] and was also cultured in HG medium.
To determine the proliferation rate of NG and HG cells, 1 × 103 cells were seeded into 96 well plates in their respective media. At the indicated time, 110 μL of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (Invitrogen, Carlsbad, CA) in PBS (0.5 mg/mL) was added and cells were further incubated at 37°C for 4 h. Formazan crystal was then dissolved in dimethylsulfoxide (DMSO). The number of viable cells was measured at 540 nm using a microplate reader (SUNRISE, Groedig, Austria).
To inhibit nuclear translocation of NF-κB, CCA cells were treated with dehydroxymethylepoxyquinomicin (DHMEQ, synthesized and kindly provided by K. Umezawa as previously described [23]) at a final concentration of 2.5 μg/mL for 48 h [24, 25]. Stattic (21.2 ng/mL; Santa Cruz Biotechnology, Santa Cruz, CA) was used to inhibit the nuclear translocation of STAT3 as previously described [11]. The control groups were treated with 0.01% DMSO.
The study protocol has been reviewed and approved by The Khon Kaen University Ethics Committee for Human Research (HE571464, December 2, 2015; HE621328, September 17, 2019) based on the Declaration of Helsinki and ICH-Good Clinical Practice Guidelines.
2.2. RNA isolation and quality control
KKU-213A-NG, -HG and -L5 cell lines were cultured in their respective medium. Confluent cells were trypsinized (Invitrogen, Carlsbad, CA) and cell pellets were preserved in RNAlater® (Invitrogen/ThermoFisher Scientific, Waltham, MA) at −80 °C. Total RNA was isolated using a combined Trizol® (ThermoFisher Scientific) and RNeasy® kit (Qiagen, Germantown, MD) protocol. Briefly, the cell pellets were lysed in TRIzol® and the phase separation was performed as per manufacturer instruction. The aqueous phase was then transferred to a silica membrane column of the RNeasy® kit for further purification. RNA concentration was measured with RiboGreen assay (Agilent Technologies, Waldbronn, Germany) and its integrity assessed with RNA Pico Chip Bioanalyzer (Agilent Technologies). Analysis revealed a non-degraded RNA with a maximal RNA Integrity Number (RIN) of 10 for all cell lines. RNA was then subjected to library preparation and sequencing.
2.3. Illumina paired-end library construction and RNA-sequencing
Strand-specific, poly-A+ RNA-seq libraries were prepared and whole-transcriptome RNA-seq was performed on the Illumina HiSeq 2000 platform. Briefly, poly-A+ mRNA was purified using Oligo(dT)25 Dynabeads (Life Technologies, Waltham, MA) followed by fragmentation. The first strand cDNA was synthesized using NEBNext RNA First Strand Synthesis Module (New England BioLabs, Ipswich, MA) and purified using Agencourt RNA Clean XP beads (Beckman Coulter, Brea, CA). A dNTP mix containing dUTP was used for the second strand cDNA synthesis, to introduce strand-specificity (NEBNext Ultra Directional RNA Second Strand Synthesis Module, New England BioLabs). The resultant cDNA was processed through end-repair and A-tailing, ligated with Illumina PE adapters, and then digested with Uracil-DNA Glycosylase (New England BioLabs). The libraries were amplified and barcoded (13 PCR cycles) using the Phusion High-Fidelity PCR Master Mix (New England BioLabs). They were purified, pooled in equimolar amounts, and loaded onto a HiSeq flow cell lane. Sequencing runs were performed in paired-end mode (2 × 100-bp reads) following amplification with Illumina’s cBot cluster generation system.
2.4. Transcriptomic analysis workflow
In each condition, pair-end reads obtained from next-generation sequencing were mapped to GRCh38 Human reference genome (http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human/) using Hierarchical Indexing for Spliced Alignment of Transcripts 2 (HISAT2, http://www.ccb.jhu.edu/software/hisat/) to generate a BAM (binary alignment/map) file [26]. Each BAM file was analyzed in normalized-count-based as well as raw-count-based. For the normalized-count-based approach, the RNA BAMs were analyzed using StringTie (http://www.ccb.jhu.edu/software/stringtie/) [27]. The expression of transcript species was reported as Fragments Per Kilobase of transcript per Million mapped reads (FPKMs). For the raw-count-based approach, the RNA BAMs were analyzed using HTSeq (https://htseq.readthedocs.io/en/master/) and EdgeR (http://bioconductor.org/packages/release/bioc/html/edgeR.html) [28, 29]. To assess the quality of the sequencing data, the External RNA Control Consortium (ERCC) spike-in was applied to all samples. Results showed a linear correlation between log2FPKM and Log2Concentration of ERCC in all samples (Supplemental Fig. S1), an indicator of high-quality data and of strong correlation between the FPKM value and the abundance of transcripts in each sample without 3’ bias. The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE137803 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE137803).
The result from the FPKMs and raw counts underwent further analyses to define the differential gene expression between HG and NG cells as a ratio. L5 cells which cultured in high glucose medium was used to confirm the upregulated genes affected by high glucose condition. The common up- or down- regulated genes in HG and L5 cells versus NG cells were analyzed using Molecular Signature Database (MsigDB) [30, 31] for their associated pathways using Hallmarks gene set collection [32].The prioritization of pathways analyses was based on the false discovery rate (FDR)-adjusted q value calculated by the online software of MsigDB, following the instruction available at https://www.gsea-msigdb.org/gsea/msigdb/index.jsp. The most significant pathway was picked as a candidate for further verification according to literature reviews.
2.5. Western blot analysis
Antibodies were used to detect vimentin (Abcam, Cambridge, MA, and Cell Signaling, Danvers, MA), β-actin (Sigma, St. Louis City, MO), MMP2, MMP7, XIAP, cyclin D1, IL-1β, pSTAT3 (Y705), pSTAT3 (S727), STAT3, NF-κB (p65), p100/52, p105/50, pIkkα, Ikkα/β, pIkBα, IkBα (Santa Cruz Biotechnology), pJAK1, JAK1, pJAK2, JAK2 (Cell Signaling), GAPDH (Merck Millipore, Darmstadt, Germany), and IL-6 (Invitrogen, Carlsbad, CA). Antibodies, manufacturers, catalog numbers, and dilution for specific assays are summarized in Supplemental Table S1.
Cells were lysed with NP-40 lysis buffer supplemented with protease and phosphatase inhibitors (Roche, Manheim, Germany). Total proteins were separated by electrophoresis on a 10% sodium dodecyl sulfate-polyacrylamide gel and transferred to a polyvinylidene fluoride (PVDF) membrane (GE Healthcare, Buckinghamshire, UK) as previously described [11]. The membrane was probed with each primary antibody at 4°C overnight and with HRP-conjugated secondary antibody (GE Healthcare) for 1 h at room temperature. The signals were detected using enhance chemiluminescence prime Western blotting detection kit (GE Healthcare). Image analysis was performed using Image Quant™ Imager and the band intensity was quantitated with ImageQuant TL software supplied by the manufacturer (GE Healthcare, Uppasala, Sweden). Three independent cultures were used for the analysis.
2.6. Immunocytofluorescent staining
CCA cells were seeded in a Matrigel (BD Bioscience, Bradford, MA) precoated slide chamber at a density of 104 cells/well and incubated at 37°C, 5% CO2 for 48 h. The cells were fixed as previously described [11] and incubated with 1:100 anti-p65, anti-p100/p52 and anti-p105/p50 (Santa Cruz Biotechnology) at 4°C overnight. After washing, PE-conjugated (Santa Cruz Biotechnology) or Alexa Fluor-568 conjugated (Invitrogen, Eugene, OR) secondary antibody (1:200) and 1:10 000 Hoechst (Invitrogen) were applied at room temperature for an hour. The fluorescent imaging was obtained using a fluorescent microscope (ECLIPSE Ni-U; Nikon, Tokyo, Japan) with Nikon NIS-Elements software. The mean fluorescent intensity of nuclear NF-κB was quantified using Image J Software (National Institute of Health, Bethesda, MD).
2.7. Cell migration and invasion
Migration and invasion assays were performed using Boyden chambers with an 8.0 μm pore size filter (Corning Inc., Corning, NY) as previously described [11]. Briefly, 3 × 104 CCA cells, suspended in serum free media, were seeded into the upper chambers of each well containing complete media as a chemoattractant in the lower chambers. For invasion assay, the cell ability to degrade the extracellular matrix and migrate, the upper chambers were precoated with 40 μg Matrigel (BD Bioscience). Cells were allowed to migrate for 9 h or invade for 24 h. After removing the non-migrating and non-invading cells, the remaining cells were stained with 0.4% sulforhodamine B and counted under light microscope, at a rate of 5 low power fields/samples.
2.8. Statistical analysis
All quantitative data were showed as mean ± SD of a representative from three independent experiments otherwise specified. Statistical analyses were performed using 2-tails Student’s t test, One-way ANOVA, or Two-way ANOVA with Turkey’s multiple comparisons test when appropriate by GraphPad Prism Ver 8.3.1 (GraphPad Software, San Diego, CA) which statistical significance was considered when p < 0.05.
3. Results
3.1. High glucose promotes proliferation of CCA cells
The effect of high glucose on CCA cells proliferation was demonstrated using MTT assay. The proliferation rates of HG cells for both KKU-213A and KKU-213B were significantly higher than those of the counterpart NG cells. The higher growth rates of HG cells were observed as early as day 2 (p < 0.05) and has increased by 3 folds for KKU-213A and by 2 folds for KKU-213B over their NG counterparts by day 5 (Fig. 1A).
Figure 1. Effect of high glucose on CCA cells proliferation and genes expression.
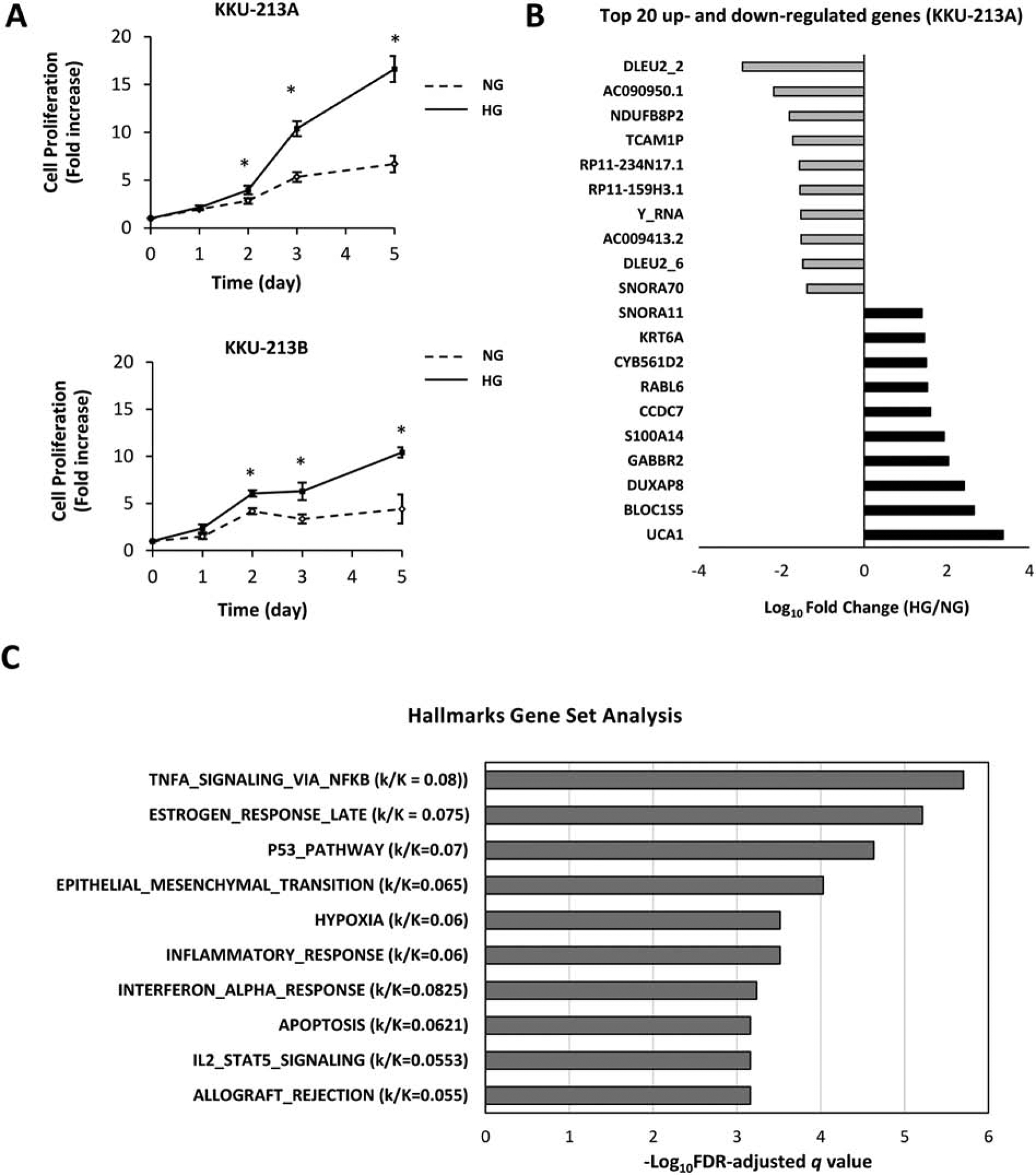
(A) KKU-213A and KKU-213B HG cells show significantly increased proliferation compared with their NG counterparts from day 2 of the culture period (*p<0.05). (B) Top 10 of the most up- and down-regulated genes by high glucose in KKU-213A as revealed by RNA-seq. (C) Pathways analysis using Molecular Signature Database-Hallmark Gene Set, the prioritization of significant pathways was based on false discover rate adjusted-q value. k/K refers to the ratio of genes in the input information (k) those overlapped to genes in genes set of each pathway (K).
3.2. RNA-seq revealed differentially expressed genes between KKU-213A -HG and -L5 and KKU-213A-NG cells,
To identify the genes and pathways underlying the high glucose-enhanced aggressiveness, RNA-seq was performed on KKU-213A NG and HG cells. KKU-213A L5 cells were included to guide the grouping of high glucose induced metastasis-associated genes. Genes with FPKM equal or lower than 2 were filtered out in all samples [33] to ascertain that only translated genes were selected. Among all annotated transcripts, 13,052 genes passed the qualification for futher analysis (Supplemental Table S2). Of these, 1,293 genes were differentially expressed in HG or L5 cells compared to NG cells (Supplemental Table S3). From this amount, 620 genes were unique to L5, possibly representing genes implicated in metastatic process (Supplemental Table S4). The remaining 674 genes were commonly differentially expressed (up or down) in HG and L5; potentially representing genes associated with high glucose condition under which these cell lines were cultured (Supplemental Table S5). The top 10 up- and down-regulated genes by high glucose are shown in Fig. 1B. Analyses using FPKMs and raw counts resulted in the same trend, thus the normalized FPKMs were used for further analysis.
3.3. NF-ĸB pathway is related to progression and is regulated by high glucose condition
The 674 differentially expressed genes in HG and L5 cells were analyzed for their interaction and pathways governing their transcription changes using MSigDB Hallmarks gene set collection. The results suggested several significant pathways under the regulation of high glucose with entities p value less than 0.05. The prioritization of suggested pathways using FDR-adjusted q value entities revealed that the TNFα signaling via NF-κB pathway was the most significant with the highest hit ratio of genes (Fig. 1C and supplemental Table S6). The NF-κB pathway was selected as a candidate pathway for further verification. The unsupervised clustering of the top 60 genes with the highest fold changes (30 up- and 30 down-regulated) confirmed a closed association between HG and L5 differentially expressed genes and a distant correlation with NG gene expression (Fig. 2A). This suggested that these NF-κB pathway-related genes were governed by the effect of glucose condition.
Figure 2. Expression of NF-κB pathway associated genes.
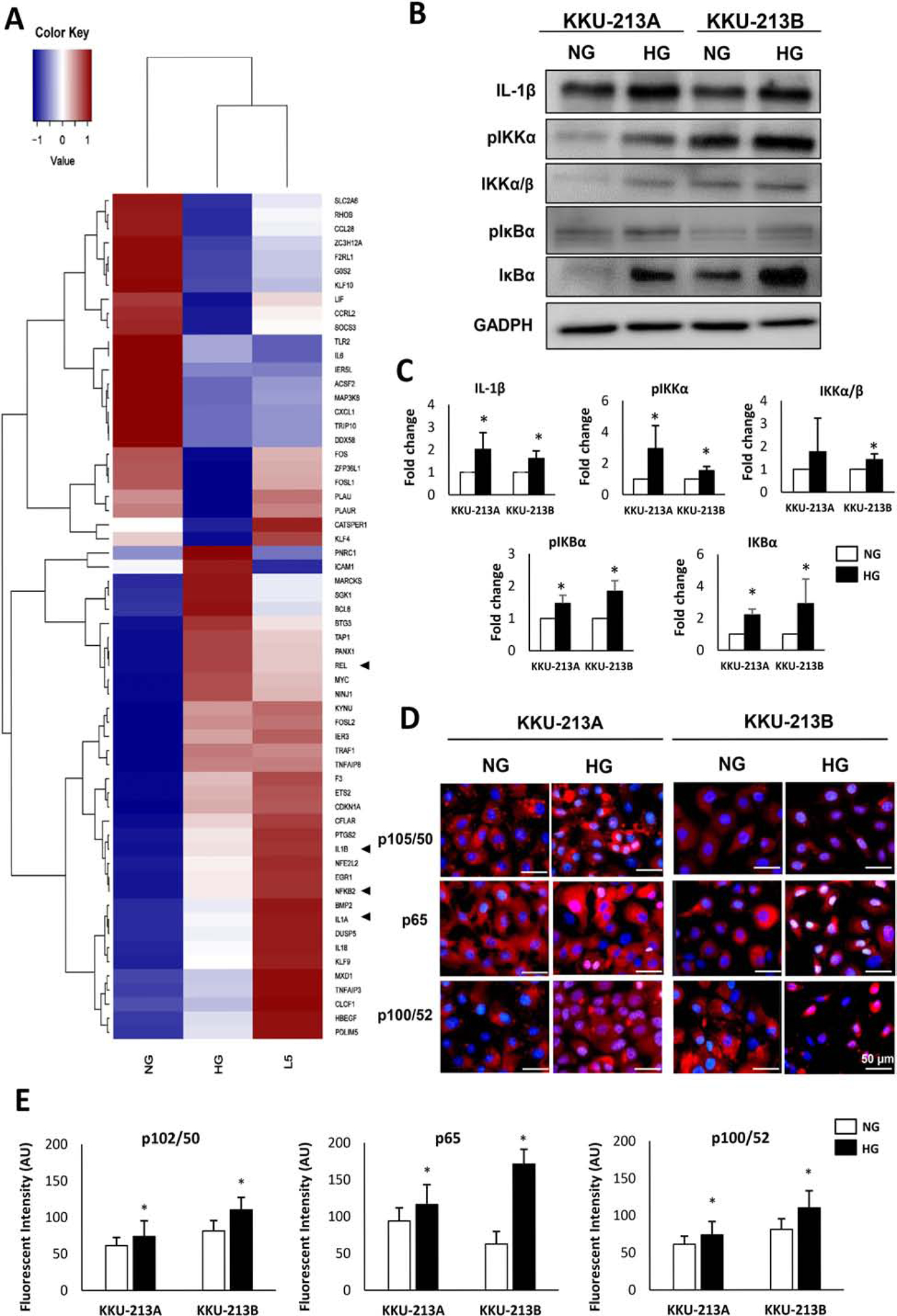
(A) Differential expression of the top 60 genes with the highest fold change associated to NF-κB pathway in L5, HG and NG as detected by RNA-seq. Arrow heads indicate genes encoded for major NF-κB transcription factor subunits (NFKB2: p100/p52, REL: c-Rel) and classical regulatory cytokines (IL1A: IL-1α, IL1B: IL-1β) (B) Western blots analysis of IL-1β, p-IKK/IKK, and p-IκB/IκB. (C) The protein bands are quantitated for their intensity and normalized using GAPDH as a loading control. The averages of relative quantification and statistical analysis are from 3 independent biological replicates. Data are shown by the ratio of targeted proteins/GAPDH and adjusted for NG group as a factor of 1 (*p<0.05). (D) Localization of NF-κB transcription factor subunit p65, P100/p52, and p105/p50 in NG and HG cells detected by immunocytofluorescence. Pink-purple color indicates nuclear localization of NF-κB transcription factors. (E) Statistical analysis of mean fluorescent intensity of nuclear NF-κB transcription factors in KKU-213A and KKU-213B NG and HG cells, data from 3 technical replicates of 2 independent experiments.
3.4. High glucose activates NF-κB signaling in CCA cell lines
The most significant pathways suggested by Hallmarks gene set was TNFα signaling via NF-κB. The expression level of TNFα between HG and NG cells was not different and did not fit in the criteria for gene selection for pathway analysis, but an increase of NF-κB was noted (Fig. 2A). In addition, the classical regulatory cytokine, interleukin 1β (IL-1β), was highly expressed in HG and L5 cells. We further investigated the role of IL-1β and NF-κB in CCA under high glucose condition. Western blotting confirmed the significant upregulation of IL-1β (average of 1.6-fold increase, p < 0.05) as well as those of the phosphorylated forms of IKKα and IκBα (average of 3.0 and 1.5-fold increase, p < 0.05, respectively) in KKU-213A HG cells (Fig. 2B and 2C). Similar results were observed in KKU-213B HG cells suggesting that the effect of high glucose on the activation of NF-κB pathway was not cell line specific (Fig. 2B and 2C).
Since the phosphorylation of IKK is an indicator of the activation of the NF-κB pathway and should result in the nuclear translocation of NF-κB transcription factors, the localizations of p50 and p65 (canonical pathway), and of p52 (non-canonical pathway) were next explored by immunocytofluorescence. As shown in Fig. 2D, the HG cells of KKU-213A showed a higher nuclear localization of p52 consistently with the increase of RNA level (RNA-seq data). On the other hand, HG cells of KKU-213B showed the greater extent of p50 and p65 localization in the nuclei (pink-purple nuclei) compared with the NG counterparts (Fig 2D). The mean fluorescent intensity of nuclear NF-κB in KKU-213A and KKU-213B HG cells was significantly higher than their NG counterparts for transcription factors in both canonical and non-canonical pathways (Fig. 2E).
3.5. NF-κB promotes migration and invasion of CCA cells
The association of NF-κB with the CCA aggressive phenotypes in high glucose condition, namely migration and invasion, was further determined by inhibiting its nuclear translocation with DHMEQ, a well-known inhibitor of NF-κB nuclear translocation in CCA as well as in other cancers [23, 24]. DHMEQ treatment significantly decreased the migration by up to 40% and the invasion by up to 50% in KKU-213A and KKU-213B cells when compared with the control cells treated with vehicle (p < 0.01) (Fig. 3).
Figure 3. Association of nuclear translocation of NF-κB and the metastatic potential of CCA cells.
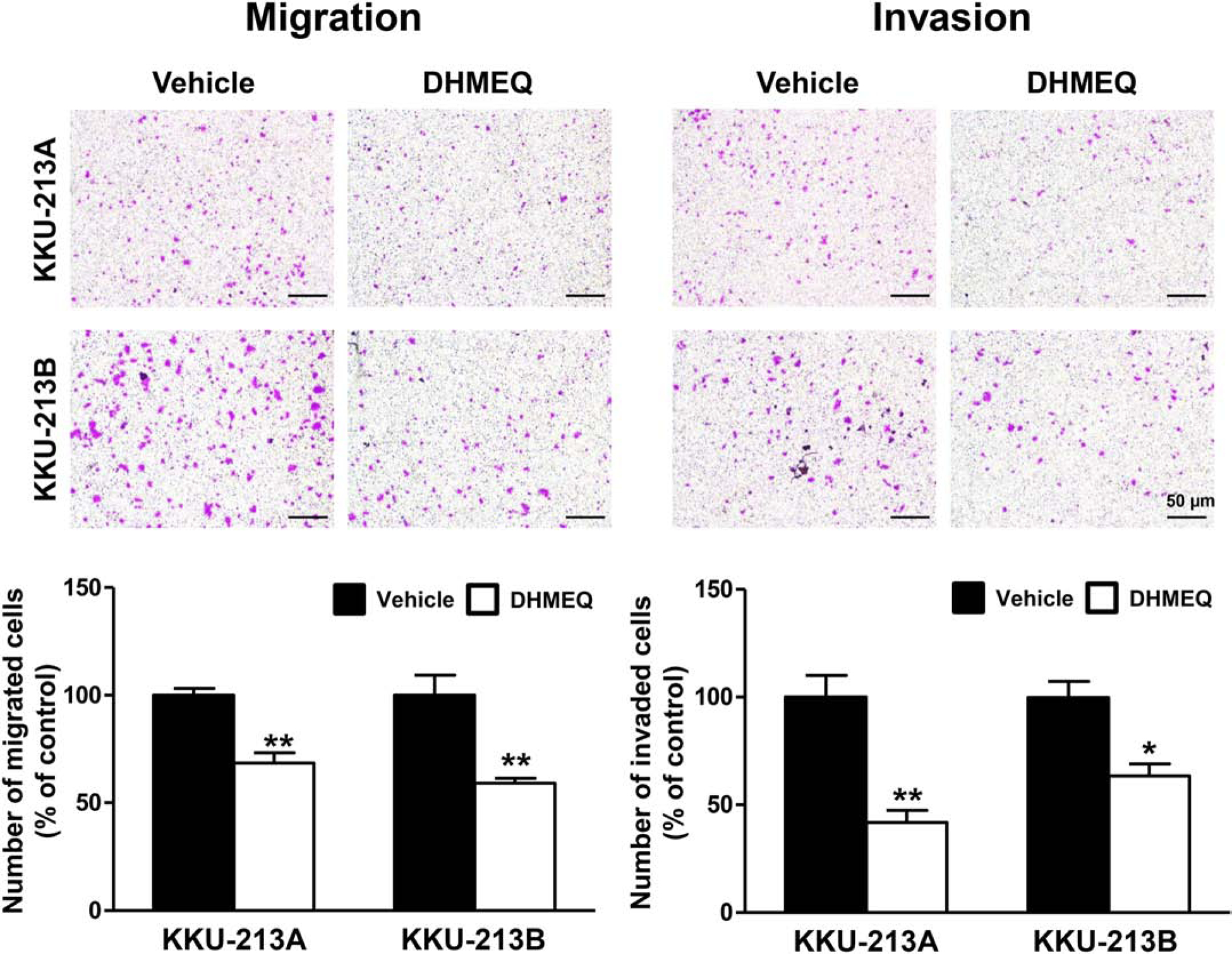
The metastatic potential of CCA cells demonstrated by migration (A) and invasion (B) of CCA cells in high glucose condition was suppressed by the inhibition of NF-κB nuclear translocation by DHMEQ treatment (* p<0.05, ** p<0.01).
To demonstrate the insight mechanism of NF-κB on aggressiveness of CCA cells, the expression of downstream targets of NF-κB related to aggressive phenotypes, i.e., MMP2, MMP7, vimentin, cyclin D1, and XIAP was analyzed. The Western blot revealed that the expression level of all downstream targets of NF-κB were significantly higher in HG than in NG cells (Fig. 4) by up to 3-fold in some cases (p<0.05).
Figure 4. High glucose enhanced the expression of aggressiveness-associated genes via NF-κB and STAT3 pathways.
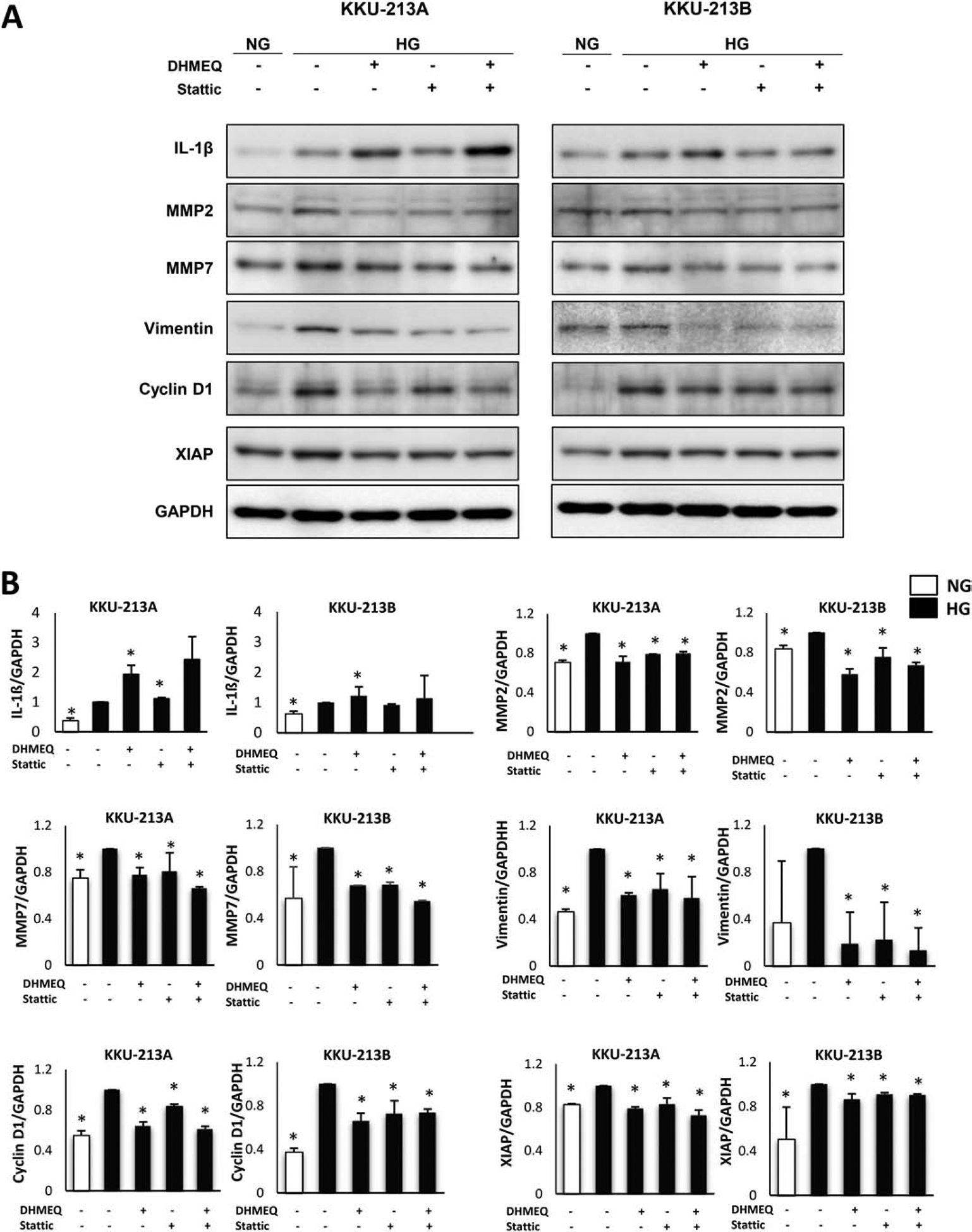
(A) Western blots of aggressiveness-associated genes namely IL-1β, MMP2, MMP7, vimentin, cyclin D1, and XIAP, indicated an upregulated expression in HG cells compared with NG cells. Treatment with DHMEQ, a NF-κB inhibitor, and Sttatic, an STAT3 inhibitor, reversed the expression of those genes in HG cells. However, the combination of these 2 inhibitors did not show any additive effect. (B) The protein band intensity was quantitated and normalized using GAPDH as a loading control. The relative quantification of expression was shown by giving the intensity of HG a factor of 1. The statistical analysis was performed using the average of band intensity from 3 independent experiments and comparing each group to HG (*p<0.05).
To confirm whether the increased expression of the NF-κB downstream targets was the consequence of NF-κB action, the target proteins were studied under DHMEQ treatment. As shown in the Western blots (Fig. 4), the expression of all downstream targets of NF-κB except IL-1β was significantly reduced in the DHMEQ treated cells (p<0.05).
3.6. NF-κB coordinates with STAT3 and contributes to the aggressiveness of CCA under high glucose condition
It is well documented that the genes involved in the aggressive phenotypes of CCA namely IL-1β, MMP2, MMP7, vimentin, cyclin D1, and XIAP, are under the regulation of both NF-κB and STAT3 [34]. Moreover, NF-κB and STAT3 also play a key role in the regulation of numerous cell functions in many physiological processes [34]. Since the STAT3 pathway was previously shown to be activated and to contribute to the high glucose induced aggressiveness of CCA cells [11], we explored if the activated STAT3 also contributed to the upregulation of those common genes under NF-κB regulation which corresponded to aggressiveness of CCA in high glucose. Stattic, an inhibitor of nuclear translocation of STAT3, was used to inhibit the activation of STAT3. The expression level of STAT3 downstream targets associated with an aggressive phenotype was determined using Western blotting. As shown in Fig. 4, the expression of all tested proteins under the control of NF-κB (at the exception of IL1-β in KKU-213A cells) was also down regulated (p<0.05).
3.7. NF-κB regulates JAK-STAT signaling via the induction of IL-6 expression
Since a STAT3 inhibitor could decrease the expression levels of several NF-κB depended gene-products, we questioned whether STAT3 and NF-κB could co-exert their actions under high glucose condition. The crosstalk between NF-κB and STAT3 plays a key role in the regulation of numerous functions of cell in many physiological processes [34]. To determine the interaction of NF-κB with STAT3 pathway under high glucose condition, the activation of STAT3, pSTAT3 (S727) and pSTAT3 (Y705) was investigated in CCA cells treated with DHMEQ and Stattic. As shown in Figs. 5A and 5B, STAT3 activation at both pSTAT3 (S727) and pSTAT3 (Y705) was higher in HG cells than in NG cells by an average of 1.2 and 2.3-fold (KKU-213A) and 2.7 and 3.3-fold (KKU-213B) (p<0.05). DHMEQ or Stattic treatment reduced the activation of pSTAT3 (Y705) by up to 50% in KKU-213A and 40% in KKU-213B (p<0.05) but not pSTAT3 (S727). The result indicated the positive regulation of NF-κB on STAT3 activation.
Figure 5. NF-κB pathway interacted with STAT3 pathway via IL-6.
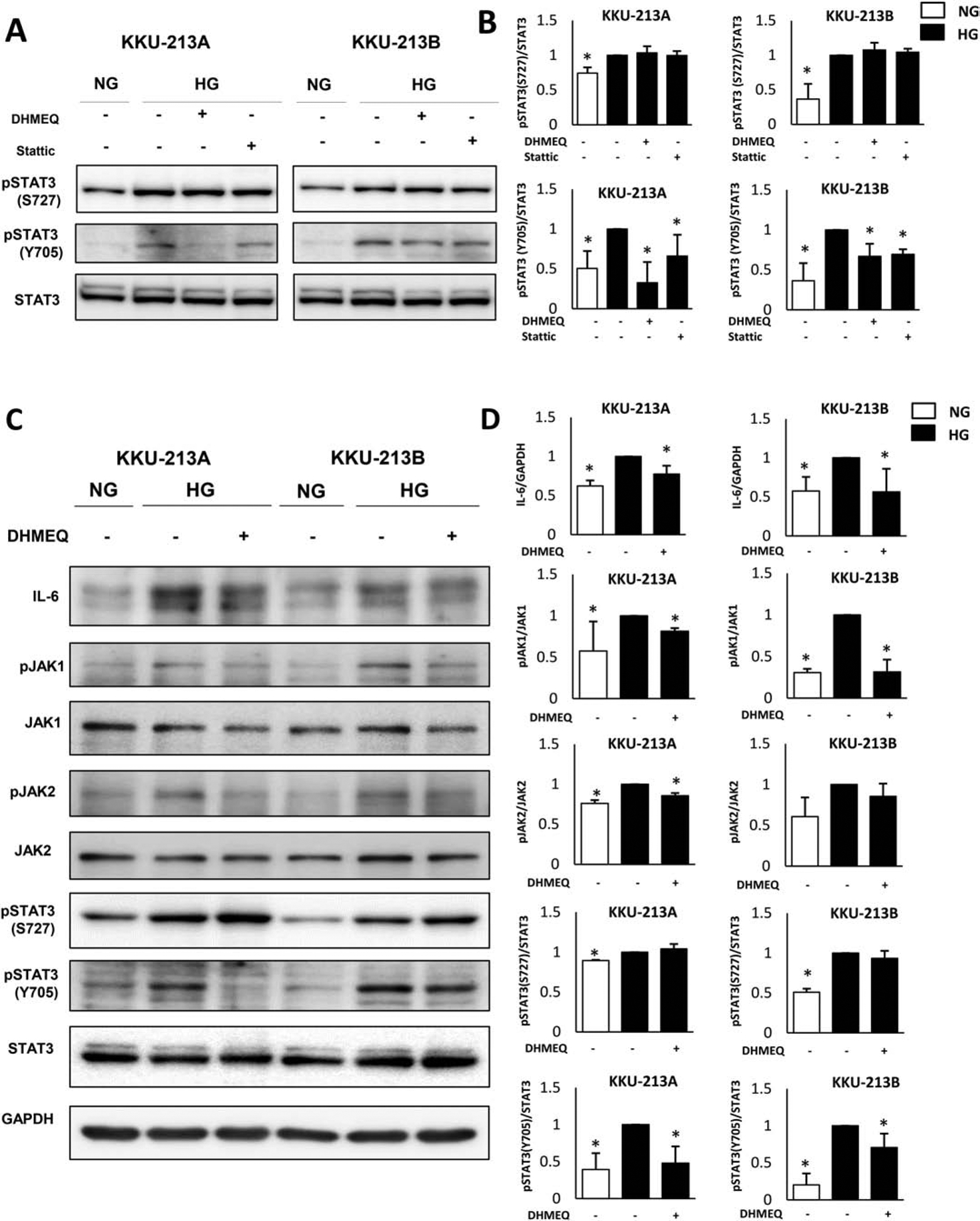
Western blots of signaling molecules in STAT3 pathway in KKU-213A and KKU-213B demonstrated that (A) The phosphorylation of STAT3 was higher in HG than NG cells and was affected by DHMEQ, a specific inhibitor of NF- κB, treatments suggesting an interaction between these two pathways. (B and D) The protein band intensity of p-STAT3 was normalized using total STAT3 as a control. The relative quantification of pSTAT3/STAT3 ratio was shown by giving the intensity of HG a factor of 1 and statistical analysis was performed and presented using the averages of band intensity from 3 independent experiments (biological replicates) (*p<0.05). (C) IL-6 expression was higher in HG than NG cells. Inhibition of NF-κB by DHMEQ reduced the expression level of IL-6 up to 25% in KKU-213A and 15% in KKU-213B and decreased the activation of STAT3 signaling cascade, pJAK1/JAK1, pJAK2/JAK2 and pSTAT3 (Y705).
The activation of STAT3 signaling under NF-κB action was further explored. Since IL-6, a cytokine under the regulation of NF-κB, is the upstream signaling of JAK1/2-STAT3 pathway, we suspected IL-6 to be a linkage between these two pathways. The expression level of IL-6, pJAK1/JAK1, pJAK2/JAK2 and pSTAT3 (S727) and pSTAT3 (Y705) in NG, HG and DHMEQ-treated HG cells were determined by Western blot. As shown in Figs. 5C and 5D, the expression of IL-6 and the entire STAT3 signaling cascade were significantly higher in HG than NG cells (p<0.05). This resulted in the activation of JAK1/2-STAT3 pathway as demonstrated by the increased level of their phosphorylated forms. The inhibition of NF-κB pathway by DHMEQ significantly decreased the expression of IL-6 which accordingly reduced the activation of JAK1/2-STAT3 pathway (p<0.05). This suggests that IL-6 is a signaling cytokine in the crosstalk of these two pathways.
4. Discussion
The correlation between diabetes and cancer reported in several epidemiological studies have drawn attention toward high glucose condition in the progression of cancer in addition to other diabetic condition [3, 4, 35]. In this study, we used a global transcriptomic approach to identify the signal pathways involved in the development of such aggressive phenotypes and we validated our results at the protein levels.
In our studies, glucose of 25 mM (450 mg/dL) was used to mimic the hyperglycemia in CCA patients with DM as: 1) this concentration exerted the optimal effect of high glucose on phenotypic and molecular alterations in the in vitro studies of several cancers [2, 11, 36]; 2) no confounding effect on the osmolarity of the culture media was observed [36, 37]; and 3) cultured CCA cells in 25 mM glucose significantly promoted CCA cells progression [11, 38]. Although the average of fasting plasma glucose (FPG) in Thai population with DM is approximately 10.5 mM (190 mg/dL), a substantial group of them had FPG more than 10.5 mM [39]. Consistently, the in vivo study of hyperglycemic effect on breast cancer indicated that the average glucose levels in fed-state type 2 diabetic mice was 22.2 mM (400 mg/dL) which is comparable to most of in vitro studies [40]. We previously showed that CCA cells under high glucose condition of 25 mM exhibited aggressive phenotypes [11, 38].
We found that one of the most significant pathways underlying high glucose enhanced CCA progression relied on NF-κB activation and that NF-κB and STAT3 exerted a coordinated action to sustain CCA aggressiveness. NF-κB plays an essential role in carcinogenesis and CCA progression [24, 25] and the common pattern of gene expression shared between our aggressive CCA cell lines further reinforced its substantial role in CCA aggressiveness. Moreover, the implication of NF-κB was supported by the increased transcription of NFKB2 (p100/p52) and IL-1β, an upstream activator [41] and downstream target of NF-κB [42, 43] which up-regulated expression was also confirmed at the protein level.
The classical NF-κB activation cascade is initiated by IL-1β induction of IKK phosphorylation followed by IκBα phosphorylation. The phosphorylation results in the dissociation of IκBα from the NF-κB transcription factors (p50, p65) which now free, migrate to the nucleus and activate the expression of several downstream genes meanwhile IκBα phosphorylated proteins are degraded [44]. In the present study, the pIKKα and pIκBα were increased in parallel with IL-1β protein level in the HG cell lines and the activation of the pathway was confirmed by NF-κB immunocytofluorescent staining as well as its nuclear localization. The highly aggressive KKU-213A HG cells exhibited high expression of NFKB2 genes and correspondingly high activation of p100/52, indicating the activation of non-canonical (alternative) NF-κB pathway. In contrast, the slightly less aggressive KKU-213B HG cells had high activation of p65 of the canonical (classical) pathway.
The nuclear translocation of the transcription factors corresponded to an increase in the protein expression level of several downstream targeted genes associated with aggressiveness (MMP2, MMP7, vimentin, XIAP, and cyclin D1) in both HG cells. Blocking the nuclear translocation of the NF-κB by DHMEQ, a synthesized derivative of epoxyquinomicin previously shown with a high potency in CCA cells [24, 25], resulted in the suppression of migration and invasion abilities of CCA cells as well as the decreased expression of proteins MMP2, MMP7, vimentin, XIAP, and cyclin D1 in HG cell lines.
Our previous study showed that STAT3 was activated in CCA cells under high glucose condition and in the tumor of CCA patients with high glucose in the plasma [11] and that the activation of STAT3 was associated with the aggressiveness of HG cells. Moreover, it is well documented that the targeted genes of NF-κB are also under the regulation of STAT3 [34]. The present study indicated that both NF-κB and STAT3 may orchestrate the intracellular signal transduction to promote the aggressiveness of CCA cells under high glucose condition: the inhibition of NF-κB as well as the inhibition of STAT3 activation and translocation similarly suppressed the expression of NF-κB downstream target genes. Since no cumulative effect could be noticed when both inhibitions were combined and since the inhibition of NF-κB also suppressed the activation of pSTAT3 (Y705), we hypothesized and confirmed that NF-κB exerts its action on STAT3 via IL-6, a key cytokine under the regulation of NF-κB and a potent upstream signaling of JAK1/2-STAT3 pathway through the following chain of events: the upregulated IL-6 expression in HG cells was suppressed with NF-κB inhibition; the decrease of IL-6 expression lowered the level of phosphorylated JAK1/2, the upstream kinase of STAT3 phosphorylation; and the phosphorylation of STAT3 at Y705 was substantially suppressed. We illustrated the coordinated actions of NF-κB and STAT3 in Fig. 6. Supporting such hypothesis, is the higher level of plasma IL-6 in CCA patients (221 times than in normal controls) previously reported by Sripa et al. [45]. Moreover, significantly higher IL-6 levels were also reported in sera from patients with type 2 DM compared to normal subjects [46]. In the present study, IL-6 mRNA level was lower in HG than in NG cells, in contrast to the protein levels. This suggests that high glucose might be involved in the protein stability rather than a direct interaction at the transcriptional level. The discrepancy between IL-6 mRNA and protein levels was also observed in streptozotocin-induced diabetic mice [47]. Compared with euglycemic mice, IL-6 mRNA levels showed a decrease tendency the first day after wounding in hyperglycemic mice, followed by unnoticeable difference between the two groups. The levels of IL-6 protein, however, were increased and remained significantly higher in hyperglycemic mice until the end of the study.
Figure 6. Schematic summary of high glucose activated NF-κB pathway.
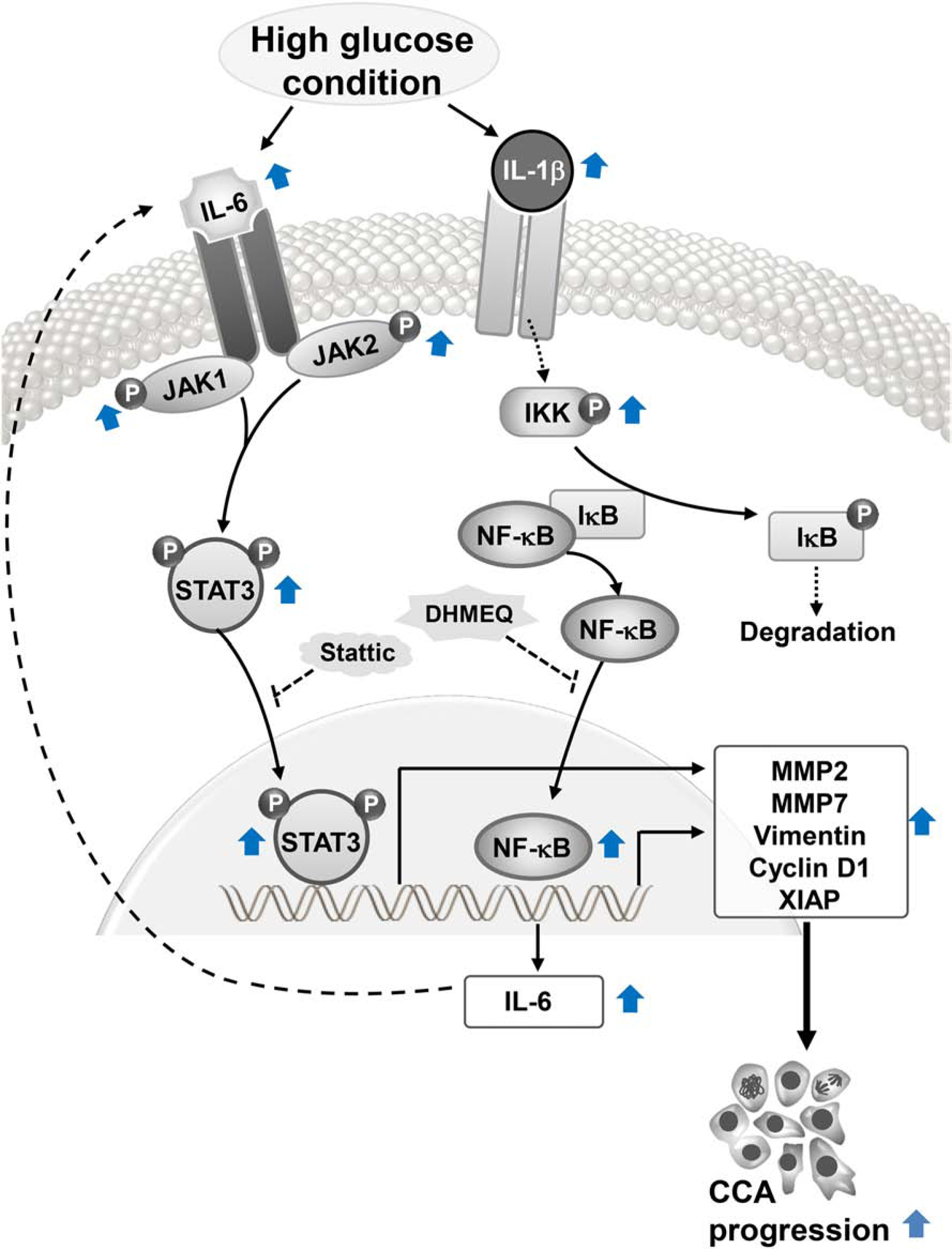
High glucose induces the expression of IL-1β, the upstream activator of NF-κB pathway and, hence, increases p-IKK/IKK in CCA cells. The activated IKK phosphorylates IκB resulting in the release and translocation of NF-κB transcription factor to the nucleus. The NF-κB activation results in the transcription of its downstream target including IL-1β which acts as positive feedback for the pathway stimulation and IL-6 which act as a signaling cytokine in a crosstalk with STAT3 pathway. Both NF-κB and STAT3 orchestrate and contribute to the effect of high glucose-induced aggressiveness of CCA. (Dash lines represent the mechanisms proven in other reports.)
IL-1β is a pro-inflammatory cytokine that cooperates with various cytokines and several cellular mediators to mediate a variety of cellular activities, such as cell proliferation, differentiation and apoptosis. High glucose induced IL-1β expression was observed in many cell types, e.g., bovine retinal endothelial cells [48], human macrophages [49], human pancreatic islets [50] and aortic endothelial cells [51]. The present study is the first to demonstrate the increase of IL-1β expression in CCA cells under high glucose. IL-1β plays important roles in cancer and cancer environment: polymorphisms in the susceptibility for breast cancer development [52]; direct contribution of IL-1β to the pathogenesis of human and mouse colon cancer cell lines by increasing pSTAT3 and decreasing pAMPK expression [53]. High glucose condition also favored a pro-inflammatory environment that stimulated the epithelial migration by stimulating IL-1β epithelial expression [54].
IL-1β occupies a dual position in the NF-κB pathway as an upstream activator as well as a downstream target. In the present study, expression of IL-1β was increased under NF-κB inhibition, in contrast with the decreased expression of the NF-κB downstream gene targets. In fact, IL-1β expression is regulated by the NF-κB and STAT3 pathways [42, 43]. However, in the present model, expression of IL-1β was increased by either NF-κB or STAT3 inhibitors, suggesting some uncovered mechanisms that counteract with NF-κB and STAT3 in regulating IL-1β expression. Noteworthy, higher level of IL-1β was observed in HG cells and was increased after treated with NF-κB and STAT3 inhibitors. This observation suggests the clinical application of using IL-1β inhibitor in CCA treatment, especially in patients with DM. Further studies are needed to verify the efficiency of the combination of these inhibitors on CCA cells.
The effect of high glucose on promoting NF-κB signaling was reported in several diabetic complication models, such as diabetic cardiomyopathy [55] and nephropathy [56]. High glucose also increases the expression of RelA (p65) mRNA by promoting epigenetic change and causing hyperglycemic memory in endothelial cells [57]. By contributing to the inflammatory process, NF-κB plays several roles and interacts with other inflammatory signaling pathways especially STAT3 [58, 59]. The association of NF-κB and STAT3 was demonstrated many times and considered a dangerous liaison [34]. The effect of high glucose on NF-κB was demonstrated in many diabetic complications, but we report for the first time the induction of NF-κB signaling by high glucose in cancer cells. The study suggests that NF-κB and STAT3 coordinate to promote progression of CCA. Metformin, an anti-diabetic drug, exhibiting the inhibitory effect on the nuclear translocation of both STAT3 and NF-κB could suppress the proliferation and metastatic potential of CCA cells in high glucose condition [20]. This reinforces the significance of both pathways in CCA and the possibility of a novel therapeutic approach.
This study, nevertheless, has some limitations. Firstly, CCA cells used in the experiments were derived from liver fluke-associated patients. As the genetic backgrounds and gene expression profiles of the fluke- and non-fluke associated CCA are different [60, 61], our results should be verified in another CCA subgroup. Secondly, besides hyperglycemia, other factors related with DM, e.g., hyperinsulinemia, insulin-like growth factor, ketone bodies, and proinflammatory cytokines [1] may also act as tumor promoting factors. Lastly, the in vitro findings shown in the current study should be validated in an animal model. The functional roles and regulatory mechanisms of IL-1β under high glucose observed in this study should be further clarified.
5. Conclusion
In summary, RNA-seq screening revealed the global differential expression of genes that may govern several signaling pathways in CCA cell lines cultured in high glucose condition. One of these pathways, NF-κB, was activated via the upregulation of IL-1β/IKK cascade and coordinated with the STAT3 pathway via the expression of a key cytokine, IL-6, to enhance the aggressiveness of CCA cells. The crosstalk of NF-κB/STAT3 pathway opens an opportunity for the intervention of new therapeutic strategy. Targeting both pathways in hyperglycemic CCA patients should be considered for a more effective outcome.
Supplementary Material
Acknowledgments:
C. Saengboonmee and S. Supabphol are scholars of Prince Mahidol Award Youth Program of Prince Mahidol Award Foundation under the Royal Patronage of HM the king of Thailand. This work was co-supported by research grants from Khon Kaen University for W. Seubwai (601802) and S. Wongkham (I62-01-02) and from NIHGRI (U54 HG003273) for the Human Genome Sequencing Center-Baylor College of Medicine (R.A. Gibbs and MC. Gingras). The funding sources have no role in study design; collection, analysis and interpretation of data; writing of the report; and in the decision to submit the article for publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest: The authors declare that there are no conflicts of interest.
References
- [1].Vigneri P, Frasca F, Sciacca L, Pandini G, Vigneri R, Diabetes and cancer, Endocr Relat Cancer. 16 (2009) 1103–1123. 10.1677/erc-09-0087. [DOI] [PubMed] [Google Scholar]
- [2].Chocarro-Calvo A, Garcia-Martinez JM, Ardila-Gonzalez S, De la Vieja A, Garcia-Jimenez C, Glucose-induced beta-catenin acetylation enhances Wnt signaling in cancer, Mol Cell. 49 (2013) 474–486. 10.1016/j.molcel.2012.11.022. [DOI] [PubMed] [Google Scholar]
- [3].Garcia-Jimenez C, Garcia-Martinez JM, Chocarro-Calvo A, De la Vieja A, A new link between diabetes and cancer: enhanced WNT/beta-catenin signaling by high glucose, J Mol Endocrinol. 52 (2014) R51–66. 10.1530/jme-13-0152. [DOI] [PubMed] [Google Scholar]
- [4].Ryu TY, Park J, Scherer PE, Hyperglycemia as a risk factor for cancer progression, Diabetes Metab J. 38 (2014) 330–336. 10.4093/dmj.2014.38.5.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Gupta C, Tikoo K, High glucose and insulin differentially modulates proliferation in MCF-7 and MDA-MB-231 cells, J Mol Endocrinol. 51 (2013) 119–129. 10.1530/jme-13-0062. [DOI] [PubMed] [Google Scholar]
- [6].Lopez R, Arumugam A, Joseph R, Monga K, Boopalan T, Agullo P, et al. , Hyperglycemia enhances the proliferation of non-tumorigenic and malignant mammary epithelial cells through increased leptin/IGF1R signaling and activation of AKT/mTOR, PLoS One. 8 (2013) e79708 10.1371/journal.pone.0079708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Li W, Ma Q, Li J, Guo K, Liu H, Han L, et al. , Hyperglycemia enhances the invasive and migratory activity of pancreatic cancer cells via hydrogen peroxide, Oncol Rep. 25 (2011) 1279–1287. 10.3892/or.2011.1150. [DOI] [PubMed] [Google Scholar]
- [8].Liu H, Ma Q, Li J, High glucose promotes cell proliferation and enhances GDNF and RET expression in pancreatic cancer cells, Mol Cell Biochem. 347 (2011) 95–101. 10.1007/s11010-010-0617-0. [DOI] [PubMed] [Google Scholar]
- [9].Ma YS, Yang IP, Tsai HL, Huang CW, Juo SH, Wang JY, High glucose modulates antiproliferative effect and cytotoxicity of 5-fluorouracil in human colon cancer cells, DNA Cell Biol. 33 (2014) 64–72. 10.1089/dna.2013.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hosokawa T, Kurosaki M, Tsuchiya K, Matsuda S, Muraoka M, Suzuki Y, et al. , Hyperglycemia is a significant prognostic factor of hepatocellular carcinoma after curative therapy, World J Gastroenterol. 19 (2013) 249–257. 10.3748/wjg.v19.i2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Saengboonmee C, Seubwai W, Pairojkul C, Wongkham S, High glucose enhances progression of cholangiocarcinoma cells via STAT3 activation, Sci Rep. 6 (2016) 18995 10.1038/srep18995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Florio AA, Ferlay J, Znaor A, Ruggieri D, Alvarez CS, Laversanne M, et al. , Global trends in intrahepatic and extrahepatic cholangiocarcinoma incidence from 1993 to 2012, Cancer. 126 (2020) 2666–2678. 10.1002/cncr.32803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Vatanasapt V, Tangvoraphonkchai V, Titapant V, Pipitgool V, Viriyapap D, Sriamporn S, A high incidence of liver cancer in Khon Kaen Province, Thailand, Southeast Asian J Trop Med Public Health. 21 (1990) 489–494. [PubMed] [Google Scholar]
- [14].Sripa B, Pairojkul C, Cholangiocarcinoma: lessons from Thailand, Curr Opin Gastroenterol. 24 (2008) 349–356. 10.1097/MOG.0b013e3282fbf9b3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ben-Menachem T, Risk factors for cholangiocarcinoma, Eur J Gastroenterol Hepatol. 19 (2007) 615–617. 10.1097/MEG.0b013e328224b935. [DOI] [PubMed] [Google Scholar]
- [16].Faramnuayphol P, Chongsuvivatwong V, Pannarunothai S, Geographical variation of mortality in Thailand, J Med Assoc Thai. 91 (2008) 1455–1460. [PubMed] [Google Scholar]
- [17].Aungkulanon S, Tangcharoensathien V, Shibuya K, Bundhamcharoen K, Chongsuvivatwong V, Post universal health coverage trend and geographical inequalities of mortality in Thailand, Int J Equity Health. 15 (2016) 190 10.1186/s12939-016-0479-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Saengboonmee C, Seubwai W, Wongkham C, Wongkham S, Diabetes mellitus: Possible risk and promoting factors of cholangiocarcinoma: Association of diabetes mellitus and cholangiocarcinoma, Cancer Epidemiol. 39 (2015) 274–278. 10.1016/j.canep.2015.04.002. [DOI] [PubMed] [Google Scholar]
- [19].Yokota J, Tumor progression and metastasis, Carcinogenesis. 21 (2000) 497–503. 10.1093/carcin/21.3.497. [DOI] [PubMed] [Google Scholar]
- [20].Saengboonmee C, Seubwai W, Cha’on U, Sawanyawisuth K, Wongkham S, Wongkham C, Metformin Exerts Antiproliferative and Anti-metastatic Effects Against Cholangiocarcinoma Cells by Targeting STAT3 and NF-kB, Anticancer Res. 37 (2017) 115–123. 10.21873/anticanres.11296. [DOI] [PubMed] [Google Scholar]
- [21].Sripa B, Seubwai W, Vaeteewoottacharn K, Sawanyawisuth K, Silsirivanit A, Kaewkong W, et al. , Functional and genetic characterization of three cell lines derived from a single tumor of an Opisthorchis viverrini-associated cholangiocarcinoma patient, Hum Cell. 10.1007/s13577-020-00334-w (2020). 10.1007/s13577-020-00334-w. [DOI] [PubMed] [Google Scholar]
- [22].Uthaisar K, Vaeteewoottacharn K, Seubwai W, Talabnin C, Sawanyawisuth K, Obchoei S, et al. , Establishment and characterization of a novel human cholangiocarcinoma cell line with high metastatic activity, Oncol Rep. 36 (2016) 1435–1446. 10.3892/or.2016.4974. [DOI] [PubMed] [Google Scholar]
- [23].Matsumoto N, Ariga A, To-e S, Nakamura H, Agata N, Hirano S, et al. , Synthesis of NF-kappaB activation inhibitors derived from epoxyquinomicin C, Bioorg Med Chem Lett. 10 (2000) 865–869. 10.1016/s0960-894x(00)00114-1. [DOI] [PubMed] [Google Scholar]
- [24].Seubwai W, Wongkham C, Puapairoj A, Khuntikeo N, Pugkhem A, Hahnvajanawong C, et al. , Aberrant expression of NF-kappaB in liver fluke associated cholangiocarcinoma: implications for targeted therapy, PLoS One. 9 (2014) e106056 10.1371/journal.pone.0106056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Seubwai W, Vaeteewoottacharn K, Kraiklang R, Umezawa K, Okada S, Wongkham S, Inhibition of NF-kappaB Activity Enhances Sensitivity to Anticancer Drugs in Cholangiocarcinoma Cells, Oncol Res. 23 (2016) 21–28. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kim D, Langmead B, Salzberg SL, HISAT: a fast spliced aligner with low memory requirements, Nat Methods. 12 (2015) 357–360. 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL, StringTie enables improved reconstruction of a transcriptome from RNA-seq reads, Nat Biotechnol. 33 (2015) 290–295. 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Anders S, Pyl PT, Huber W, HTSeq--a Python framework to work with high-throughput sequencing data, Bioinformatics. 31 (2015) 166–169. 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Robinson MD, McCarthy DJ, Smyth GK, edgeR: a Bioconductor package for differential expression analysis of digital gene expression data, Bioinformatics. 26 (2010) 139–140. 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. , Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles, Proceedings of the National Academy of Sciences. 102 (2005) 15545–15550. 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP, Molecular signatures database (MSigDB) 3.0, Bioinformatics. 27 (2011) 1739–1740. 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P, The Molecular Signatures Database (MSigDB) hallmark gene set collection, Cell systems. 1 (2015) 417–425. 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Gonzàlez-Porta M, Frankish A, Rung J, Harrow J, Brazma A, Transcriptome analysis of human tissues and cell lines reveals one dominant transcript per gene, Genome Biol. 14 (2013) R70 10.1186/gb-2013-14-7-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Grivennikov SI, Karin M, Dangerous liaisons: STAT3 and NF-kappaB collaboration and crosstalk in cancer, Cytokine Growth Factor Rev. 21 (2010) 11–19. 10.1016/j.cytogfr.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Duan W, Shen X, Lei J, Xu Q, Yu Y, Li R, et al. , Hyperglycemia, a neglected factor during cancer progression, Biomed Res Int. 2014 (2014) 461917 10.1155/2014/461917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Yamamoto M, Patel NA, Taggart J, Sridhar R, Cooper DR, A shift from normal to high glucose levels stimulates cell proliferation in drug sensitive MCF-7 human breast cancer cells but not in multidrug resistant MCF-7/ADR cells which overproduce PKC-betaII, Int J Cancer. 83 (1999) 98–106. . [DOI] [PubMed] [Google Scholar]
- [37].Lin CY, Lee CH, Huang CC, Lee ST, Guo HR, Su SB, Impact of high glucose on metastasis of colon cancer cells, World J Gastroenterol. 21 (2015) 2047–2057. 10.3748/wjg.v21.i7.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Phoomak C, Vaeteewoottacharn K, Silsirivanit A, Saengboonmee C, Seubwai W, Sawanyawisuth K, et al. , High glucose levels boost the aggressiveness of highly metastatic cholangiocarcinoma cells via O-GlcNAcylation, Sci Rep. 7 (2017) 43842 10.1038/srep43842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Aekplakorn W, Stolk RP, Neal B, Suriyawongpaisal P, Chongsuvivatwong V, Cheepudomwit S, et al. , The Prevalence and Management of Diabetes in Thai Adults, The International Collaborative Study of Cardiovascular Disease in Asia. 26 (2003) 2758–2763. 10.2337/diacare.26.10.2758. [DOI] [PubMed] [Google Scholar]
- [40].Fainsod-Levi T, Gershkovitz M, Völs S, Kumar S, Khawaled S, Sagiv JY, et al. , Hyperglycemia Impairs Neutrophil Mobilization Leading to Enhanced Metastatic Seeding, Cell Rep. 21 (2017) 2384–2392. 10.1016/j.celrep.2017.11.010. [DOI] [PubMed] [Google Scholar]
- [41].Weber A, Wasiliew P, Kracht M, Interleukin-1 (IL-1) pathway, Sci Signal. 3 (2010) cm1. 10.1126/scisignal.3105cm1. [DOI] [PubMed] [Google Scholar]
- [42].Hiscott J, Marois J, Garoufalis J, D’Addario M, Roulston A, Kwan I, et al. , Characterization of a functional NF-kappa B site in the human interleukin 1 beta promoter: evidence for a positive autoregulatory loop, Mol Cell Biol. 13 (1993) 6231–6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Samavati L, Rastogi R, Du W, Huttemann M, Fite A, Franchi L, STAT3 tyrosine phosphorylation is critical for interleukin 1 beta and interleukin-6 production in response to lipopolysaccharide and live bacteria, Mol Immunol. 46 (2009) 1867–1877. 10.1016/j.molimm.2009.02.018. [DOI] [PubMed] [Google Scholar]
- [44].Hoesel B, Schmid JA, The complexity of NF-kappaB signaling in inflammation and cancer, Mol Cancer. 12 (2013) 86 10.1186/1476-4598-12-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Sripa B, Thinkhamrop B, Mairiang E, Laha T, Kaewkes S, Sithithaworn P, et al. , Elevated plasma IL-6 associates with increased risk of advanced fibrosis and cholangiocarcinoma in individuals infected by Opisthorchis viverrini, PLoS Negl Trop Dis. 6 (2012) e1654 10.1371/journal.pntd.0001654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Menegazzo L, Ciciliot S, Poncina N, Mazzucato M, Persano M, Bonora B, et al. , NETosis is induced by high glucose and associated with type 2 diabetes, Acta Diabetol. 52 (2015) 497–503. 10.1007/s00592-014-0676-x. [DOI] [PubMed] [Google Scholar]
- [47].Lee EG, Luckett-Chastain LR, Calhoun KN, Frempah B, Bastian A, Gallucci RM, Interleukin 6 Function in the Skin and Isolated Keratinocytes Is Modulated by Hyperglycemia, J Immunol Res. 2019 (2019) 5087847 10.1155/2019/5087847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kowluru RA, Odenbach S, Role of interleukin-1beta in the pathogenesis of diabetic retinopathy, Br J Ophthalmol. 88 (2004) 1343–1347. 10.1136/bjo.2003.038133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Shashkin PN, Jain N, Miller YI, Rissing BA, Huo Y, Keller SR, et al. , Insulin and glucose play a role in foam cell formation and function, Cardiovasc Diabetol. 5 (2006) 13 10.1186/1475-2840-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Maedler K, Sergeev P, Ris F, Oberholzer J, Joller-Jemelka HI, Spinas GA, et al. , Glucose-induced beta cell production of IL-1beta contributes to glucotoxicity in human pancreatic islets, J Clin Invest. 110 (2002) 851–860. 10.1172/jci15318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Asakawa H, Miyagawa J, Hanafusa T, Kuwajima M, Matsuzawa Y, High glucose and hyperosmolarity increase secretion of interleukin-1 beta in cultured human aortic endothelial cells, J Diabetes Complications. 11 (1997) 176–179. [DOI] [PubMed] [Google Scholar]
- [52].Lee KM, Park SK, Hamajima N, Tajima K, Choi JY, Noh DY, et al. , Genetic polymorphisms of interleukin-1 beta (IL-1B) and IL-1 receptor antagonist (IL-1RN) and breast cancer risk in Korean women, Breast Cancer Res Treat. 96 (2006) 197–202. 10.1007/s10549-005-9079-6. [DOI] [PubMed] [Google Scholar]
- [53].Moon HS, Mantzoros CS, Adiponectin and metformin additively attenuate IL1beta-induced malignant potential of colon cancer, Endocr Relat Cancer. 20 (2013) 849–859. 10.1530/erc-13-0240. [DOI] [PubMed] [Google Scholar]
- [54].Kallens V, Tobar N, Molina J, Bidegain A, Smith PC, Porras O, et al. , Glucose Promotes a Pro-Oxidant and Pro-Inflammatory Stromal Microenvironment Which Favors Motile Properties in Breast Tumor Cells, J Cell Biochem. 118 (2017) 994–1002. 10.1002/jcb.25650. [DOI] [PubMed] [Google Scholar]
- [55].Min W, Bin ZW, Quan ZB, Hui ZJ, Sheng FG, The signal transduction pathway of PKC/NF-kappa B/c-fos may be involved in the influence of high glucose on the cardiomyocytes of neonatal rats, Cardiovasc Diabetol. 8 (2009) 8 10.1186/1475-2840-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Wei M, Li Z, Xiao L, Yang Z, Effects of ROS-relative NF-kappaB signaling on high glucose-induced TLR4 and MCP-1 expression in podocyte injury, Mol Immunol. 68 (2015) 261–271. 10.1016/j.molimm.2015.09.002. [DOI] [PubMed] [Google Scholar]
- [57].Siebel AL, Fernandez AZ, El-Osta A, Glycemic memory associated epigenetic changes, Biochem Pharmacol. 80 (2010) 1853–1859. 10.1016/j.bcp.2010.06.005. [DOI] [PubMed] [Google Scholar]
- [58].Bollrath J, Greten FR, IKK/NF-kappaB and STAT3 pathways: central signalling hubs in inflammation-mediated tumour promotion and metastasis, EMBO Rep. 10 (2009) 1314–1319. 10.1038/embor.2009.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Whitley SK, Balasubramani A, Zindl CL, Sen R, Shibata Y, Crawford GE, et al. , IL-1R signaling promotes STAT3 and NF-κB factor recruitment to distal cis-regulatory elements that regulate Il17a/f transcription, J Biol Chem. 293 (2018) 15790–15800. 10.1074/jbc.RA118.002721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Chan-On W, Nairismägi ML, Ong CK, Lim WK, Dima S, Pairojkul C, et al. , Exome sequencing identifies distinct mutational patterns in liver fluke-related and non-infection-related bile duct cancers, Nat Genet. 45 (2013) 1474–1478. 10.1038/ng.2806. [DOI] [PubMed] [Google Scholar]
- [61].Jinawath N, Chamgramol Y, Furukawa Y, Obama K, Tsunoda T, Sripa B, et al. , Comparison of gene expression profiles between Opisthorchis viverrini and non-Opisthorchis viverrini associated human intrahepatic cholangiocarcinoma, Hepatology. 44 (2006) 1025–1038. 10.1002/hep.21330. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


