Abstract
Macrophages (MΦ) and dendritic cells (DC) play a fundamental role in shaping immune responses by sensing a plethora of Pathogen Associated Molecular Patterns (PAMPs), phagocytosis and antigen presentation to T lymphocytes. These important biological processes require efficient cell movement and an intact cellular morphology for dynamic interaction. The role of microRNAs (miRs) in this regard, however, is not well understood. In the present study, we show that miR-30b and miR-142–3p regulate migration and morphology of MΦ and DC. Transient overexpression of miR-30b and miR-142–3p attenuates migration and these cells display unique morphological deformities observed under electron microscopy. In addition, miR-142–3p overexpression in MΦ impaired phagocytosis of FITC-conjugated latex beads using live microscopy imaging. Interestingly, live cell imaging and F-actin staining revealed marked changes in the cell polarity and actin polymerization status, respectively. To identify miR-142–3p regulated pathways, we profiled global transcriptome changes in miR-142–3p or control mimic transfected DC. Expression of several genes were differentially altered by miR-142–3p and were associated with pathways related to cell movement, cell adhesion, and cytoskeletal rearrangement. Bioinformatics analysis identified a significant subset of downregulated genes with one or more predicted miR-142–3p binding sites in their 3′UTR strongly suggesting direct post-transcriptional impact of these miRNAs on multiple transcripts. Using dual luciferase assays, novel miR-142–3p binding sites were validated for three genes (Vinculin, Dab2 and Skap2) directly associated with cytoskeletal rearrangement and cell movement. In summary, our results show that miR-30b and miR-142–3p acts as negative regulators of myeloid cell cytoskeletal homeostasis and morphology.
Keywords: MicroRNA, cell migration, phagocytosis, cytoskeletal network, myeloid cells
Introduction
Macrophages (MΦ) and dendritic cells (DC) are components of the mononuclear phagocyte system (MPS). They are highly specialized Antigen Presenting Cells (APC) performing key functions such as antigen recognition, uptake, processing, and presentation, processes that lead to the activation of innate and adaptive immune responses (1–3). Critical to their functions is their ability to respond, under the influence of certain cytokines or factors, and migrate to inflammatory sites. This is attributed to their immense potential to rapidly and transiently modify their cellular morphology (4–6). Evidently, pathways associated with cytoskeletal homeostasis play crucial roles in mediating these migratory events and in eliciting adept immune responses (7). However, the role of miRNAs in these biological pathways remain poorly studied.
MicroRNAs (miRNA) are short (~21 nts), non-protein coding RNAs that control translational output of cell by post-transcriptional regulation of transcripts. Mature miRNA interacts with the mRNA harboring complementary sequences leading to translation inhibition or transcript degradation mediated by RNA-protein complex called miRNA-induced silencing complex (mi-RISC) (8–11). By virtue of their ability to regulate expression of hundreds of transcripts, miRNAs are critical regulators of several cellular processes including cytoskeleton homeostasis, cell migration, proliferation, apoptosis, immune response, etc. (12–17). To date, more than 1900 mature miRNAs are reported in human (miRBase v.22.1) and are predicted to modulate the expression of more than 60% human protein-coding (Friedman et al., 2009). Identifying a complete repertoire of every miRNA-targeted genes (direct or indirect) can provide us with a thorough understanding of molecular networks regulated by these noncoding RNAs and that information can be used to manipulate miRNA functions for therapeutic purpose.
By virtue of their ability to regulate expression of hundreds of transcripts, miRNAs are critical regulators of several cellular processes including cytoskeleton homeostasis, cell migration, proliferation, apoptosis, immune response, etc. (12–17). Indeed, miRNA regulation of cellular motility, migration, and endosomal trafficking has been reported in multiple cell types. Not surprisingly, altered expression of miRNAs or mutations in their ‘seed’ region can result in adverse effects on their regulatory cellular functions and has been implicated in various diseases (19–23). Imbalance in miRNA levels can promote cell proliferation, enhance oncogenic potential of transformed cells and facilitate tumor metastasis. For instance, members of miR-200 family target epithelial to mesenchymal transition (EMT) promoting zinc finger E-box-binding homeobox (ZEB1 and ZEB2) transcription factors that suppress critical epithelial marker E-cadherin expression as well as miR-200 by transcriptional silencing (24, 25). This feedback loop entails dynamic interaction between miRNA and their targets at multiple levels in order to control cellular transformation. miR-138 directly regulates Ras homolog gene family, member C (RhoC) and Rho-associated, coiled-coil containing protein kinase 2 (ROCK2) and suppress cell migration and invasion in tongue squamous carcinoma cells (26, 27). Cytoskeleton regulation by miR-24 is mediated by regulation of various genes including direct silencing of PAK4, Tks5, and ArhGAP19 leading to reduced cell migration (28). Most of the studies examining miRNA-mediated impact on cell migration and movement focused on cell lines or tumor cells. However, equally significant is identification of specific miRNA and their associated mechanisms regulating cell movement in primary cells. This is especially important for myeloid inflammatory cells, key components of host defense that constantly patrol portals of pathogen/antigen entry and depend on dynamic and efficient cytoskeletal rearrangements to perform their crucial functions including migration and interaction with other cells.
In our previous studies, several differentially expressed miRNAs were identified during monocyte to MΦ or DC differentiation (29, 30). In particular, expression levels of mature miRNAs, miR-30b and miR-142–3p were significantly reduced during the differentiation process and preceded changes in differentiation markers indicating that miRNA downregulation may be required for efficient function of differentiated MΦ/DC (29). Indeed, enforced expression of miR-30b or miR-142–3p induces defects in phagocytosis and consequently interferes with antigen processing and presentation by MΦ or DC during innate as well as adaptive immune responses (17, 30). Significantly, all coordinated processes are dependent on cell motility, which in turn is closely associated with the cytoskeletal network. In this study, we examined the impact of miR-30b and miR-142–3p on myeloid cell migration, cell morphology and cytoskeletal rearrangement. Global transcriptome profiling and in silico analysis identified molecular pathways and direct gene targets of miR-142–3p associated with cell movement and cytoskeletal arrangement. Our results demonstrate both miRs, but predominantly miR-142–3p, acts as a negative regulator of myeloid cell cytoskeletal homeostasis and morphology signifying its importance in acquiring robust myeloid function.
Material and Methods
Primary human monocyte isolation and differentiation
Freshly prepared buffy coats were collected from healthy donors (Sylvan N. Goldman Oklahoma Blood Institute, Oklahoma City, OK, USA) and CD14+ monocytes were separated by density gradient centrifugation and magnetic bead isolation as described previously (17, 29, 30). In brief, PMBCs were purified using Ficoll Paque™ (GE Healthcare, Princeton, NJ, USA)-based density centrifugation. PBMCs were incubated with magnetic labeled CD14 beads (Miltenyi Biotech, Auburn, CA, USA) according to manufacturer’s instructions. The purity of CD14+ cells was >95% as determined by flow cytometry (29, 30). For MΦ differentiation, monocytes were plated at 2×106/ml in DMEM supplemented with penicillin (100U/ml) and streptomycin (100μg/ml). After 2h the media was substituted with media containing 10% FBS (Life Technologies, Grand Island, NY, USA), and rhM-CSF (50 ng/mL; Peprotech, Rocky Hill, NJ, USA). For DCs, monocytes were cultured in RPMI-1640 supplemented with rhGM-CSF (1000U/ml) and rhIL-4 (500U/ml) (both from Peprotech). Media was replaced every 72h. At day 7, cells were harvested and differentiation confirmed by flow cytometric analysis as described earlier (29, 30).
Cell culture
Human monocytic cell line, THP-1 (ATCC®TIB-202™) and human embryonic kidney cell line, HEK293 (ATCC®CRL1573™) were grown in RPMI media 1640 and DMEM (both from Gibco™- ThermoFisher Scientific, Grand Island, NY, USA) respectively, containing 10% fetal bovine serum. Culture media were supplemented with 2 mM glutamine, 1% non-essential amino acids, penicillin (100 U/ml), streptomycin (100 μg/ml) and amphotericin B (2.5 μg/ml). Cells were maintained in a humidified atmosphere at 37°C and 5% CO2. After cultures became 80% confluent (usually after 3 days), cells were trypsinized, centrifuged, and suspended in fresh medium. All cells used for experiments displayed >95% viability. All experiments were carried out in duplicate and repeated at least three times.
Transient miRNA transfections
miR-30b, miR-142–3p and control miRNA mimic were purchased from Qiagen (Gaithersburg, MD, USA). Transient miRNA overexpression was performed by transfecting miRNA mimics using Lipofectamine 2000 (Invitrogen-Life Technologies Corporation, Carlsbad, CA, USA) according to the manufacturer’s instructions. Day 7-differentiated MΦ or DC were transfected at a final concentration of 50 nM. Red siGLO oligos (ThermoFisher Scientific, Grand Island, NY, USA) were used to determine transfection efficiency.
Scratch-wound healing assay
Primary MΦ were grown on 48-well plates to a nearly confluent monolayer. After 36h post-transfection with miR-30b, miR-142–3p or control mimic, the confluent monolayers were scratched to form a “wound” using a sterile needle (20 gauge x 1.5 inch). Cellular debris were removed by washing with PBS. The images were recorded for a period of 48h to monitor the migration of cells into the wounded area using a light photomicroscope at 10X magnification. At the end of assay, cells were fixed, stained with H&E and images captured. The number of cells migrating into the wound area were counted. Each assay was performed in triplicate.
Cell migration assay
Cell migration assay was performed using the Radius ™ Cell Migration Assay Kit (Cell Biolabs, San Diego, CA, USA) following the manufacturer’s instructions. PMA (Phorbol myristate acetate; 50 nM final concentration) differentiated THP-1 monocyte-derived macrophages were seeded in 24 well plate at a density of 100,000/well and transfected with miScript miRNA or control miRNA mimics (Qiagen). After 36 h of transfection, the biocompatible hydrogel spot was removed and the cell-free area was exposed for cell migration. Cells were monitored across 72 h and images were captured on EVOS microscope (ThermoFisher Scientific, Grand Island, NY, USA) at 4X magnification. Image-J software was used to quantify would healing changes. The wound scratch area at 0 h for control mimic was arbitrarily assigned as 100% and the percentage of wound healing for control and miR-142–3p mimic transfected cells at different time points was calculated compared to control mimic at 0 hr. Each assay was performed in triplicate.
Phagocytosis assay and imaging
MΦ cells were seeded at a density of 300,000/well (96-well plate) and transfected on day 7 with miScript miRNA and control miRNA mimics (Qiagen) as described above. After 36 h of transfection, phagocytosis assays were performed using Phagocytosis Assay kit containing latex beads conjugated with FITC-labelled IgG (Cayman, Ann Arbor, MI, USA) 1.5 ×106 latex beads (1:5 cells to beads ratio) coated with fluorescently-labeled rabbit IgG were incubated with cells and live imaging was performed using Leica DMI 600B Live Cell Microscope with HCX/PL Fluotar L 20X/0.40 Corr PH1objective (Leica Microsystems Inc, Buffalo Grove, IL, USA), capable of measuring FITC fluorescence (ex/em 485 nm/535 nm). Las X software (Leica Microsystems Inc) was used to acquire a time-lapsed movie during 1 hour where every frame was captured per minute. Images from five randomly selected fields were captured for each donor (n=3).
Scanning Electron microscopy
Monocyte-derived DC were transfected with miR-142–3p or control mimics. After 48 h, cells were fixed with 2.5% glutaraldehyde, followed by 1% OsO4 (Electron Microscopy Sciences, Hatfield, PA, USA). After drying, samples were mounted onto aluminum stubs (Electron Microscopy Sciences), then sputter coated with 3.5 nm gold/palladium (Auto 108; Cressington, Watford, UK). At least 25 randomly selected fields were captured for four independent donors on a JSM-6330F electron microscope (JEOL, Peabody, MA, USA). To measure the differences in the cell morphology, we used ImageJ to assess the cell diameter and area in mimic, inhibitor or control transfected DC. Images were uploaded to ImageJ software and the area were measured by drawing a ROI (Region of Interest) around of the cell periphery by selecting wand tool using the following command: Choose Analyze>Set Measurements>Choose area and click in Analyze. Area data are showing means ± SEM of images from four independent donors. Mean value of the control mimic transfected cells were used to normalize the percent change in cell area of miR-30b or miR-142–3p mimic or inhibitor. For cell diameter measurement, scale bar of each image was used to create a line across the longest diameter of the cell to obtain the exact cell diameter. Mean ± SEM of images from four independent donors was measured for control, miR-30b and miR-142–3p mimic transfected DC.
Immunofluorescence microscopy analysis
To analyze the F-actin distribution in MΦ, we transfected the cells with miR-142–3p or control mimic. After 36 h, the culture media was discarded, cells are washed thrice in PBS and fixed with 4% paraformaldehyde/PBS at room temperature for 20 mins. MΦ were permeabilized with 0.5% Triton-X100 in PBS for 4 min, rinsed thrice in PBS and incubated in blocking solution (20% goat serum in 2% BSA in PBS) for 30 min. After washing thrice in PBS, F-actin was labelled using Texas-red phalloidin (1:200 dilution; ThermoFisher Scientific, Grand Island, NY, USA) for 1 hour. Finally, cells were rinsed thrice with PBS and F-actin distribution was visualized under Leica DMI 600B Live Cell Microscope with HC PL Apo 63x/1.40–0.60 oil objective (Leica Microsystems Inc.). Images from five randomly selected fields were captured for each donor (n=3) by Las X software and Image-J software analysis was used to quantify the size of the MΦ.
Microarray Analysis
Dendritic cells transfected with miR-142–3p or control mimic were harvested after 36 h and total RNA was isolated using the miRNeasy kit (Qiagen). For transcriptome profiling, we used Affymetrix HTA-2_0 arrays (Santa Clara, CA, USA). RNA samples were labeled and hybridized according to standard WT (whole transcript) plus target labeling protocol recommended by the manufacturer. After hybridization, each image was analyzed for the following quality metrics: average signal present, signal intensity of species-specific housekeeping genes, relative signal intensities of labeling controls, and absolute signal intensities of hybridization controls. Data was processed using Genomics Suite 6.6 Statistical Package (Partek, Inc., St. Louis, MO, USA). The following parameters were applied for hybridization signal processing: Algorithm: RMA; Background Correction: RMA Background Correction; Normalization: Quantile Normalization; Log Probes using Base: 2; Probeset Summarization: Median Polish. ANOVA test was used to calculate significance of the differential expression in each comparison (miR-142–3p vs control). Differentially expressed transcripts were annotated according to Affymetrix ‘NetAffx Analysis Center’. The significance cut-off was at 0.05. Array data were in compliance with Minimum Information About a Microarray Experiment (MIAME) guidelines and deposited in the Gene Expression Omnibus public database under Accession Number GSE151262.
Pathway Analysis Using Reactome Pathway Database
Reactome Pathway Database software ((https://reactome.org/) was used to predict the networks and canonical pathways that are associated with the significantly expressed genes. This software collects information from a database built from published relationships between genes and their biological functions, mechanisms, canonical pathways, and networks. Significantly over- or under-expressed genes (p values <0.05) with relative expression values of ±1.25-fold change and their corresponding gene identifiers were imported into Reactome and analyzed with the pathway browser tool which displays an overview of all Reactome pathways. Pathways were organized hierarchically and often found to have sub-pathways. At the highest hierarchical level, all pathways were represented in an Overview. At intermediate levels, pathways were often represented as interactive illustrations, with selectable regions that act as links to the lower levels, which are represented as detailed Pathway Diagrams. Reactome’s Overview uses a unique graphical visualization of the hierarchy, representing pathways at the uppermost hierarchical level as central nodes (circles) with subpathways arranged concentrically (in rings) around them. Many have further concentric rings of ‘child’ subpathways. Pathway nodes were connected to their subpathways by edges (lines). Selected Reactome pathways with FDR corrected p-value of ≤0.05 are shown or included in the list. If multiple pathways have the same p-value, they were ranked by the number of identifiers in the query that match the pathway. The number of molecules matched/total number of molecules and FDR values were added to the right side of pathway names in the Hierarchy Panel.
Immunoblotting
Cells transfected with miR-142–3p or control mimic were harvested after 36 h with cold PBS and centrifuged at 300 × g, for 5 minutes at 4°C. The cell pellet was incubated for 15 minutes on ice with 300 μl of cell lysis buffer (Cell Signaling Technology Incorporated, Danvers, MA, USA) with protease and phosphatase inhibitor cocktail (ThermoFisher Scientific, Grand Island, NY, USA) and centrifuged at 10,000 x g for 15 minutes at 4°C. Total protein concentration was quantified by a standard Bradford assay using the colorimetric reagent from BioRad Laboratories (Hercules, CA, USA). Proteins (25 μg) were separated onto SDS polyacrylamide gels using a 4–12% Bis-Tris gradient gels in the BioRad Criterion System and transferred to nitrocellulose membranes, which were blocked with 3% BSA, and probed with the appropriate antibodies. For western blot analysis, the following antibodies were used: Lims1 (Pinch (5G7) mouse mAb), Vasp Rabbit Ab, Vinculin antibody, Dab2 (D709T) Rabbit mAb and p44/42 MAPK (Erk1/2); all from Cell Signaling Technology Incorporated. Skap2 polyclonal antibody and mouse anti-GAPDH were purchased from ThermoFisher Scientific, mouse IgG HRP-linked antibody and rabbit IgG HRP-linked antibody were purchased from Santa Cruz Biotechnology Inc (Dallas, TX, USA). Immunocomplexes were detected with appropriate horseradish peroxidase-conjugated secondary antibodies and detected by enhanced chemiluminescence with the ECL Plus Western Blotting Detection System or ECL Advance Western Blotting Detection Kit (GE Healthcare Life Sciences, Little Chalfont, UK). Images were captured on a ChemiDoc XRS Imaging System (BioRad).
MiRNA target prediction of differentially downregulated genes
To identify miR-142–3p gene targets with high confidence, we first selected downregulated genes. The 3′UTR of these genes were extracted using BioMart tool on Ensembl (http://www.ensembl.org/biomart/martview/aa867419c3c6fd64f94af6d4a6549d3c) as previously described (31). Briefly, we selected Ensembl Genes 87 database and Human Genes dataset (GRCh38.p7). Next, the “Filters” were selected to match the input genes list. In the “Gene”tab we set the “ID list limit” filter to “HGNC symbol(s)”. Finally, to procure the 3’UTR sequences “Attributes” were set. In the “Attributes”, “Sequences” was first selected followed by selecting 3′UTR start and 3’UTR end. “EnsemblGene ID” and “Associated Gene Name” were collected. The results were exported to using the “FASTA “and “Unique results only” attributes. miR target 3’UTR interaction was assessed by target prediction tool RNA Hybrid software (https://bibiserv2.cebitec.uni-bielefeld.de/rnahybrid?id¼rnahybrid_view_submission). The procured 3′UTR sequences and miR-142–3p sequence (extracted from miRbasev.21) were provided as input for RNA Hybrid analysis. The stringency parameters were set up for individual sequences and we opted for three hits per target to highlight any probable miRNA binding sequence present on the target. We considered the following parameters to select putative miR regulated genes: (i) There should be high sequence complementarity in the seed region (positions 2–8 nt from 5′ of miRNA), with only 1 mismatch allowed (ii). For stringency, we picked miR-target interactions where more than 11 nts of the miR sequence are involved in the interaction. (iii) If there is any mismatch in the seed regions, this should be compensated by strong binding beyond the seed region. (iv) The bulge in the interaction region should not involve more than 3 nucleotides. (v) Entropy of the miR-target interaction was set at stringent level with cut-off < 22 kcal/mol.
Dual luciferase reporter assays
Dual luciferase experiments were performed as previously described (16, 17, 30, 32). In brief, HEK293 cells were seeded at the density of 3×104 cells/well (96-well plate) in DMEM supplemented with 10% fetal bovine serum. All transfections were performed in quadruplicate using 0.5 μL Lipofectamine 2000 (Invitrogen), 120 ng dual luciferase reporter control (CmiT000001-MT06 miRNA Target clone control vector for pEZX-MT06) or plasmids containing human VCL (NM_003373.3; HmiT018467-MT06), human DAB2 (NM_001244871.1; HmiT055467-MT06) or human SKAP2 (NM_001303468.1; HmiT067400-MT06; All from GeneCopoeia Incorporated, Rockville, MD, USA) were co-transfected with a final concentration of 1 pmol, 5 pmol and 10 pmol of synthetic miR-142–3p or control mimics (10 pmol) (Qiagen). After 36 h post-transfection, cells were lysed in passive lysis buffer (Promega Corporation) and dual luciferase assays performed using the multilabel reader Victor™ x5 (PerkinElmer 2030-Perkin Elmer Health Sciences Inc, Shelton, CT, USA). For each reporter 3′UTR construct, the Rluc/Fluc value obtained was normalized to the value obtained for pEZX-MT06 no-insert control (EV) co-transfected with the same miRNA mimic. The values obtained were plotted as histograms, where EV is set at one.
Statistical Analysis
Data were analyzed on GraphPad Prism (LaJolla, CA, USA). The results are represented as standard deviation or ±SEM from three independent replicates and experiments were conducted at least thrice. P-values were calculated using Students t-test and p<0.05 were considered significant.
Results
Myeloid cell migration is attenuated by miR-30b and miR-142–3p
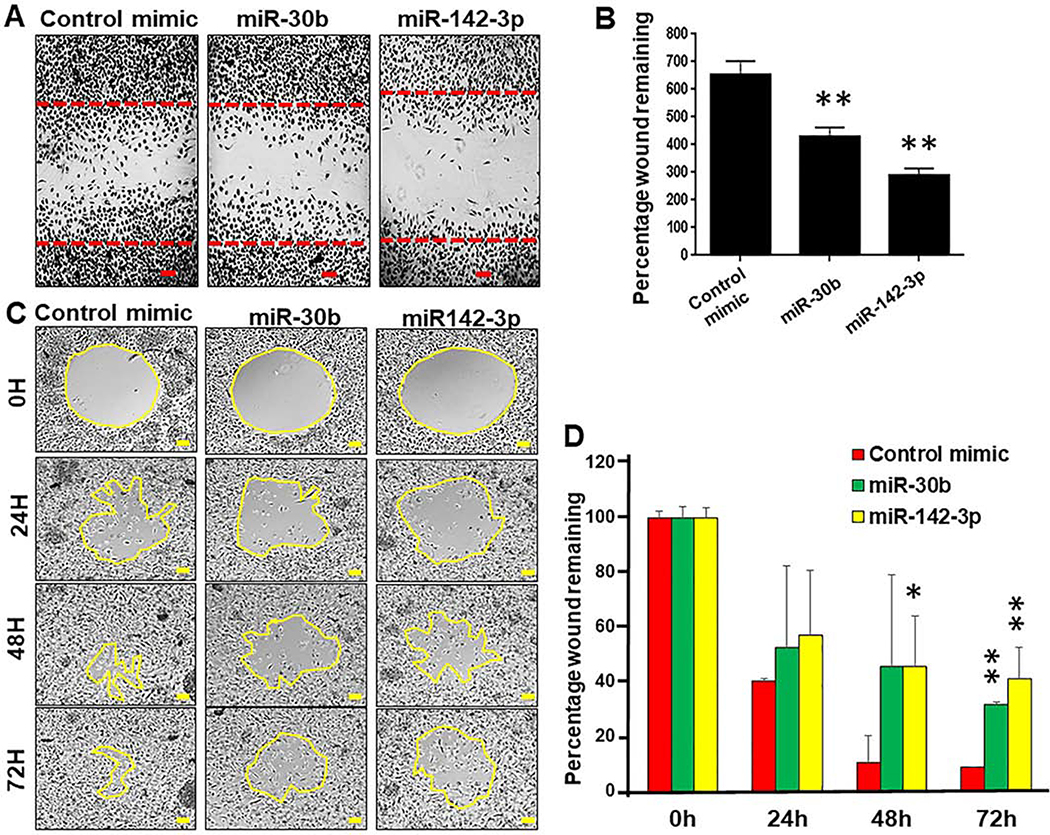
Our previous results showed impairment in particle internalization by miR-30b and miR-142–3p transfected MΦ and DC (16, 30). Further, expression of phagocytosis-associated genes was downregulated in miR-30b and miR-142–3p overexpressing MΦ and DC including members of PI3K and PKC signaling pathway that play critical role in cytoskeletal restructuring and movement (29, 30). These results prompted us to investigate if myeloid cell migration is hindered by these miRNAs. To test this hypothesis, primary MΦ and THP-1 monocyte-derived macrophages were transiently transfected with miR-30b and miR-142–3p and cell migration was monitored. Migration of miR-142–3p, miR-30b or control mimic transfected primary MΦ was assayed by the scratch-wound healing assay and images were captured after 48 h. Compared to control mimic, we observed significantly attenuated migration in macrophages overexpressing miR-142–3p and miR-30b (Figure 1A and Supplementary File 1). We counted the total number of cells migrated in the scratch zone and noticed significantly less cells in the migratory zone of miR-142–3p (~290) and miR-30b (~425) transfected MΦ compared with control mimic (~650) (Figure 1B). Compared to miR-30b, robust inhibition of cell migration was observed in miR-142–3p overexpressing MΦ.
Figure 1. Macrophages transfected with miR-30b and miR-142–3p exhibit attenuated cell migration.
(A) Macrophages were transfected with miR-30b, miR-142–3p or control mimics. After 24 h, a scratch was induced and cells were monitored for 36 h. Cells were fixed, stained with H&E and imaged with light microscopy. The red lines indicate the invasive front in the wound healing assay. (B) Total number of cells counted in the migration zone after 36 h. Data are means ± SEM of three independent experiments. Student’s t-test was conducted to calculate p-values. *p<0.05, **p < 0.01. (C) Time course analysis of cell migration was performed in THP-1 macrophages transfected with miR-30b, miR-142–3p or control mimic for 24 h. The yellow lines indicate the invasive front in the wound healing assay. ImageJ analysis was performed to quantify the wound closure for 72 h. The scratched area at control (0 h) was arbitrarily assigned as 100%. (D) Cell migration assay kit was used to analyze the inhibition of cell migration in THP-1 transfected with miR-30b, miR-142–3p mimic and control mimic for 36 h. Data are means ± SEM of three independent experiments. Student’s t-test was conducted to calculate p-values. *p<0.05, **p < 0.01. Final magnification: X40, scale bar corresponds to 100 μm.
To confirm these findings, we next performed the radius gel migration assays using THP-1 monocyte-derived macrophages. THP-1 MΦ were transfected with miR-142–3p, miR-30b or control mimic. Migration was initiated by dissolving the gel 24 h post-transfection and cells were monitored over a 72 h period. Time-lapse imaging clearly showed attenuated migration in cells overexpressing miR-142–3p or miR-30b compared to those transfected with control mimic (Figure 1C). At the 72 h time point, wound area in control mimic transfected cells was completely confluent while miR-142–3p or miR-30b transfected cells show partially filled wounds (Figure 1D). Cells transfected with miR-142–3p showed more significant delay in wound closure compared to miR-30b in THP-1 MΦ and corroborate with our observation in primary macrophages. To quantify the migration rate, the area of the migration zone was calculated for miRNA or control mimics for each time point and normalized with control mimic at the 0 h time point. At 24 h post-scratch, the percentage of wound remaining was ~ 57 and ~ 53 in miR-30b and miR-142–3p respectively, compared to the control mimic (~40) (Figure 1D). After 48 h, both miR-30b and miR-142–3p showed ~46% wound remaining compared to the control mimic (~11%). Finally, at 72h of migration monitoring course, miR-30b and miR-142–3p transfected cells showed ~32% and 41% wound remaining, which is statistically significant compared to the control mimic (9% wound remaining) (Figure 1D).
Another remarkable result to be considered was the loss of cellular polarity during the migration course. Live microscopy imaging revealed that MΦ transfected with control mimic move rapidly migrate towards the direction of wound area and eventually fill the zone in 48 h (Supplementary File 1). However, we noticed that miR-142–3p or miR-30b transfected MΦ move slowly and migrate in both forward and backward direction indicating a loss of cellular polarity. Together, the migration assays unequivocally show that enforced expression of miR-142–3p and miR-30b significantly impair migration rate and cellular polarity of myeloid cells.
MΦ transfected with miR-30b and miR-142–3p exhibit impaired phagocytosis
MΦ and DC are myeloid inflammatory cells capable of recognizing a plethora of microorganisms via conserved pathogen-recognition receptors, antigen uptake and subsequent presentation. Ligation of these pathogen-recognition receptors results in a pattern of altered gene expression that enhances pathogen recognition, uptake, and removal (33). Our previous results showed that MΦ and DC transfected with miR-142–3p and miR-30b negatively affected the phagocytic uptake of various antigens including bacteria, IgG coated latex beads or ovalbumin (16, 17, 29, 30). However, we did not evaluate changes in real-time using live cells.
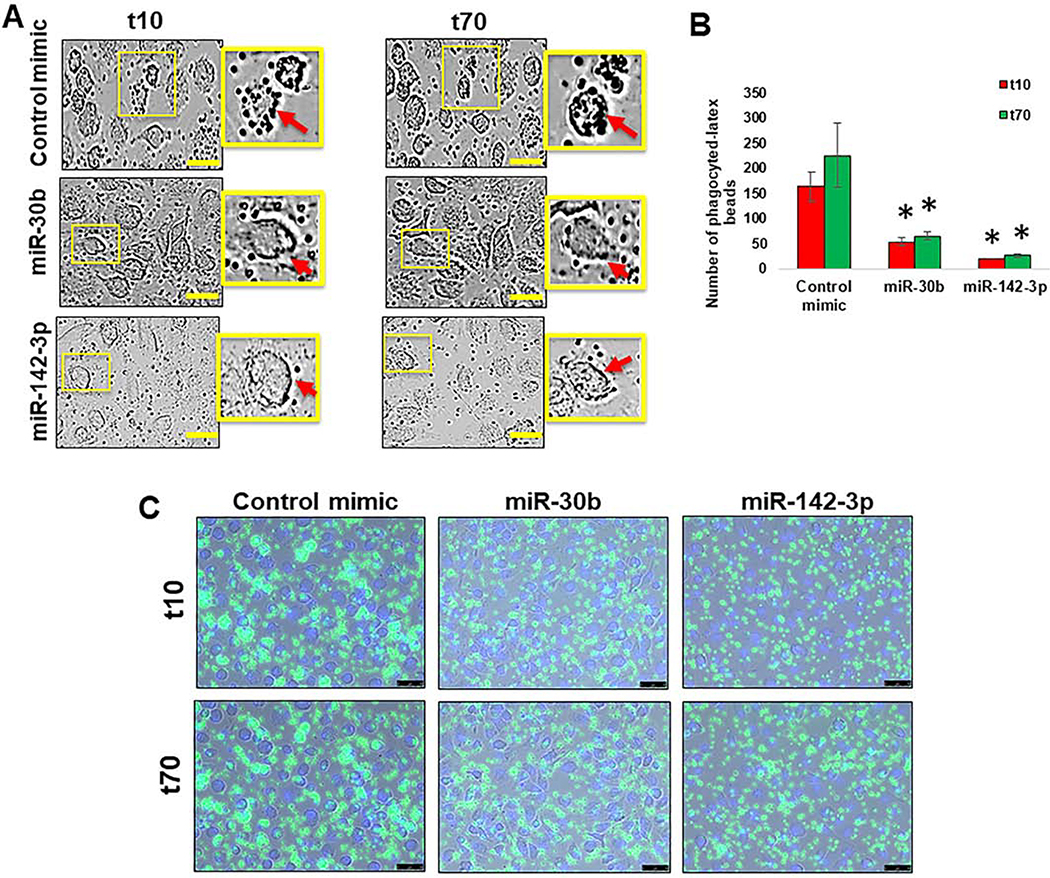
Cell motility is a highly dynamic process and time-lapse examination of live cells will provide a better understanding of how miRNA transfected myeloid cells behave in the presence of antigens. We therefore decided to analyze phagocytic activity in miR-142–3p and miR-30b transfected-MΦ by live imaging. As shown in Figure 2A, the ability of phagocytose IgG-conjugated, FITC-labeled, latex beads was attenuated in miR-142–3p or miR-30b transfected-MΦ compared to control mimic transfected cells. The number of intracellular FITC-beads were significantly higher in control mimic transfected-MΦ than miR-142–3p and miR-30b transfected cells (Figure 2B). Live cell imaging of phagocytosis process further shows impairment of bead uptake by miR-142–3p or miR-30b (Supplementary File 2). Together, these results demonstrate that miR-30b and miR-142–3p are negative regulators of phagocytosis in MΦ. These findings indicate that phagocytic and/or endocytic machinery and pathways are inefficient in miR-30b and miR-142–3p transfected cells.
Figure 2. miR-30b and miR-142–3p transfected cells show impaired phagocytosis.
(A) Macrophages were transfected with miR-30b, miR-142–3p or control mimics and phagocytosis assay was performed after 36 h by incubating cells with FITC-conjugated IgG-coated latex beads. Representative live, contrast-phase microscopy images of phagocytosed-latex beads coated with IgG-FITC in MΦ transfected with miR-30b, miR-142–3p or control mimic were captured at 10 min and 70 min after incubation. Data are means ± SEM of three independent experiments (*p <0.05, compared with the control mimic for same time point). Scale bar- 50 μm. (B) Histograms showing number of phagocytosed beads quantified in MΦ transfected with miR-30b or miR-142–3p mimics compared with the t10 of the corresponding control mimic, and (C) a decrease of intensity and number of FITC beads were performed by live imaging in MΦ transfected with miR-30b and miR-142–3p after 36 h.
Structural defects in myeloid cells overexpressing miR-30b and miR-142–3p
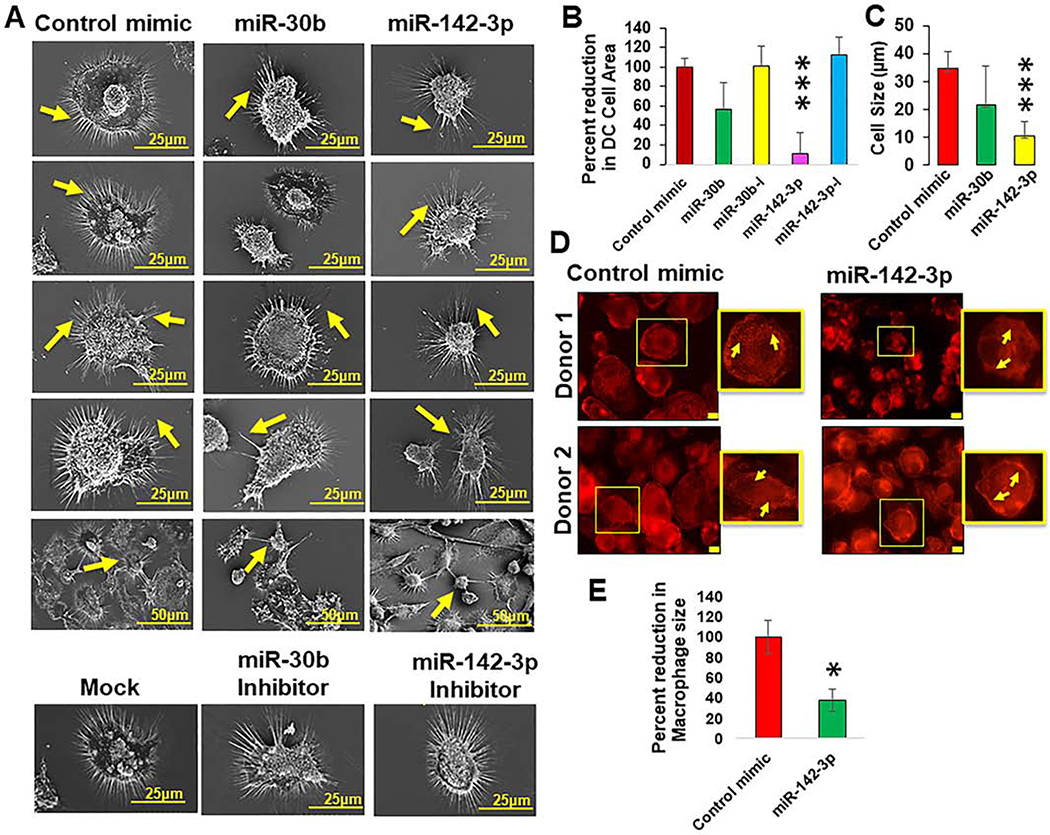
Antigen presenting cells (DC and MΦ) are highly structured myeloid cells that form cytoplasmic protrusions necessary to establish the physical immunological synapses required for efficient antigen presentation (34, 35). The defects observed in our cell migration assays prompted us to assess the impact of miRNAs on DC surface phenotype. The topology of miRNA transfected DC was examined using surface electron microscopy (SEM). Compared with control mimics, we noticed marked structural deformations in DC transfected with miRNA mimics but not with corresponding inhibitors (Figure 3A). Control or miRNA inhibitors display normal surface morphology similar to mock transfected cells as observed by long dendrites, higher density of dendrites and large cell bodies (Figure 3A; lower panel). To examine the changes in cell size, we measured the area of cells using ImageJ analysis and normalized to control mimic. We observed reduction in the cell area in miR-142–3p (~70%) and miR-30b (~25%) mimic transfected DC, however, significant differences were observed for miR142–3p but not miR-30b mimic (Figure 3B). Corresponding inhibitors of miR-30b or miR-142–3p transfected DC did not display marked difference compared to control and showed antagonistic phenotype with respect to the miRNA mimics (Figure 3B). We further measured cell length (diameter) in control or miRNA mimic transfected DC and observed that miR-142–3p overexpressing cells were significantly smaller (10.6±5.3 μm in diameter) in length compared with control transfected cells (34.9±5.9 μm in diameter). In miR-30b transfected cells, no significant changes in cell length (21.9±13.7) were observed with respect to control mimic (34.9±5.9 μm in diameter) (Figure 3C). This is in line with our previous confocal microscopy experiments where we noted the relatively small size of both miR-142–3p transfected MΦ (30). On the other hand, miR-30b transfected DC exhibited a wide spectrum of defects predominantly associated with changes in dendrite density, shape and length (Figure 3A). The differences in dendrite length between miR-30b and control mimic were visually evident and were not measured using ImageJ. The density and length of dendrites in miR-142–3p mimic transfected DC were comparable to their respective inhibitor or control mimic. These morphological changes were not associated with comprised cell viability, as we did not observe significant difference in the cell death due to miR-30b or miR-142–3p transfection (Data not shown). These results clearly indicate that enforced expression of miR-142–3p and miR-30b induces gross morphological changes in myeloid cells that may negatively affect their ability to perform key cellular functions.
Figure 3. Myeloid cells overexpressing miR-30b and miR-142–3p exhibit structural defects and impaired cytoskeleton homeostasis.
(A) Macrophages were transfected with miR-30b, miR-142–3p mimic, inhibitor or control mimics and the structural defects were visualized by surface electron microscopy after 48 h. Scale bar corresponds to 25 or 50 μm. Measurement of DC (B) cell area and (C) cell diameter in miRNA mimic or inhibitor transfected cells compared with control mimic using ImageJ software. Area under the Region of Interest (ROI) was measured and normalized to control mimics. At least 25 randomly selected fields were captured for six independent donors. Data are means ± SEM of fours independent donors. Student’s t-test was conducted to calculate p-values. ***p < 0.001. Representative images of F-actin labeled with Texas-red phalloidin in MΦ transfected with miR-142–3p performed by immunofluorescence microscopy shows (D) Impairment in the actin polymerization status and (E) a reduced size of cells. Image J analysis was performed to quantify the size of MΦ. Data are means ± SEM of three independent experiments (*p <0.05, compared with the control mimic). Final magnification: X630, scale bar corresponds to 10 μm.
miRNA-142–3p impairs cytoskeletal rearrangement in myeloid cells
Based on cell migration experiments, phagocytosis assay and SEM imaging, miR-142–3p overexpressing cells displayed pronounced alterations of cellular morphology and therefore we performed subsequent experiments with miR-142–3p. We next focused on assessing the impact of miR-142–3p overexpression on actin distribution and polymerization status. Primary macrophages transfected with miR-142–3p or control mimic were stained for F-actin using Texas-Red Phalloidin. Immunofluorescence staining showed a non-homogeneous distribution of F-actin (Figure 3D) and a smaller cell size in miR-142–3p transfected cells compared to control transfected cells (Figure 3E). These findings strongly support the notion that miR-142–3p directly or indirectly reduced length of actin fibers. This may cause reduction in cell size that also corroborates with the electron microscopy data. These results clearly demonstrate that miR-142–3p can profoundly alter the cytoskeleton homeostasis in MΦ, which can interfere with their function and interaction with other immune cells such as T-cells.
Global transcriptome profiling identified a large repertoire of miR-142–3p regulated genes
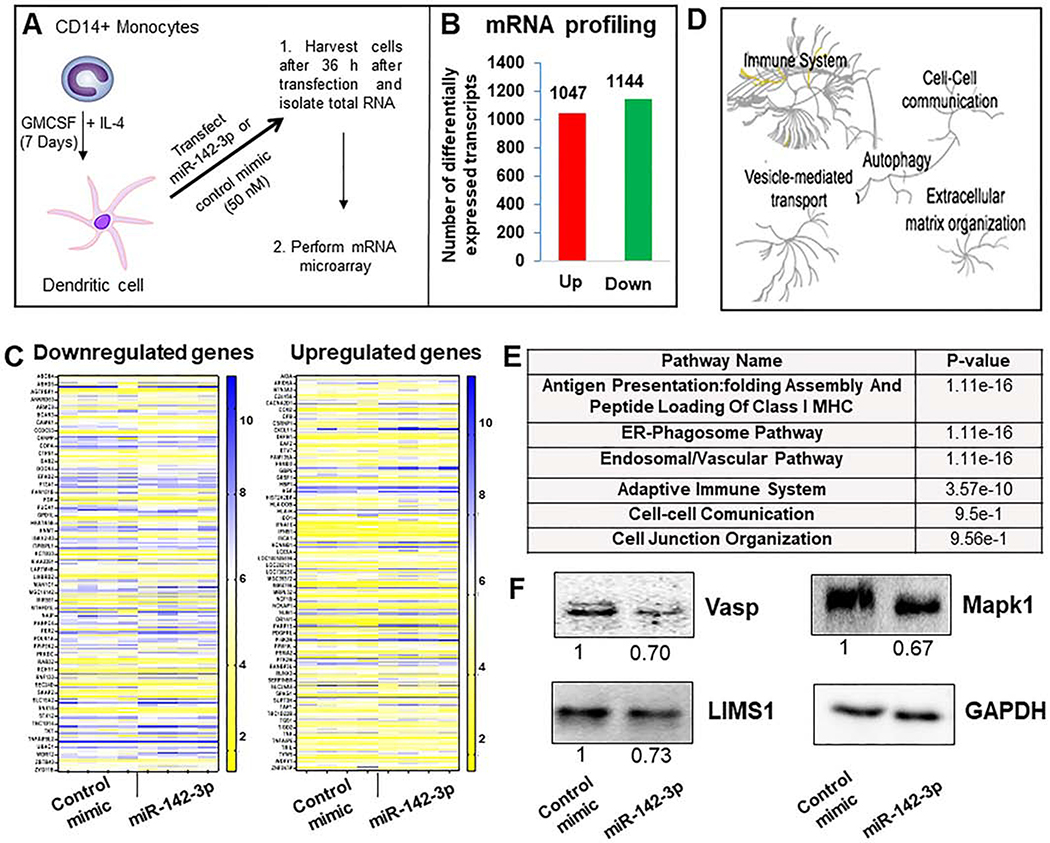
Our previous miRNA profiling data showed numerous miRNAs differentially expressed in monocyte-to-MΦ and monocyte-to-DC differentiation over the course of 7 days. Expression of miR-142–3p (and its isoform, miR-142–5p) was downregulated significantly during monocyte-to-DC differentiation strongly suggesting that miR-142–3p suppression is required for DC functionality (29, 30). Indeed, overexpression of this miRNA interfered with phagocytosis and antigen processing and presentation by DC (16, 17, 30). Next, we examined the transcriptome-wide impact of miR-142–3p overexpressed in DC to identify which mRNAs and pathways are globally perturbed by miR-142–3p. DC were transfected with miR-142–3p or control mimic and total RNA was isolated 36 h post-transfection for mRNA microarray analysis (Schematic in Figure 4A). Our results show that miR-142–3p transfected cells exhibit altered expression of several transcripts indicating that it can profoundly affect the transcriptome. In miR-142–3p transfected cells, 2193 transcripts were differentially expressed among which 1047 were upregulated and 1144 were downregulated compared to the control mimic (Figure 4B and Supplementary File 3). After merging the transcript variants, 866 genes were considered significantly altered by miR-142–3p among which 464 mRNAs were upregulated and 402 were downregulated. Figure 4C shows a heat map of differentially expressed (upregulated and downregulated) genes between control and miR-142–3p transfected DC.
Figure 4. Transcriptome analysis of miR-142–3p overexpression in dendritic cells.
(A) Schematic graph shows as DC after 7 days of monocytes differentiation. DC are transfected with corresponding control mimic and miR-142–3p during 36 h. Thereafter, RNA was isolated and transcriptome analysis performed. (B) Number of differentially expressed upregulated and downregulated transcripts in miR-142–3p overexpressing DC compared to control mimic. (C) Heat maps showing expression profiles of downregulated and upregulated genes based on a fold change (a cut off between −1.25 and 1.25) and p value (<0.05). (D) Global pathway analysis overview by Reactome analysis. The center of each of the circular “bursts” is the root of one of a top ranked pathway. Each step away from the center represents the next level lower in the pathway hierarchy. The color code denotes over-representation of that pathway in your input dataset. Light grey signifies pathways that are not significantly over-represented. (D) Table shows several relevant pathways identified in Reactome analysis that are related to cell movement, cell adhesion and cytoskeletal rearrangement. (E) Expression of several downregulated genes in MΦ overexpressing miR-142–3p were analyzed by western blot. Day 7 differentiated cells were transfected with miR-142–3p mimic or control mimic. Cell lysates were prepared after 36 h of transfection and Vasp, LIMS1 and Mapk1 levels were detected by immunoblotting. The expression level of GAPDH was used as loading control. The corresponding densitometric analysis by Image Lab software is also shown. Data are means ± SEM of three independent experiments (*p <0.05, compared with the control mimic)
To investigate miR-142–3p regulated pathways, we performed global pathway analysis of differentially expressed genes using Reactome pathway database. We identified a large array of pathways impacted by miR-142–3p overexpression in DCs. Some of the key pathways related to the miR-142–3p-mediated impairment of myeloid functions included immune response, extracellular matrix organization, vesicle-mediated transport, cell-cell communication pathways and autophagy (Figure 4D). Genome-wide overview of pathways and most significantly altered pathways identified from Reactome pathway database are provided in Supplementary File 4. Further dissecting the analysis, we noted that a large number of upregulated genes were involved in antigen presentation: folding, assembly and peptide loading of class I MHC (75 genes), ER-phagosome pathway (80 genes), endosomal/vascular pathway (73 genes), and adaptive immune system (98 genes) (Figure 4E and Supplementary File 4). Interestingly, several miR-142–3p downregulated genes were involved in regulating various aspects related to intracellular cytoskeletal rearrangement. For instance, vasodilator-stimulated phosphoprotein (VASP) and vinculin (VCL) are involved in the cytoskeleton homeostasis pathway and anchoring F-actin to the membrane with roles in cell morphology and locomotion, respectively. LIM Zinc Finger Domain Containing 1 (LIMS1) is involved in cell-cell communication pathway by shaping cell junction organization, while DAB adaptor protein 2 (DAB2) is required for clathrin-mediated endocytosis and is involved in myeloid cell differentiation and can induce macrophage adhesion and spreading. Two important regulators of cytoskeletal signaling cascades viz., Src Kinase Associated Phosphoprotein 2 (SKAP2) and Mitogen-Activated Protein Kinase 1 (MAPK1) respectively control immune cell activation and cell growth, adhesion, survival and differentiation (Figure 4E; Supplementary File 4).
To validate our microarray data, we analyzed the expression of several downregulated genes (Vasp, LIMS1 and MAPK1) in MΦ overexpressing miR-142–3p by immunoblotting. Compared to control mimic, marked reduction (~27–33%) was observed in the protein levels of Vasp, LIMS1 and MAPK1 in MΦ overexpressing miR-142–3p (Figure 4F). Our immunoblot results corroborate with the transcriptome profiling data. Together, our findings clearly demonstrate that miR-142–3p is a critical post-transcriptional regulator of mRNAs involved pathways controlling cellular architecture, cytoskeletal rearrangement and cell migration and movement. Thus, altered expression of miR-142–3p can perturb these pathways and hence multiple cellular functions.
miR-142–3p directly targets genes involved in cytoskeleton homeostasis
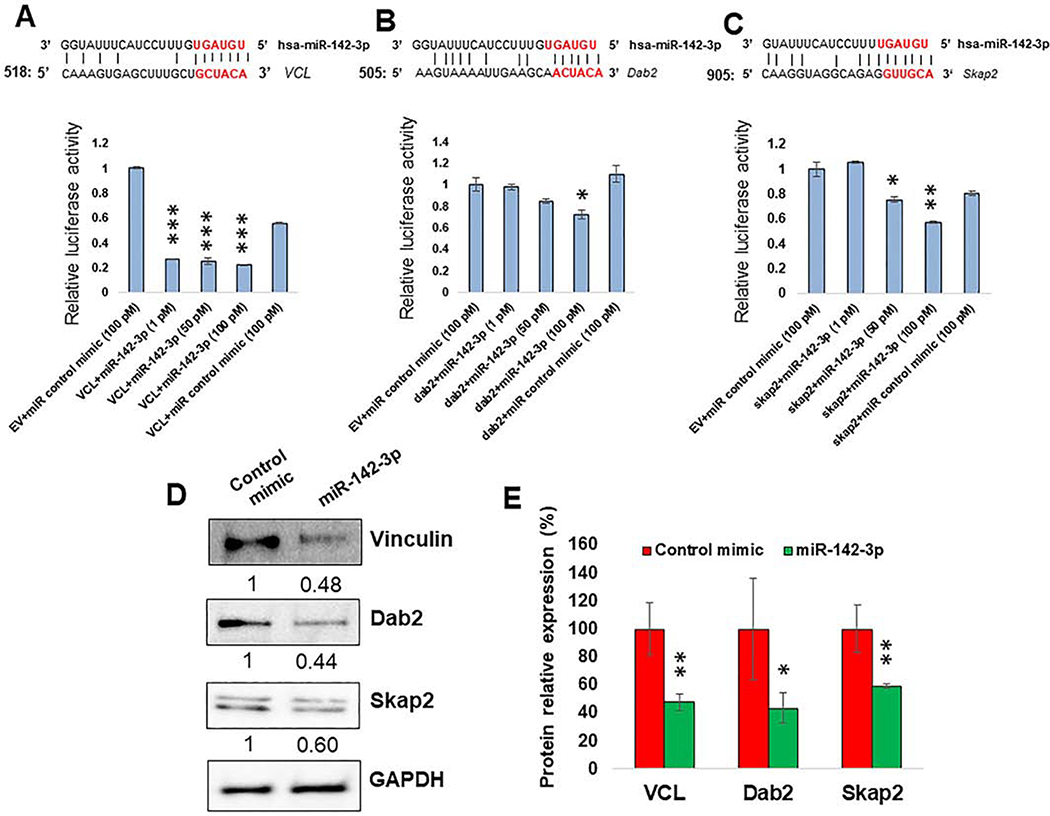
MiRNA can simultaneously bind to multiple transcripts and induce transcript degradation or translational repression (8–11). In general, miRNA exert their function by binding to the 3′UTR of target transcripts (9, 10). Depending on the evolution of miRNA:mRNA interaction, miRNA may have one or more sites on the same 3′UTR so that the presence of multiple functional miRNA binding sites can act synergistically to efficiently repress target transcripts. Moreover, changes in a subset of direct miRNA targets can also have an indirect effect on downstream pathways causing widespread perturbations in gene expression both at the transcriptional or post-transcriptional level. Our microarray data showed both upregulated and downregulated transcripts. However, we specifically focused on the transcripts downregulated in miR-142–3p overexpressing cells that may be potential direct targets of miR-142–3p. For this, all the downregulated genes (402) were submitted to ENSEMBL database to procure their 3′UTR sequences. The ENSEMBL database released 3′UTR of 395 downregulated genes that were submitted to RNAhybrid 2.2 for the prediction of miR-142–3p binding site(s). A complete list of all the miR-142–3p binding sites on the downregulated genes predicted based on our criteria (mentioned in the Material and Methods section) are listed in the Supplementary File 5. While most of the genes exhibit single miR-142–3p binding site, some genes showed more than one binding sites. Supplementary File 5 lists selected gene (VCL, Dab2, Vasp, LIMS1, MAPK1 and Skap2) target interactions obtained using stringent parameters demonstrating miR-142–3p and selected target gene sequence alignment, position of binding sites on the target, and minimum free energy (mfe) values. For VCL gene, our in silico analysis predicted four different miR-142–3p binding sites at the nucleotide positions 518, 937, 1380 and 1199. For Dab2 gene, the software detected four binding sites 505, 656, 858 and 1244 positions) and for Skap2 gene, three (314, 905 and 2190) with perfect matched seed sequence were predicted.
Next, we decided to validate the predicted miR-142–3p binding site(s) on the 3′UTR of downregulated genes. To this end, we selected genes showing strong complementarity between miRNA and mRNA and their association in pathways related to cytoskeleton remodeling and cell polarity such as cell morphology, macrophage adhesion, spreading, lamellipodia and filopodia dynamics and cell migration. Based on our criteria, vinculin, Dab2 and Skap2 were selected for this analysis. Figure 5A (upper panel) shows the sequence alignment between miR-142–3p and candidate gene 3′UTR. Interestingly, these interaction sequences are highly conserved across mammals (8–11). To validate the predicted miRNA-target interaction, we performed dual luciferase assays. Compared to control mimic, miR-142–3p transfected HEK293 cells (at 1, 5 and 10 pmol final concentration) showed more than 80% reduction of renilla luciferase activity when cells were co-transfected with plasmid containing 3’UTR of Vinculin (Figure 5A; lower panel). Increasing concentrations of miR-142–3p had no further significant effect on luciferase activity indicating a strong interaction and suppression of vinculin. In contrast, HEK293 cells co-transfected with miR-142–3p and Dab2 or Skap2 demonstrated concentration dependent inhibition of luciferase activity. At 5 pmol and 10 pmol concentrations of miR-142–3p, we observed ~30% and ~40% reduction in the luciferase activity for Dab2 and Skap2, respectively, compared to control mimic (Figure 5A). These results demonstrate that miR-142–3p can regulate multiple transcripts by directly interacting with high affinity complementary sequences in the 3′UTR leading to efficient silencing of the reporter gene.
Figure 5. miR-142–3p directly regulate genes involved in cell migration, phagocytosis and cell polarity.
(A-C; Upper panel) Sequence alignment of predicted miR-142–3p binding sites in the 3′UTR of VCL, Dab2 and Skap2. Only the binding sites with mfe <−20 kcal/mol are shown. (A-C; Lower panel) HEK293 cells were co-transfected with dual luciferase reporter plasmids containing 3′UTR of VCL, Dab2 and Skap2 or control vector and miR-142–3p or control mimic. After 36 h, cell lysates were prepared to measure renilla and firefly luciferase activity. Renilla activity was normalized to firefly activity and the ratios were subsequently normalized to empty vector transfected with miR-142–3p mimic set as 1. Data are expressed as ± SEM of four independent transfections. Student’s t-test was conducted to calculate p-values. *p<0.05, **p<0.01, ***p<0.001. (D) Expression of VCL, Dab2 and Skap2 in MΦ by Western blot. Day 7 differentiated cells were transfected with miR-142–3p mimic or control mimic. Cell lysates were prepared after 36 h of transfection and VCL, Dab2 and Skap2 levels were detected by immunoblotting. The expression level of GAPDH was included as loading control. (E) The corresponding densitometric analysis by Image Lab software is also shown. Data are means ± SEM of three independent experiments (*p <0.05, **p<0.01; compared with the control mimic).
To confirm the dual luciferase results, we further analyzed the impact of miR-142–3p on protein levels of Vinculin, Dab2 and Skap2 in MΦ. Cell transfected with miR-142–3p showed significantly reduced protein expression compared with the control mimic (Figure 5C,D). These results corroborate with the luciferase reporter assays. Taken together, these results show the miR-142–3p is an important post-transcriptional regulator of cytoskeleton homeostasis by directly targeting multiple genes involved in the pathway. Figure 6 summarizes the regulation of myeloid cell functions by miR-142–3p by direct inhibition of multiple genes involved in the pathways related to cell migration, cell polarity, phagocytosis, cytoskeletal homeostasis.
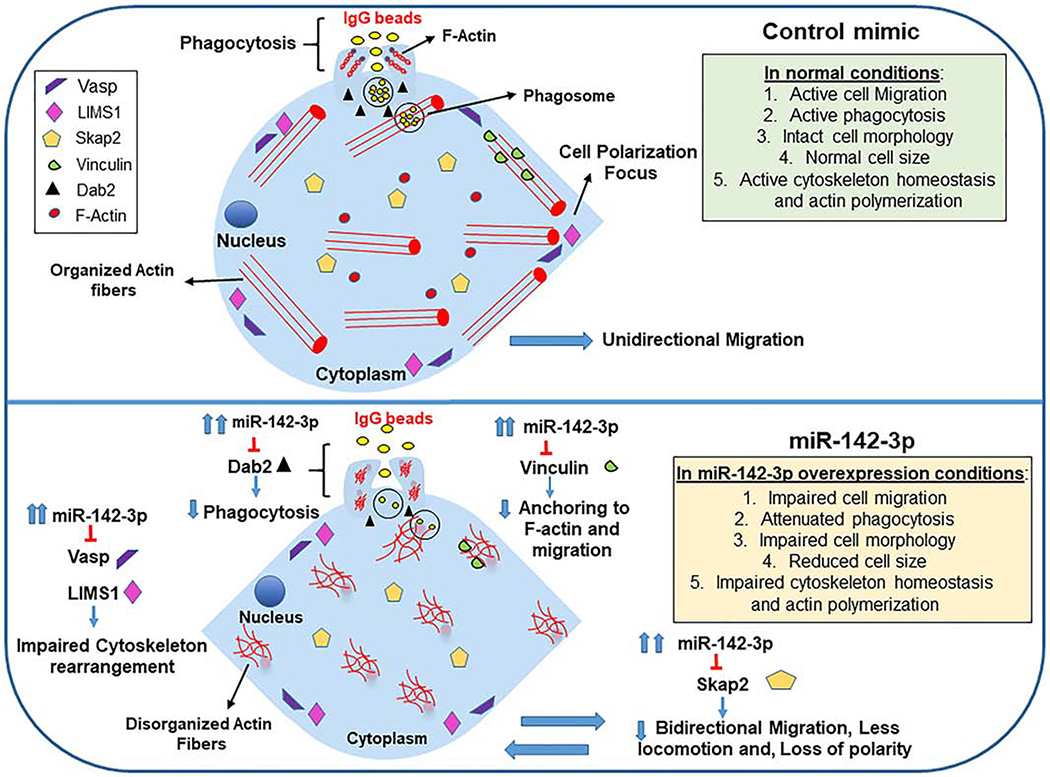
Figure 6. Schematic showing miR-142–3p-mediated regulation of myeloid cell pathways related to cell migration, cell polarity, cytoskeletal homeostasis and phagocytosis.
Macrophages or DC transfected with miR-142–3p mimics display impaired cellular functions compared to control mimic. miR-142–3p-mediated downregulation of multiple genes including vinculin, Dab2, Skap2 (novel validated targets), Vasp and LIMS1 can suppress cellular functions related to cell migration/motility, cell polarity, phagocytosis, and cytoskeletal rearrangement.
Discussion
Functional integrity of APC relies heavily on their ability to detect, uptake and degrade antigens. This dynamic process requires migration of APC in constant search of antigens for presentation to T cells (35). Evidently, regulators of cytoskeletal homeostasis are critical for efficient immune responses. However, the role of miRNAs in the regulation of myeloid cells migration is not well understood. In this study, we demonstrate functional roles of two microRNAs, viz., miR-30b and miR-142–3p in regulating migration and cytoskeletal arrangement in myeloid cells. Both miR-30b and miR-142–3p negatively regulate migration and structural morphology of MΦ and DC suggesting their integral role in cell migration and polarity. Notably, expression of miR-30b and miR-142–3p was drastically reduced during MΦ and DC differentiation. This downregulation may be required for cellular expansion as the size of monocytes increased from ~10 μm to ~25–40 μm as they differentiate to MΦ and DC (data not shown). Thus, downregulation of miR-30b and miR-142–3p expression correlates with the expansion of cytoskeletal networks. Conversely, enforced expression of these miRNAs attenuate migration of myeloid cells as observed by two different wound closure assays and likely impair the cell expansion. Our scanning electron and confocal microscopy images unequivocally show a marked reduction in myeloid cell size (both area and diameter) in miR-142–3p overexpressing cell compared to control mimic. This may reflect perturbation in actin rearrangement or organization and corroborates with a robust attenuation of cell migration noticed in miR-142–3p transfected cells compared with control or miR-30b mimics. This suggests that miR-142–3p specifically regulate genes involved in cytoskeletal homeostasis related pathways.
Recent studies clearly demonstrate that miR-142–3p regulates actin organization by targeting several genes, including cofilin-2, Wasl, Bod1, Twf1, Itgav and Brlf, which collectively control actin filament homeostasis (36). Similarly, Bettencourt et al. showed that intracellular pathogen Mycobacterium tuberculosis induced expression of miR-142–3p leads to targeting of N-Wasp and blocks actin polymerization required for bacteria persistence suggesting that pathogens target this cellular mechanism to disrupt cytoskeletal homeostasis to facilitate their persistence (37). Consistent with these observations, miR-142–3p is downregulated in CD4+ T cells collected from subjects with arteriosclerosis obliterans compared with healthy counterparts (38). This allows higher expression of miR-142–3p which targets Ras-related C3 botulinum toxin substrate 1 (RAC1) and Rho Associated Coiled-Coil Containing Protein Kinase 2 (ROCK2), causing enhanced migration of CD4+ T cells to vascular walls (38). Cell migration defects associated with miR-30b are reported through targeting of key genes in cell cytoskeleton homeostasis. For instance, miR-30 family control cell migration by silencing Vimentin and insulin receptor substrate 2 (IRS2) levels (39, 40). Although migration assays showed highly similar defects in miR-30b or miR-142–3p overexpressing myeloid cells, SEM images in contrast show distinct morphological differences in terms of cell size and dendrite density. These results indicate that these miRNAs target overlapping yet exclusive pathways. Indeed, our previous results show that these miRNAs impact expression of various genes associated with phagocytosis (30). Many of these regulate cellular structural dynamics, including Phospholipase C (PLC) and Protein Kinase C (PKC) family members. Taken together, these results support findings that miR-30b and miR-142–3p negatively regulate various critical components of cytoskeleton dynamics leading to altered morphology of cells as observed for DCs, and which may significantly impair their capacity to activate T-cells.
Expression of both miR-30b and miR-142–3p is downregulated in various cancers including non-small cell lung cancer (NSCLC) (41), colorectal cancer (CRC) (42), gastric cancer (43), breast cancer (44), pancreatic adenocarcinoma (45), periodontal disease (46), and arteriosclerosis obliterans (38). Downregulation of these miRNA is associated with aggressive forms of cancers. In our previous microarray analysis of differentiating macrophages and DC, all members of the miR-30 family and miR-142–3p (also 5p isoform) exhibit downregulated expression patterns suggesting that they share biological functions. miR-30 family is ubiquitously expressed and consists of five members: miR-30a, -b, -c, -d, -e., while miR-142–3p is predominantly expressed in hematopoietic cells. Their role in cell development and various biological processes is well reported (16, 17, 29, 30, 36–42). While extensive studies on cancer biopsies have shown that miR-30 family and miR-142–3p acts as tumor suppressors, their role in myeloid cell migration and movement is largely unexplored. By targeting critical genes involved in cell migration, miR-30b and miR-142–3p are shown to regulate tumor initiation, development and metastasis. For instance, downregulation of miR-30b is mediated by SIX1 (SIX homeobox 1), a member of the SIX homeodomain family of transcription factors and an EMT (epithelial-mesenchymal transition)-promoting gene (42). In NLSCC, miR-30b is downregulated leading to upregulation of collagen triple helix repeat containing 1 (Cthrc1), a gene associated with the development of some forms of cancer (41). miR-142 also acts as a tumor suppressor gene by regulating cell cycle progression, cell migration, apoptosis, and invasion. miR-142–3p and its target estrogen receptor 1 (ESR1), a positive regulator of cell growth, proliferation and differentiation, exhibit antagonistic expression in ER-positive breast cancer (44). In line with published studies, our findings unequivocally demonstrate that miR-30b and miR-142–3p inhibit monocyte-derived MΦ and -DC migration and induce structural defects causing impairment of key cellular and biological functions.
miR-142–3p-mediated impairment of cell migration and motility occur by disrupting actin homeostasis and cytoskeletal structure causing loss of cell polarity. Live cell imaging of MΦ transfected with miR-30b and miR-142–3p showed multidirectional movements instead of predominantly unidirectional movement (as observed in control mimic transfected cells) to close the healing wound thus demonstrating a loss of cell polarity. Indeed, F-actin in MΦ transfected with miR-142–3p shows a non-homogeneous distribution, disrupted actin fibers and a cell size reduction. Schwickert et al. showed that miR-142–3p reduced the expression of many factors related to cytoskeletal function, such as WASL (N-WASP), CFL2, RAC1 and ROCK2 by direct interaction that resulted in membrane reduction, cell shape changes, reduction in cell size and a significant inhibition of breast cancer cell invasiveness (47). In this context, the cytotoxic drug, OSW-1, with antitumor effects used against several types of malignant cells has been reported to regulate miR-142–3p expression. Higher levels of miR-142–3p were observed in OSW-1+doxorubicine treated cells compared to doxorubicine alone (48). Deregulation of miR-142 by OSC-1 can impair cell proliferation, differentiation, apoptosis, cell adhesion, migration, polarity and mesenchymal transition (48). In another study, miR-142–3p was demonstrated to control actin cytoskeleton by targeting of small GTPase translation in neutrophils at the wound sites (49). Neutrophils from miR-142−/− mice exhibit altered phagocytosis as a consequence of dysregulated chemotactic behavior, including enhanced F-actin assembly, disturbed cell polarity, and increased cell motility (49).
Global transcriptome profiling identified a large subset of genes regulated by miR-142–3p. Among these, cell migration, movement, endocytosis, and antigen presentation-related pathways were among the top hits. Indeed, a number of these key genes, such as LIMS1, VASP, VCL, DAB2, SKAP2, CLTC and MAPK1 are related to cytoskeleton homeostasis signaling pathways. These findings are in line with transcriptome analysis in miR-142 KO mice (36). Cytoskeletal pathway related genes are specifically targeted by miR-142 that generate 3p and 5p mature miRNAs; however, 3p isoform is more predominant and hence may play a critical role in regulating migration and actin homeostasis. Based on our functional assays, transcriptome analysis, bioinformatics prediction of miR-142–3p targets and their validation, a role of miRNA in regulating myeloid cell migration/motility, cell polarity, phagocytosis, cytoskeletal homeostasis is shown in schematic (Figure 6). Concurrent suppression of multiple genes (including LIMS1, VASP, VCL, DAB2, SKAP2) by miR-142–3p allows it to modulate various biological networks. Besides cell migration related biological processes, several pathways related to immune response were identified in global pathway analysis. Studies from our lab and others have reported an integral role of miR-142 in shaping innate and adaptive immune responses. Expression of miR-142 is highly responsive to various stimuli including bacterial challenge, LPS, IgG coated beads and several immune-mediated diseases and cancers and is generally downregulated (16, 17, 29, 30). In conclusion, our results show that in addition to immune response, miR-142–3p also negatively regulates cytoskeletal homeostasis.
Supplementary Material
Supplemental File 3. Transcriptome profiling in miR-142-3p overexpressing dendritic cells. Microarray was performed for global expression of transcripts impacted by miR-142-3p overexpression. Excel sheet shows the transcriptome analysis in DC transfected with control or miR-142-3p mimic. The list of all the genes detected in the microarray are included in sheet labelled “Complete Microarray Data”. In this sheet, Transcript ID, Gene assignment, Gene symbol, RefSeq, GO Biological process, GO Cellular component, GO Molecular function, Pathway, P value (miR-142-3p mimic vs Control), Fold change (miR-142-3p vs Control) is provided for each gene. In sheet labeled as “Up and Down regulated genes”, the list of differentially expressed up- (blue highlighted) and down (yellow highlighted) -regulated genes with p value <0.05 and a cut-off between −1.25 and 1.25 are listed.
Supplemental File 4. Global pathway analysis of differentially expressed transcripts using Reactome Pathway. Reactome Pathway Database software was used to predict the networks and canonical pathways that are associated with the differentially expressed genes in miR-142-3p transfected dendritic cells compared to control mimic. List of 25 most relevant pathways regulated by differentially expressed genes is provided.
Supplemental File 5. List of predicted miR-142-3p binding sites on gene targets. Predicted miR-142-3p binding sites on the downregulated genes identified using RNA Hybrid analysis. Sequence alignment of selected potential miR-142-3p binding sites with mfe<−20 kcal/mol are shown. Interactions highlighted in yellow are selected as they fulfilled all the criteria listed in the results and methods.
Highlights.
miR-30b and miR-142–3p regulate myeloid cell migration and phagocytosis.
Myeloid cells overexpressing miR-30b and miR-142–3p exhibit defects in structural morphology and cytoskeletal rearrangement.
Global transcriptome profiling in dendritic cells identified ~850 genes regulated by miR-142–3p.
miR-142–3p directly targets genes involved in cytoskeleton homeostasis supporting its role in cell motility, polarity, and migration pathways.
Acknowledgements
Part of this work was funded by the NIH/NIDCR R01 DE027980, R21 DE026259-01A1 and R03 DE027147 (ARN) and R01 DE021052 (SN).
Footnotes
Supplementary material
Supplemental File 1. Impaired cell migration and loss of the polarity in macrophages overexpressing miRNA-30b and miR-142-3p. Primary human macrophages transfected with (A) control, (B) miR-30b or (C) miR-142-3p mimic and a scratch was introduced after 24 h. Time course analysis of cell migration was performed for 48 h. Yellow lines indicate the invasive front in the wound healing assay. ImageJ analysis was performed to quantify the wound closure for 72 h. The scratched area at control (0 h) was arbitrarily assigned as 100%.
Supplemental File 2. Live cell imaging of bacterial phagocytosis in miR-30b and miR-142-3p overexpressing macrophages. Movies were captured using live imaging microscopy during 60 min in MΦ transfected with (A) control, (B) miR-30b or (C) miR-142-3p mimic and incubated with latex beads coated with IgG FITC. Final magnification: X200, scale bar corresponds to 50 μm.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. Development of monocytes, macrophages and dendritic cells. Science. 2010; 327:656–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hartenstein V, Martinez P. Phagocytosis in cellular defense and nutrition: a food-centered approach to the evolution of macrophages. Cell Tissue Res 2019; 377:527–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guilliams M, Ginhoux F, Jakubzick C, et al. Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny. Nat. Rev. Immunol 2014; 14:571–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Serbina NV, Jia T, Hohl TM, Pamer EG. Monocyte-Mediated Defense Against Microbial Pathogens. Annu Rev Immunol 2008; 26:421–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol 2011; 11:723–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding P, Wu H, Fang L, Wu M, Liu R. Transmigration and Phagocytosis of Macrophages in an Airway Infection Model Using Four-dimensional Techniques. Am J Respir Cell Mol Biol 2014; 51:1–10. [DOI] [PubMed] [Google Scholar]
- 7.Rougerie P, Miskolci V, Cox D. Generation of membrane structures during phagocytosis and chemotaxis of macrophages: role and regulation of the actin cytoskeleton. Immunol Rev 2013; 256:222–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004; 116: 281–297. [DOI] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009; 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs”. Ann Rev Biochem 2010; 79:351–379. [DOI] [PubMed] [Google Scholar]
- 11.Naqvi AR, Islam MN, Choudhury NR, Haq QM. The fascinating world of RNA interference. Int J Biol Sci 2009; 5:97–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miska EA. How microRNAs control cell division, differentiation and death. Curr Opin Genet Dev 2005; 5: 563–568. [DOI] [PubMed] [Google Scholar]
- 13.Subramanian S, Steer CJ. MicroRNAs as gatekeepers of apoptosis. J Cell Physiol 2010; 223:289–298. [DOI] [PubMed] [Google Scholar]
- 14.Liu M, Zhang Y, Zhang J, et al. MicroRNA-1253 suppresses cell proliferation and invasion of non-small-cell lung carcinoma by targeting WNT5A. Cell Death Dis 2018; 9:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mayoral RJ, Deho L, Rusca N, et al. MiR-221 influences effector functions and actin cytoskeleton in mast cells. PLoS One. 2011; 6:e26133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naqvi AR, Fordham JB, Nares S. MicroRNA target Fc receptors to regulate antibody-dependent antigen uptake in primary macrophages and dendritic cells. Innate Immun 2016; 22: 510–521. [DOI] [PubMed] [Google Scholar]
- 17.Naqvi AR, Fordham JB, Ganesh B, Nares S. miR-24, miR-30b and miR-142–3p interfere with antigen processing and presentation by primary macrophages and dendritic cells. Sci Rep 2016; 6:32925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009; 19:92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and downregulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002; 99:15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol 2012; 6:590–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saunders MA, Liang H, Li WH. Human polymorphism at microRNAs and microRNA target sites. Proc. Natl Acad. Sci. USA. 2007; 104:3300–3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen K, Rajewsky N. Natural selection on human microRNA binding sites inferred from SNP data. Nat Genet 2006; 38:1452–1456. [DOI] [PubMed] [Google Scholar]
- 24.Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 2008; 10:593–601. [DOI] [PubMed] [Google Scholar]
- 25.Burk U, Schubert J, Wellner U, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep 2008; 9:582–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang L, Liu X, Kolokythas A, et al. Downregulation of the Rho GTPase signaling pathway is involved in the microRNA-138-mediated inhibition of cell migration and invasion in tongue squamous cell carcinoma. Int J Cancer. 2010; 127:505–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koch U, Radtke F. Notch and cancer: a double-edged sword. Cell Mol Life Sci 2007; 64:2746–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amelio I, Lena AM, Viticchiè G, et al. miR-24 triggers epidermal differentiation by controlling actin adhesion and cell migration. J Cell Biol 2012; 199:347–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fordham JB, Naqvi AR, Nares S. Regulation of miR-24, miR-30b, and miR-142–3p during macrophage and dendritic cell differentiation potentiates innate immunity. J Leukoc Biol 2015; 98:195–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naqvi AR, Fordham JB, Nares S. miR-24, miR-30b, and miR-142–3p regulate phagocytosis in myeloid inflammatory cells. J. Immunol 2015; 194:1916–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naqvi AR, Seal A, Shango J, Shukla D, Nares S. In silico prediction of cellular gene targets of herpesvirus encoded microRNAs. Data Brief. 2018; 19:249–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naqvi AR, Fordham JB, Khan A, Nares S. MicroRNAs responsive to Aggregatibacter actinomycetemcomitans and Porphyromonas gingivalis LPS modulate expression of genes regulating innate immunity in human macrophages. Innate Immun 2014; 20:540–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soehnlein O, Lindbom L. Phagocyte partnership during the onset and resolution of inflammation. Nat Rev Immunol 2010; 10:427–439. [DOI] [PubMed] [Google Scholar]
- 34.Thauland TJ, Parker DC. Diversity in immunological synapse structure. Immunology. 2010; 131:466–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hivroz C, Chemin K, Tourret M, Bohineust A. Crosstalk between T lymphocytes and dendritic cells. Crit Rev Immunol 2012; 32:139–155. [DOI] [PubMed] [Google Scholar]
- 36.Chapnik E, Rivkin N, Mildner A, et al. miR-142 orchestrates a network of actin cytoskeleton regulators during megakaryopoiesis. Elife. 2014; 3:e01964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bettencourt P, Marion S, Pires D, et al. Actin-binding protein regulation by microRNAs as a novel microbial strategy to modulate phagocytosis by host cells: the case of N-Wasp and miR-142–3p. Front Cell Infect Microbiol 2013; 3:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J, Li W, Wang S, et al. MiR-142–3p attenuates the migration of CD4+ T cells through regulating actin cytoskeleton via RAC1 and ROCK2 in arteriosclerosis obliterans. PLoS One. 2014; 9:e95514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng CW, Wang HW, Chang CW, et al. MicroRNA-30a inhibits cell migration and invasion by downregulating vimentin expression and is a potential prognostic marker in breast cancer. Breast Cancer Res Treat. 2012; 134:1081–1093. [DOI] [PubMed] [Google Scholar]
- 40.Yang SJ, Yang SY, Wang DD, et al. The miR-30 family: Versatile players in breast cancer. Tumour Biol 2017; 39:1010428317692204. [DOI] [PubMed] [Google Scholar]
- 41.Chen S, Li P, Yang R, et al. microRNA-30b inhibits cell invasion and migration through targeting collagen triple helix repeat containing 1 in non-small cell lung cancer. Cancer Cell Int 2015; 15:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao H, Xu Z, Qin H, Gao Z, Gao L. miR-30b regulates migration and invasion of human colorectal cancer via SIX1. Biochem J. 2014; 460:117–125. [DOI] [PubMed] [Google Scholar]
- 43.Qiao F, Zhang K, Gong P, Wang L, Hu J, Lu S, Fan H. Decreased miR-30b-5p expression by DNMT1 methylation regulation involved in gastric cancer metastasis. Mol Biol Rep 2014; 41:5693–5700. [DOI] [PubMed] [Google Scholar]
- 44.Mansoori B, Mohammadi A, Gjerstorff MF, et al. miR-142–3p is a tumor suppressor that inhibits estrogen receptor expression in ER-positive breast cancer. J Cell Physiol 2019. 10.1002/jcp.28263 [DOI] [PubMed] [Google Scholar]
- 45.Godfrey JD, Morton JP, Wilczynska A, Sansom OJ, Bushell MD. MiR-142–3p is downregulated in aggressive p53 mutant mouse models of pancreatic ductal adenocarcinoma by hypermethylation of its locus. Cell Death Dis 2018; 9:644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Naqvi AR, Brambila MF, Martínez G, Chapa G, Nares S. Dysregulation of human miRNAs and increased prevalence of HHV miRNAs in obese periodontitis subjects. J Clin Periodontol 2019; 46:51–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwickert A, Weghake E, Brüggemann K, et al. microRNA miR-142–3p Inhibits Breast Cancer Cell Invasiveness by Synchronous Targeting of WASL, Integrin Alpha V, and Additional Cytoskeletal Elements. PLoS One. 2015; 10:e0143993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jin JC, Jin XL, Zhang X, Piao YS, Liu SP. Effect of OSW-1 on microRNA expression profiles of hepatoma cells and functions of novel microRNAs. Mol Med Rep 2013; 7:1831–1837. [DOI] [PubMed] [Google Scholar]
- 49.Tanaka K, Kim SE, Yano H, et al. MiR-142 Is Required for Staphylococcus aureus Clearance at Skin Wound Sites via Small GTPase-Mediated Regulation of the Neutrophil Actin Cytoskeleton. J Invest Dermatol 2017; 137:931–940. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental File 3. Transcriptome profiling in miR-142-3p overexpressing dendritic cells. Microarray was performed for global expression of transcripts impacted by miR-142-3p overexpression. Excel sheet shows the transcriptome analysis in DC transfected with control or miR-142-3p mimic. The list of all the genes detected in the microarray are included in sheet labelled “Complete Microarray Data”. In this sheet, Transcript ID, Gene assignment, Gene symbol, RefSeq, GO Biological process, GO Cellular component, GO Molecular function, Pathway, P value (miR-142-3p mimic vs Control), Fold change (miR-142-3p vs Control) is provided for each gene. In sheet labeled as “Up and Down regulated genes”, the list of differentially expressed up- (blue highlighted) and down (yellow highlighted) -regulated genes with p value <0.05 and a cut-off between −1.25 and 1.25 are listed.
Supplemental File 4. Global pathway analysis of differentially expressed transcripts using Reactome Pathway. Reactome Pathway Database software was used to predict the networks and canonical pathways that are associated with the differentially expressed genes in miR-142-3p transfected dendritic cells compared to control mimic. List of 25 most relevant pathways regulated by differentially expressed genes is provided.
Supplemental File 5. List of predicted miR-142-3p binding sites on gene targets. Predicted miR-142-3p binding sites on the downregulated genes identified using RNA Hybrid analysis. Sequence alignment of selected potential miR-142-3p binding sites with mfe<−20 kcal/mol are shown. Interactions highlighted in yellow are selected as they fulfilled all the criteria listed in the results and methods.