Abstract
The innovative discovery of aptamers was based on target-specific treatment in clinical diagnostics and therapeutics. Aptamers are synthetic, single-stranded oligonucleotides, simply described as chemical antibodies, which can bind to diverse targets with high specificity and affinity. Aptamers are synthesized by the SELEX technique, and possess distinctive properties as small size (10–50 kDa), higher stability, easy manufacture and less immunogenicity. These oligonucleotides are easily degraded by nucleases, so require some important modifications like capping and incorporation of modified nucleotides. RNA aptamers can be modified chemically on 2′ positions using -NH3, -F, -deoxy, or -OMe groups to enhance their nuclease resistance. Aptamers have been employed for multiple purposes, as direct drugs or aptamer–drug conjugates targeted against different diseased cells. Different aptamer-conjugated nanovehicles (e.g., micelles, liposomes, silica nano-shells) have been designed to transport diverse anticancer-drugs like doxorubicin and cisplatin in bulk to minimize systemic cytotoxicity. Some drug-loaded nanovehicles (up to 97% loading capacity) and conjugated with specific aptamer resulted in more than 60% tumor inhibition as compared to unconjugated drug-loaded nanovehicles which showed only 31% cancer inhibition. In addition, aptamers have been widely used in basic research, food safety, environmental monitoring, clinical diagnostics and therapeutics. Different FDA-approved RNA and DNA aptamers are now available in the market, used for the treatment of diverse diseases, especially cancer. These aptamers include Macugen, Pegaptanib, etc. Despite a good progress in aptamer use, the present-day chemotherapeutics and drug targeting systems still face great challenges. Here in this review article, we are discussing nucleic acid aptamers, preparation, role in the transportation of different nanoparticle vehicles and their applications as therapeutic agents.
Keywords: Nucleic acid aptamers, SELEX, Aptamosomes, Micelles, Drug delivery, Therapeutics
Introduction
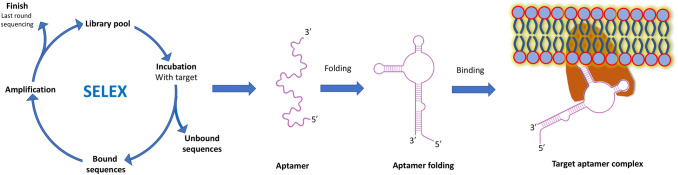
The approach of constant search for innovative pharmacological and diagnostic agents against different diseases also led to the discovery of aptamers. The oligonucleotide aptamers are relatively small single-stranded RNA, DNA or XNA (xeno nucleic acid) molecules (20–100 bases), which can selectively bind with almost any associated target like ions, small molecules, peptides, proteins, or even whole cells (Mascini 2009; Hoppe-Seyler et al. 2004). The complex three-dimensional shape of an aptamer by its folding allows it to fit and bind with the specific target with great specificity and affinity (Hermann and Patel 2000). Aptamers possess the same level of target binding affinity like monoclonal antibodies with a dissociation constant (Kd) usually varies from 0.1 to 50 nm (Fig. 1) (Hianik et al. 2007).
Fig. 1.
A schematic presentation of SELEX, aptamer folding and its binding with a final target protein. The aptamer folds into a specific three-dimensional structure and binds with a target molecule (e.g., a protein) which results in the formation of a target-aptamer complex
Aptamers have several advantages over their competitors like monoclonal antibodies, as they possess low toxicity, less immunogenicity, easier synthesis and efficient stability against harsh environmental conditions. These molecules are cheaper and also possess a good shelf life, strong tolerance against surfactants, low batch to batch variation, and their chemical modifications are easily performed (Chen and Yang 2015). Unlike antibodies, which require animals or cell lines for their production, the aptamers synthesis is fully accomplished by in vitro environment which minimizes the contamination risks by viruses or bacteria. These characteristics of aptamers promote them as smart therapeutic agents and efficient substitutes for antibodies (Osborne et al. 1997). Due to some stability limits of native RNA and DNA aptamers in cellular environments and biological fluids, a growing interest has been focused on their backbone modifications, which has resulted in the formation of xeno nucleic acids (XNA). These modified nucleic acids are not readily recognised and degraded by nucleases and, thus, are well-suited for in vivo applications (Ma et al. 2016; Torres et al. 2018).
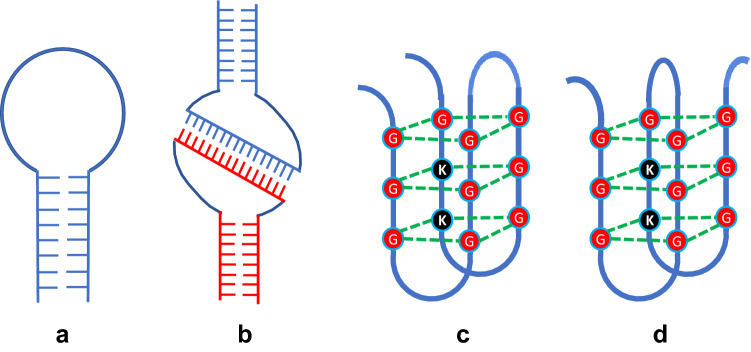
The oligonucleotide aptamers implement different three-dimensional structures like hairpin (stem) loop, kissing complexes, and quadraplexes. These distinctive structures help the aptamers to bind with diverse targets and enhances their strong specificity and activity. (Miao and Westhof 2017) (Fig. 2).
Fig. 2.
Aptamer three-dimensional structures a hairpin (stem) loop; b kissing complex; c quadraplex (side by side); d quadraplex (cross-over)
The hairpin (stem) loop is formed when two portions of the same nucleic acid strand are complementary to each other and can form Watson–Crick nitrogenous base pairing. A more advanced structure occurs by kissing complex formation, which is obtained when a few unpaired nitrogenous bases of one hairpin loop can base pair with the complementary unpaired nitrogenous bases in another hairpin loop. This type of structure usually occurs in RNA aptamers. In addition to these two structures, quadraplexes are formed either side by side or in cross-over fashion, when a guanine-rich sequence with four guanine nitrogenous bases associates with each other through hydrogen bonding (Fig. 2) (Zhou and Rossi 2017).
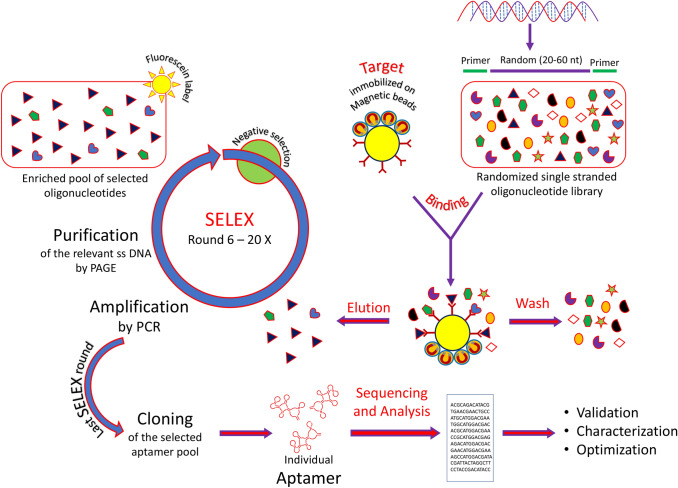
Aptamers are synthesized in vitro by a well-defined technique known as Systematic Evolution of Ligands by EXponential enrichment (SELEX) method (Turek and Gold 1990). This procedure is used to generate both single-stranded DNA and RNA aptamers that fold into a definite three-dimensional structures which can specifically bind to a target ligand (Figs. 1, 3).
Fig. 3.
Diagrammatic illustration of SELEX starting with ssDNA or RNA library of random oligonucleotides. After binding with the target of interest, the unbound oligonucleotides are washed from the bound sequence, eluted and amplified by PCR/RT-PCR. The subsequent enriched DNA/RNA pool is taken for the next round of selection
The earliest reported SELEX procedures have been difficult, laborious, and time-consuming methods. But since the last three decades, the recent development in this technology has been reported to reduce the processing time from months to a few hours (Zhuo et al. 2017). The comprehensive reduction of SELEX cycle timing has drastically revolutionized the aptamer market and it is now efficiently used in basic research, pharmaceutical industries and environmental monitoring. The overall striking properties of aptamers have led them to be used as promising tools with great potential in biomedical applications, including specific targeting ligands, diagnostics, biosensing, biotechnology, therapy and biomarker discovery and even genetic control devices (Radom et al. 2013). Globally the aptamer market was expected to be at almost $107 million in 2015 which may touch roughly $244 million in 2020 (Zhu et al. 2015). In this review article, we are fully focusing on nucleic acid aptamers, types, preparation and their role in clinical applications.
Oligonucleotide aptamers and its types
The primary choice in oligonucleotide aptamers selection remains whether to take RNA, DNA or XNA aptamers for further research (Ulrich et al. 2004). The earlier studies about aptamers were mainly focused on RNA as it forms more distinct and complicated three-dimensional structures with a greater number of conformations as compared to DNA aptamers.
Even though, both the DNA and RNA oligonucleotide aptamers have been certified to be effective in different applications, still there remains some basic differences between them, which help us to decide for the selection of each aptamer for a specific application. Functionally, these two aptamers are similar but vary in accessibility and stability (Min et al. 2008). RNA aptamers are unstable due to the reactive −OH group at the 2′ position of ribose sugar (Prakash 2011). This group gets easily deprotonated especially in an alkaline medium which further results in the breakdown of phosphodiester linkage, leading to RNA hydrolysis. In comparison to DNA aptamers, RNA aptamers need extra chemical modifications to improve their chemical durability (Wang et al. 2011a, b).
The initial decision about the selection of a nucleic acid library provides a crucial starting point for SELEX. For DNA aptamer requirements, the primary library should be ssDNA and for RNA aptamer, a dsDNA library is transcribed to generate the starting RNA library. So for RNA aptamers, a reverse transcription step is mandatory in each cycle of PCR and succeeding transcription for the following cycles of SELEX. This interconversion step amid RNA and DNA during SELEX cycles makes the task more expensive and time-consuming for the preparation of RNA aptamers (Bugaut et al. 2006). The DNA aptamers form larger structures as compared to RNA aptamers, even bearing the equal number of monomers. Recently, several two- and three-dimensional structures have been evaluated, that assist the stability and specific binding of DNA aptamers. These include internal loop, bulge loop, hairpin, and G-quadruplex folds (Sundaram et al. 2013) (Fig. 2). Herein, we summarize the different properties of DNA, RNA and XNA aptamers and their possible chemical modifications to improve their varied features.
DNA aptamers
Native DNA aptamers are comparatively more stable and less reactive as compared to RNA aptamers. The in vitro half-life of DNA aptamer is reported to be 30–60 min in plasma as compared to RNA aptamers which possess only a few seconds half-life (White et al. 2000). In comparison to RNA aptamers, DNA aptamers possess enhanced stability against biodegradation, which brands them superior for clinical applications (Orava et al. 2010).
The inherent advantage in stability of DNA aptamers over its RNA counterparts is because of the C–H bonds at the 2′ position of the deoxyribose sugar of DNA nucleotide. For the purpose of getting additional complicated structures, the DNA aptamers are generally selected from the libraries of longer randomized regions (Ravelet et al. 2006).
One of the biggest challenges which researchers face during the applications of DNA aptamers is its less stability when treated with complicated biological samples. The degradation of nucleic acid aptamers by nucleases is still a challenge that limits its use (Ma et al. 2015). A novel strategy to enhance the stability of DNA aptamers against nucleases is by capping at its one end (Shoji et al. 2007). The incorporation of some unusual nucleotides like locked nucleic acids (LNA) is also a good approach to increase its stability against nucleases. The synthesis of spiegelmers, also known as mirror aptamers, is also a new approach to overcome the nuclease attacks (Singh et al. 1998).
DNA aptamers have been found to be quite useful for clinical purposes. These aptamers are easier to manufacture and are comparatively stable in harsh conditions. These aptamers can be conjugated to different nanomedicine-based formulations including liposomes. In addition to this, the concentration of DNA aptamers can be controlled by inhibition with complementary DNA sequences to avoid their overdose during drug delivery.
RNA aptamers
RNA aptamers are a class of oligonucleotides which bind with high specificity and affinity with a definite target, almost like antigen–antibody reaction. These aptamers are thermodynamically more stable as compared to peptide aptamers. Like antibodies, RNA aptamers possess a unique three-dimensional structure that improves its specificity and form more tighter binding with the target molecule (Ulrich, 2006).
Different types of RNA aptamers have been synthesized and used as therapeutic, diagnostic and prognostic tools in a broad range of applications like macular degeneration, viral infections, cancer etc. (Kang et al. 2012). Due to their smaller size as compared to DNA aptamers, even with the same number of nucleotides, it becomes easier for it to enter the cells of interest.
RNA aptamers are also synthesized by the method of SELEX, but the method is longer than DNA synthesis. RNA aptamers are manufactured in a good amount and possess almost no immunogenicity as compared to antibody synthesis. Besides this, the method of preparation works in a controlled manner and the structure and stoichiometry remains properly in order. In addition, RNA aptamers can be further modified chemically on 2′ positions using -NH3, -F, -deoxy, or -OMe groups. These modifications greatly improve the stability against RNAases, commonly found in the blood stream (Soukup et al. 2000; Wang et al. 2011a, b).
In plasma, the half-life of an RNA aptamer is only a few seconds as compared to DNA aptamers, which possess 30–60 min life. Due to the presence of the 2′-OH group, the RNA aptamer is unstable and this particular group makes RNA prone to hydrolysis (Goringer et al. 2008). Furthermore, this group leads to RNA strand scission by endoribonucleases like RNase A. So for these limitation of RNA aptamers, it becomes mandatory to chemically modify this molecule to enhance its life time. Most RNA aptamers are modified through pyrimidine present at 2′-position, to upgrade its chemical stability. Once modified, these RNA molecules even surpass the DNA aptamer stability. However, the higher reactivity of RNA aptamers and certain non-Watson–Crick base pairings, it leads the RNA aptamers to form complex and more varied three-dimensional structures, which is a good choice to have higher affinity and more specificity aptamers (Yan et al. 2004).
XNA aptamers
The practice of chemical modifications in pentose sugar moieties in DNA and RNA oligonucleotides leads to their outstanding resistance against enzymatic breakdown. These chemically modified nucleic acids are known as xenobiotic nucleic acids (XNA) (Pinheiro and Holliger 2012). XNAs vary in their backbone structure significantly from natural DNA and RNA structures. The enzymatic synthesis and enrichment of XNA libraries has been practical, due to the discovery of specialized XNA polymerases (Rangel et al. 2018). These XNA polymerases are capable of reading a DNA template to synthesize XNA sequences. However, till now, only a few chemical modifications have been successful for in vitro synthesis of XNA (Eremeeva et al. 2019).
XNA have played a great role as aptamers to combat some drastic diseases like cancer. The different modifications at the 2′-OH group of RNA pentose sugar led to the generation of Pegaptanib, which is an anti-human VEGF165 aptamer. The backbone of this aptamer is chemically modified during or post-SELEX cycles with alternate 2′-OMe-purine and 2′-F-pyrimidine nucleotides. The earliest report of XNA manufacture and transcription of OMe-modified aptamers with DNA polymerase, was engineered to bind with neutrophil elastase (Barman 2015).
An innovative XNA aptamer showing high affinity with HIV-1 reverse transcriptase was synthesized using 2′-deoxy-2′-fluoroarabino nucleic acid (Ferreira-Bravo et al. 2015). In addition to this, octahedron A and human thrombin were found to have high affinity with threose nucleic acid (TNA) aptamer. In addition to the modifications at the 2′ position of pentose sugar, the whole furanose ring has been replaced with six-membered hexose sugar in DNA (Mei et al. 2018). This led to the generation HNA (1,5-anhydrohexitol) aptamer which has been found to form stable duplexes with natural DNA or RNA. The HNA possesses minimal toxicity in cell lines as well as remarkable alkaline, acidic, and nuclease resistance. Random HNA libraries, were used to make aptamers, with a dissociation constant in the nanomolar scale, to bind with hen egg lysozyme and HIV trans-activation response RNA (Lipi et al. 2016).
For a quick comprehension about different oligonucleotide aptamers, their comparative characteristics are presented in tabular form (Table 1).
Table 1.
Characteristics of different oligonucleotide aptamers
| Characteristics | DNA aptamer | RNA aptamer | XNA aptamer |
|---|---|---|---|
| Backbone structure and nucleobases | Phosphate-pentose sugar, regular nucleobases (A, G, C, T) | Phosphate-pentose sugar (regular nucleobases (A, G, C, T, and U) | Phosphate-modified sugars (regular nucleobases (A, G, C, T or U) |
| Method of preparation | SELEX using a DNA polymerase, Amplification by PCR so easiest to synthesize | SELEX using a DNA polymerase, Amplification by RT-PCR, so challenging to synthesize | SELEX using XNA polymerase (e.g. Phi29 DNA polymerase) so challenging to synthesize |
| The nucleic acid library used at the start of SELEX | Native DNA library, SELEX cycle is standard and requires no additional steps | Native RNA library, SELEX cycle is longer as reverse transcriptase is required in each cycle, | Xeno nucleic acid library, and post selection modifications of DNA or RNA bases |
| Post selection modifications | Capping at the ends, addition of locked nucleic acids | Ribose 2ʹ position modification with -NH3, -F, -deoxy, or -OMe groups | Initial native DNA or RNA library whose nucleotides are replaced with 2′ OMe or 2′F nucleotides) |
| Stability and shelf life in different environments | Native DNA aptamer is more chemically and biologically stable (30–60 min), so the selection and application is easier | The native aptamer is unstable due 2ʹ-OH group (only a few seconds). RNA aptamer achieves its good stability after modifications | Outstanding stability |
| Nucleases sensitivity | Very sensitive | Very sensitive | Superb stability |
| Cost and time of the manufacture | DNA-based SELEX is the most cost-effective and least time-consuming | RNA-based SELEX is more costlier than DNA SELEX and time-consuming | Direct selection of aptamer from the XNA library is less expensive and less time-consuming. Post selection modification is expensive and time-consuming |
| Transportation within the cell | Larger in size, cannot enter the cells, only with the help of aid e.g., patch clamping, nanoparticles, liposomes | Smaller in size, so can enter the cells easily or with the help of nanocarriers | Smaller in size, require nanocarriers to be utilized within the cells |
| Important examples | sgc8, AS1411 | Pegaptanib, A15 | HNA, FANA, 2ʹ-fluoro-DNA, TNA (e.g., Macugen, |
Preparation of nucleic acid aptamers
Aptamers have been synthesized in vitro by Tuerk and Gold in 1990 by choosing RNA ligands for T4 DNA polymerase and independently by Ellington and Szostak against organic dyes in the same year (Du et al. 2015). The first group named the technique as SELEX while the second group coined the term aptamer. SELEX procedure is started with a randomly sequenced desired library (1014–1016) of single-stranded nucleic acid sequences (20–100 bases) (Stoltenburg et al. 2012). Each oligonucleotide consists of a fixed sequence on both the terminals and a middle portion with a random region (Fig. 3). These randomly sequenced oligonucleotides are allowed to bind with a purified protein or other compounds like small molecules, nucleic acids, peptides, or even cells usually associated with diseased states (Gopinath 2007). The bound nucleic acids are separated from the one which do not show any binding activities by different methods like chromatography (size-exclusion or affinity), electrophoresis on polyacrylamide gels or by nitro-cellulose binding (Ahirwar and Nahar 2015). The nucleic acids are then separated from their bound counterparts and are further amplified by PCR as DNA molecules.
The earliest SELEX technology was mainly focused to synthesize RNA aptamers as these forms were supposed to from more complex three-dimensional structures as compared to DNA aptamers. The synthesis of RNA aptamers can be started directly from ssRNA or ssDNA library, which are directly incubated with the target. The unbound complexes are separated from the bound complexes. The target bound oligonucleotides are eluted and amplified RT-PCR to synthesize RNA aptamers. It requires T7 RNA polymerase, reverse transcriptase and protection from RNAases before PCR cycle (Ouellet et al. 2015).
After several cycles of selection, the resulting oligonucleotides, known as an aptamer, possess a high specificity and affinity, are enriched and sequenced. To generate superior aptamers, several modifications are introduced in it as well, which improve its binding capacity to different targets or with the immobilization matrix (Tao et al. 2016) (Figs. 1, 3).
XNAs vary from their natural counterparts (DNA and RNA) in at least one chemical constituent moiety: sugar, phosphate, or nucleobase. The backbone modified XNAs are not generally processed by natural DNA polymerase for further processing. This hurdle has been solved by utilizing Phi29 DNA polymerase, a replicative enzyme of B. subtilis bacteriophage Phi29. This DNA polymerase has been efficiently used for the production of various types of XNAs carrying some backbone modifications like 2ʹ-fluoro-2ʹ-deoxyribonucleic acid (2ʹ-fluoro-DNA), 1,5-anhydrohexitol nucleic acid (HNA), and 2ʹ-deoxy-2ʹ-fluoro-arabinonucleic acid (FANA) (Pinheiro and Holliger 2014).
Since the emergence of the SELEX technique, the aptamer development has mainly focused on perfecting the SELEX process further to develop optimized aptamers that can compete with antibodies in all aspects of the pharmaceutical industry. It has now been almost 30 years since then, and more than 25 different SELEX processes have been developed which are briefly summarized as in Table 2.
Table 2.
Brief summary of different SELEX methods and the principles developed with the year of the invention
| S. no. | Year of invention | SELEX technique | Brief principle | References |
|---|---|---|---|---|
| 1 | 1992 | Negative SELEX | Exclusion of non-specific aptamers by mixing the aptamer library with a matrix | Ellington and Szostak (1992) |
| 2 | 1994 | Counter SELEX | Removal of cross-reactive species by mixing with closely related species | Jenison (1994) |
| 3 | 1997 | Genomic SELEX | An easy SELEX method that can identify the genomic sequence binding motif of the organism itself | Singer et al. (1997) |
| 4 | 1997 | In vivo SELEX | Aptamer selection under in vivo conditions made within the cell | Coulter et al. (1997) |
| 5 | 1998 | Chimeric SELEX | Preparation of recombinant aptamers that binds with two different targets | Burke and Willis (1998) |
| 6 | 1999 | Multi-stage SELEX | Aptamers designated against dissimilar targets fused and reselected against such targets through a multi-step process | Wu and Curran (1999) |
| 7 | 1999 | Cell-SELEX | Aptamer preparation targeting the markers of the whole cell | Homann and Goringer (1999) |
| 8 | 2000 | Indirect SELEX | Preparation of special aptamers directed to bind with the native protein in the presence of a metal ions | Kawakami et al. (2000) |
| 9 | 2000 | Photo-SELEX | Light-sensitive nucleotide insertion to design the aptamers targeted directly after UV irradiation | Golden et al. (2000) |
| 10 | 2001 | Toggle SELEX | The synthesis of cross-reactive aptamers while targeting toggling during selective preparation | Bianchini et al. (2001), White et al. (2001) |
| 11 | 2003 | Tailored SELEX | The short and primer free aptamer selection sequences using cleavable primer-hybridization sites | Vater et al. (2003) |
| 12 | 2004 | CE-SELEX | Aptamer selection by the application of electrophoretic mobility shift standards | Mendonsa and Bowser (2004) |
| 13 | 2005 | FluMag-SELEX | The use of fluorescence tagged targets that allows cleaner aptamer selection | Stoltenburg et al. (2005) |
| 14 | 2006 | TECS-SELEX | Recombinant proteins expressed on modified cells used for aptamer selection | Ohuchi et al. (2006) |
| 15 | 2006 | NECEEM (non-SELEX) | Aptamer selection based on non-equilibrium electrophoresis | Berezovski et al. (2006) |
| 16 | 2007 | Nano selection-based SELEX | Atomic force and fluorescence microscopy integration to purify aptamer in a single step | Peng et al. (2007) |
| 17 | 2007 | MonoLEX | Specific sequence selection through chromatography column fragmentation and pyrosequencing | Nitsche et al. (2007) |
| 18 | 2010 | Microfluidic SELEX | The purification of aptamers with the help of microfluidic chips integrating at different steps on a single surface | Cho et al. (2010); Huang et al. (2010) |
| 19 | 2011 | High-throughput SELEX | The evaluation of high-throughput DNA sequencing by the combination of bioinformatic analysis | Hoon et al. (2011) |
| 20 | 2014 | Particle display SELEX | Flow cytometry-based selection of aptamer particles | Wang et al. (2014a, b, c, d) |
| 21 | 2015 | Hi-fidelity SELEX | Digital PCR used to enhance the SELEX progress by the inclusion of fixed sequence blocking elements | Ouellet et al. (2015) |
| 22 | 2016 | Isogenic cell SELEX | The selection of overexpressed isogenic cell line as SELEX target and downregulating it by micro RNA-mediated silencing for the counter selection | Takahashi et al. (2016) |
CE-SELEX capillary electrophoresis SELEX, FluMag fluorescent/magnetic beads, TECS target expressed on cell surface, NECEEM non-equilibrium capillary electrophoresis of equilibrium mixtures, MonoLEX affinity chromatography SELEX
Role of aptamers in the transportation of drugs
The use of specific native drugs for a particular disease confronts different challenges before it reaches to its actual site of action. Some of these challenges include early chemical breakdown, complex biological barriers and lessening of pharmacological activity while reaching the site of action (Yang et al. 2017). Previously, these obstacles were being resolved inappropriately using large doses of drugs either as a single dose or by repetitive use. These tactics often lead to increased healthcare cost, off-target toxicity, and delayed cure (El-Say and El-Sawy 2017). All these problems have forced the healthcare researchers, to design the drugs which can be specially transported and properly targeted at the site of action, while minimizing its build-up at non-targeted normal tissues. Aptamers have proven to be good candidates as drug transporters, as they can be easily conjugated with different drugs directly or by the use of a linker.
Aptamers are designed to recognize and attach with high specificity and affinity with cognate targets like tumor surface receptors (Hicke et al. 2001; Yin et al. 2012). The novel approach of engineering aptamer–drug conjugates, as an efficient drug delivery systems, briefly involves the attachment of different drugs with specific aptamers, either covalently or non-covalently. It may also involve the use of some specific linkers between the drugs and the aptamer. This system has been used to deliver general therapeutic and chemotherapeutic agents, photosensitizer agents and even protein/peptide molecules. Some of the examples of aptamers attached to specific drugs and targeted against some disease sites are listed in Table 3.
Table 3.
Examples of some aptamers directly conjugated with different drugs and targeted to specific diseased sites
| Aptamer and its type | Drug | Target sites | References |
|---|---|---|---|
| DNA aptamer (P19) | MMAE | Anti-Pancreatic ductal adenocarcinoma | Yoo et al. (2017) |
| Long repetitive drug intercalating dsDNA | Doxorubicin | antitumor efficacy in leukemia mouse xenograft tumor model | Zhu et al. (2013) |
| NucA aptamers | MMAE and MMAF | Tf and EGF receptor binding on pancreatic cancer cell lines | Kratschmer and Levy (2018) |
| Nucleolin aptamer | PTX | Human ovarian cancer xenografted mouse model | Li et al. (2017) |
| DNA aptamer sgc8 | Doxorubicin | protein tyrosine kinase 7 overexpressed on many types of cancers | Meng et al. (2010) |
| DNA sgc8 aptamer | N-heterocyclic carbene–gold complexes | CCRF-CEM leukemia cells | Niu et al. (2016) |
| DNA strand P19 | 5-fluorouracil | pancreatic cancer cell line PANC-l cells | Kruspe and Hahn (2014) |
|
dsDNA/dsRNA sgc8, AS1411 |
Anthracycline drugs (Dox, Epirubicin, Daunorubicin), cisplatin | Tumor growth, leukemia, liver cancer xenograft, | Zhu et al. (2012a) |
| RNA aptamer A10 | Doxorubicin | PMSA targeting, anticancer drug delivery against cancer and leukemia xenograft model | Bagalkot et al. (2006) |
| Sgc8 + sgd5a | Anticancer drug delivery | Cancer detection | Zhu et al. (2012b) |
| Phosphoramidite linker with sgc8 | 5-flurorouracil | Colorectal and pancreatic cancer treatment | Wang et al. (2014a, b, c, d) |
MMAE monomethyl auristatin E, MMAF monomethyl auristatin F, Tf transferrin, EGF epidermal growth factor, PTX paclitaxel
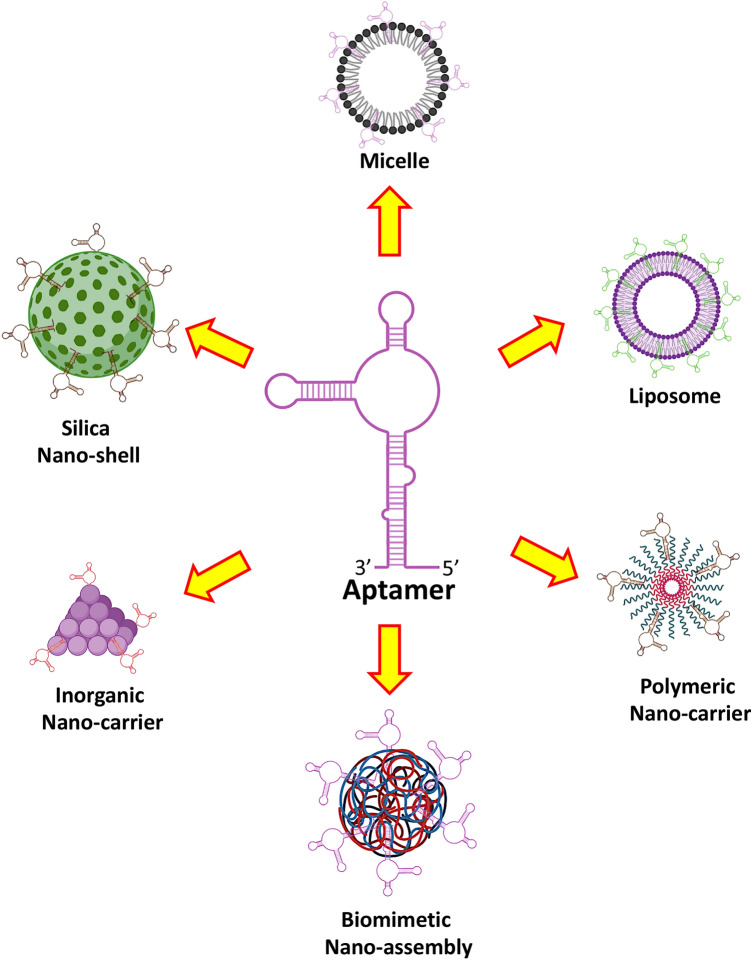
Due to the unique intrinsic properties of aptamers, different aptamer-mediated nanovehicles have been designed to transport in bulk diverse anticancer drugs against various tumor cells to minimize systemic cytotoxicity (Fig. 4). Despite a good progress in synthesis and selection of aptamers to conjugate with different nano-vehicles, the present-day chemotherapy and drug targeting systems still face a great challenge (Zhou and Rossi 2017). Some of the most common nano-vehicles carrying the anticancer drugs which are conjugated with different aptamers are discussed here.
Fig. 4.
Some common examples of nanocarriers loaded with specific drugs and conjugated with definite aptamers to enhance their targeted delivery to minimize their non-specific cytotoxicity
Liposomes (Aptamosomes)
Liposomes are spherical lipid bilayer vesicles that have been used as the most efficient drug nanocarriers (Khan et al. 2020; Brandl 2001). Different types of liposomes have been tagged with aptamers by direct conjugation or by post-insertion of an aptamer attached to lipid anchor (Cao et al. 2009). Liposomes conjugated with aptamers are also known as aptamosomes, which are used to deliver chemotherapeutic molecules like doxorubicin and cisplatin, imaging agents like Actinium derivatives (Ac225) and Gadolinium (Gd3+), and nucleic acids like siRNA (Baek et al. 2014).
The role of Anti-nucleolin aptamer (AS1411) has been studied both as in vitro and in vivo, by conjugation with doxorubicin-loaded liposomes. These aptamosomes have been tested on MCF cells and a mouse model of xenograft human breast cancer (Hua et al. 2013). It has been observed that doxorubicin-loaded AS1411-aptamosomes show higher cytotoxic effects as compared to control liposomes, liganded with some other non-specific DNA aptamer. Similarly, AS1411 aptamosomes carrying cisplatin were used against NCL positive MCF-7 cells and NCL negative LNCaP cells (Vandghanooni et al. 2018). The cytotoxic activity was reported to be more in MCF-7 cells as compared to LNCaP cells. Some more examples of aptamers, drug-loaded aptamosomes and the target cells are given in Table 4.
Table 4.
Application of different types of aptamosomes loaded with specific drugs and liganded with varied aptamers
| S. no. | Aptamer | Drug used | Application of aptamosome | References |
|---|---|---|---|---|
| 1 | DNA (sgc8) | FITC-dextran | Used against CEM-CCRF acute leukemia cells targeting the protein tyrosine kinase receptor | Kang et al. (2010) |
| 2 | DNA (AraHH001) | Anti-angiogenesis drugs | Used against mouse tumor endothelial cells (mTEC) cells | Ara et al. (2012) |
| 3 | DNA (anti-PMSA) | doxorubicin | PMSA+ cell line (LNCaP), a mouse model of xenograft human prostate cancer | Baek et al. (2014) |
| 4 | DNA (AS1411) | siRNA | B-Raf oncoproteins for the treatment of malignant melanoma | Li et al. (2014a, b) |
| 5 | RNA aptamer | siRNA | Human transferrin expressing cells | Wilner et al. (2012) |
| 6 | RNA (Apt1) | siRNA | CD44+ receptor cancer cell line, (MDA-MB-231 breast cancer cells and A549 lung cancer cells) | Alshaer et al. (2015) |
| 7 | DNA (AS1411) | Doxorubicin and ammonium bicarbonate | Breast cancer cells (MCF-7/MDR) | Liao et al. (2015) |
| 8 | DNA (AS1411) | Doxorubicin | MDA-MB 231 breast cancer cell line | Zhang et al. (2015a, b, c) |
| 9 | AS1411 and Anti MUC-1 (S2.2) | Docetaxel and ammonium chloride | Dual targeting and tumor suppression | Zhao et al. (2017) |
CEM-CCRF leukemia lymphoblasts, PMSA prostate-specific membrane antigen, LNCaP androgen-sensitive human prostate adenocarcinoma cells, siRNA small interfering RNA
Aptamers are polyanionic and cannot pass the cellular membranes independently, so this limitation has been overcome by patch-clamp pipetting technique (Lennarz et al. 2015). This method also does not require a special delivery system like liposomes, nanoparticles, viral transfection, so this technique is straightforward. Patch clamp technique is a standard method to study the role of some aptamers directly in neuronal cells to work out the variation in biological activities of the target molecules in real time (Wolter and Mayer 2017).
Polymeric micelles
Micelles are self-assembled lipid nanoparticles with diameters 5–100 nm, prepared from amphiphilic lipid molecules. The hydrophilic heads stabilize the hydrophobic core of the micelle, which is formed from the hydrocarbon tails, and it acts as a reservoir for carrying hydrophobic drugs or imaging agents (Leonhard et al. 2012). Micelles are favourite drug carriers due to simple core manipulation and simpler drug encapsulation. The hydrophobic anticancer drugs, e.g., docetaxel and paclitaxel, have been efficiently encapsulated in the polymeric micelles. The polymeric micelles are synthesized by enhancing their concentration above the critical micelle concentration (Liu et al. 2011).
SKOV3 ovarian cancer cells, present in mice, have been found to have enhanced cytotoxicity due to the higher uptake of paclitaxel, both in vivo and in vitro. Similar results have been found in a xenograft model of human ovarian cancer with enhanced cytotoxicity, tumor growth reduction and myelosuppression (Zhou et al. 2007). The efficacy of active targeting has been further enhanced using polymeric micelles tagged with aptamers, e.g., AS1411 aptamer, which is docked on the surface of composite micelle. This micelle is made up of poloxamer and β-cyclodextrin-linked poly(lactide)-copoly(ethylene glycol) (PLA-PEG), which is used to encapsulate doxorubicin. The anti-PMSA aptamer covers the surface of the micelle loaded with doxorubicin. The micelle is made from hyperbranched (H40 polymer) polymer molecules and the shell arms are made from PLA-PEG (Alizadeh et al. 2020).
Some examples of micelle-based transportation vehicles are NK105® (paclitaxel encapsulate), SP1049C® (doxorubicin encapsulated, pluronic® based polymeric micelle) NK911® (doxorubicin encapsulated), and Genexol®-PM NK911(paclitaxel-encapsulated) which is in phase II stage of approval (Tsukioka et al. 2002; Wang et al. 2016). The enhancement of these transportation strategies is further achieved by aptamer conjugation, to further improve the drug efficacy. This includes TD05 aptamer, produced by cell SELEX method and it binds receptor of antibody heavy μ chain, present on B-cell lymphoma ramose cells (Phillips et al. 2008). This aptamer was effectively conjugated with lipid PEG linker micelles. Further, two cell lines were selected to test these aptamer-conjugated micelles as HL60 cells (negative cells) and ramose (target cells). The binding affinity constant was observed to be 750 times more for TD05-conjugated micelles as compared to free TD05 aptamer. This illustrates the significance of multivalent interactions on general affinity for the target cells. In addition to this, 80-fold internalization was observed by aptamer-TD05-micelles by ramose cells as compared to unconjugated micelles (Wu et al. 2010).
AS1411 aptamer has also been used to generate more complexed pH-sensitive polymeric micelles having d-α-tocopheryl polyethylene glycol-block-poly-(β-amino ester) (TPGS-b-PBAE, TP), encapsulated with paclitaxel. At pH 7.4, this micelle possessed higher stability and efficiently released paclitaxel at lower pH (pH 5.5) (Zhang et al. 2015a, b, c).
Polymeric nanocarriers
Polymeric nanocarriers are constructed from polymers like polyesters, poly(lactic-co-glycolic acid) (PLGA), poly(lactic acid) (PLA), and polyalkylcyanoacrylate. Polymeric nanocarriers can also be synthesized from cationic polymers like polyethyleneimine (PEI). Further, polymeric nanocarriers can also be synthesized from natural polymers, such as polysaccharides (dextran, chitosan) or proteins (collagen, albumin) (Plapied et al. 2011; Rytting et al. 2008). These particles are routinely used as drug carriers and usually display size ranging from 50 to 200 nm. Different methods are involved to deliver their payloads like diffusion through polymer and bulk erosion and swelling procedures (Fleige et al. 2012). Polymeric nanocarriers usually comprise (i) nanospheres, generally formed by hydrophobic core of polyester, which is enclosed by a hydrophilic PEG layer, and (ii) polyelectrolyte-based nanocarriers, which are formed by the association of anionic and cationic macromolecules (Beloqui et al. 2016). Different aptamer conjugation strategies are used to enhance its transportation and delivery efficiency. These strategies are commonly based on PLGA-b-PEG copolymer, as PLGA is easily biodegradable. These nanoparticles entrap different types of small molecules, which are mostly hydrophobic (Ho et al. 2015).
Anti-PSMA A10 aptamer has been conjugated by carbodiimide coupling to PLGA-b-PEG nanoparticles. This has resulted in improved docetaxel anticancer activity found in xenograft pancreatic cancer models (Zhou and Rossi 2009). A similar strategy of aptamer-conjugation, PLGA-based nanocarriers have been utilized for the drug delivery in different types of cancers using various types of aptamers like anti-NCL aptamer (AS1411), anti-MUC1 aptamer and EpCAM aptamer (D'Avino et al. 2016). Generally, PLGA-b-PEG nanocarriers have been encapsulated with doxorubicin with efficient loading capacity (~ 90%), conjugated with EpCAM aptamer. These nanocarriers showed tumor inhibition up to ~ 61% as compared to uncoupled PLGA-b-PEG nanocarriers (~ 31%), when used in mouse xenograft model of human small lung cancer (Alibolandi et al. 2015).
A15 is a well-known RNA aptamer, used to bind CD133 cancer stem cell (CSC) markers, which has been efficiently conjugated with PLGA-b-PEG nanocarriers carrying the salinomycin. This is a hydrophobic antibiotic that efficiently kills CSCs. This aptamer-conjugated nanoparticle presented almost fivefold higher in vitro cytotoxicity against CD133+ Saos-2 osteosarcoma CSCs as compared to free salinomycin and non-conjugated nanoparticles which showed only about twofold cytotoxicity (Long et al. 2008).
Biomimetic nanocarriers
Biomimetic nanocarriers have been used quite efficiently for biomedical applications, as they can help in precise drug delivery, specific to diseased cells. These nanoparticles are hybrid structures with the outer most layer similar to a cell membrane. The bacterium, E. coli is engineered to produce bacteriophage MS2 genome-free protein coat, which consists of 180 sequences of identical monomers, arranged as a sphere. This MS2 viral capsid nanostructure is non-toxic, robust and biodegradable, with pores enabling to trap active drugs and imaging molecules (Mastico et al. 1993). An aptamer known as sgc8c which targets PTK-7, has been fused to this genome-free MS2 product. Porphyrins have been used to modify the interior of the capsid and after conjugation with sgc8c aptamer, it has been used to target Jurkat leukemia T cells. Further, almost 76% of the cancer cells were killed after the illumination with UV irradiation of porphyrins (Leitner et al. 2017).
Self-assembled DNA monomers make efficient nanostructures that are biocompatible and biodegradable. These biomimetic nanocarriers have controlled size, shape, and position of functional groups which conjugate with specific targeting ligands (Sun et al. 2016). Aptamers, such as AS1411, have been successfully conjugated with these DNA biomimetic nanocarriers. The conjugation of AS1411 to these DNA pyramids has led to aptamer-decorated nanocarriers, which possessed enhanced nucleases resistance (Ai et al. 2014). In addition, this aptamer-conjugated nanocarrier displayed increased selective uptake, depending upon the number of conjugated aptamers. In this regard, six anti-MUC1 aptamers have been liganded to icosahedral 3D DNA nanostructures for specific targeting to MCF-7 breast cancer cells to deliver doxorubicin (Rui et al. 2017). In parallel, anti-PMSA aptamer (A9) has also been liganded to unmethylated CpG motifs, and has been used to functionalize dendrimers. This system was followed by doxorubicin docking which showed improved prostate cancer chemotherapy (Levy-Nissenbaum et al. 2008). In addition to this, a construct has been made with multiple copies of anti-PTK7 aptamers (sgc8) to three-armed Y-DNA, which has been found to enhance the binding as compared to the monovalent aptamer.
Other nanocarriers
Metal and silica-based nanocarriers have also been investigated as aptamer-guided drug delivery systems. Even though such nanocarriers are concerned with their biodegradation and toxicity complications, they have a proper degree of control over their physicochemical properties. One of the major concerns with the metallic nanocarriers is their difficulty in elimination from circulation and, therefore, normal tissues suffer from their long-term toxicity.
A promising strategy has been reported by these nanoparticles liganded with aptamers, for the delivery of imaging molecules and drugs (Mody et al. 2010). In an experiment, twelve cancer cell lines have been checked by gold nanoparticles complexed with AS1411 aptamer. Overall, almost 17% of increased cell death has resulted from these conjugated gold nanoparticles as compared to free AS1411 aptamers. In addition to this, the active trafficking of AS1411-conjugated gold nanoparticles possessed active transportation across the nuclear membrane and induced apoptosis accordingly (Dam et al. 2014).
The aptamer complexed to metallic nanocarriers increases the binding affinity of these particles. This binding affinity has been checked on CCRF-CEM cells, using Au–Ag nanoparticles, complexed with sgc8c aptamer. This complexing of aptamer has enhanced the binding affinity by 26 times as compared to the dye-labelled aptamer alone (Huang et al. 2008).
Silica nanoshells have also been reported to be conjugated with several aptamers to achieve different goals. In this regard, HER2 aptamer (HB5) conjugated with silica–carbon nanoparticles were loaded with doxorubicin, which has been used for chemo-photothermal therapy of HER2+ breast cancer cells. The outcome demonstrated a greater synergistic cytotoxicity by this combined therapy as compared to individual photo- or chemotherapy alone (Wang et al. 2015).
A redox sensitive, mesoporous nanocarrier was loaded with doxorubicin and was sealed with cytochrome and conjugated with AS1411 aptamer for specific targeting. This type of nanoparticle was found to exhibit triple therapeutic effects to tumors as cytochrome helps in protease activation, doxorubicin enhances the apoptosis and AS1411 stabilizes the anti-apoptotic protein BCL-2 (Zhu et al. 2014).
In addition to the above-discussed nanoparticles, it is worth mentioning that carbon-nanotubes are non-biodegradable. An aptamer (hnRNP) has been conjugated with carbon nanotubes, and has been used to bind with multiple tumor cell lines. These hnRNP-nanotubes minimize the tumor proliferation by inhibiting heterogenous nuclear ribonucleoprotein (hnRNP) A2/B1 (Zhang et al. 2015a, b, c).
Diagnostic applications of oligonucleotide aptamers
Applications of DNA aptamers
In 1992, the first ssDNA aptamer was obtained from human thrombin and since then DNA aptamers have been applied in various fields, especially in the diagnosis and treatment of various diseases (Tucker et al. 2012; Xing et al. 2014). The DNA aptamers have been utilized independently for the diagnosis of cardiovascular complications, infectious diseases and mainly cancer (Wang et al. 2011a, b). The major convenience of cell-SELEX has led to the utilization of DNA aptamers, which are selected against cancer cells of breast, colon, gastric, and liver etc., directly without the requirement of preparation and purification of disease-specific biomarkers. Generally, it requires positive selection with tumor cells and negative selection with normal cells (Fang and Tan, 2010; Sefah et al. 2010).
The DNA aptamers are typically selected from the libraries with longer and randomized sequences to achieve more complicated structural variations. To study the role of different DNA-based aptamosomes against many types of cancer cells, several researchers are engaged in this modern and innovative field. An aptamosome was synthesized with guanosine rich 26-mer DNA aptamer called AS1411. This aptamer showed a robust binding affinity with nucleolin (bcl2 mRNA-stabilizing protein), which is overexpressed on the cell surface of different types of malignant cells, including breast cancer and leukemia (Soundararajan et al. 2008, 2009). This aptamer or its aptamosome is quite resistant against nuclease degradations. In addition, this aptamosome has been used to cargo cisplatin, which is used as a cytotoxic agent for MCF-7 cells (Vandghanooni et al. 2018). In comparison to this, no such activity was found by liposomes without this aptamer, or the liposomes liganded with a non-aptameric DNA strand.
An aptamer (AS1411) containing liposomes carrying doxorubicin drug was synthesized, which showed efficient internalization and better cytotoxicity to MCF-7 breast cancer cells (Zhang et al. 2017). In addition, A DNA aptamer sgc8 was created, possessing great affinity with CEM-CCRF leukemia cells. Using this oligonucleotide, joined with PEG as a spacer arm, an aptamosome was generated for fluorescein isothiocyanato-dextran as a model drug (áO’Donoghue 2010). In 2006, Macugen was used as the first very popular aptamer-based therapeutics, for the treatment of macular degeneration, and approved by FDA (Lee et al. 2008; Deissler et al. 2008). The role of some special DNA aptamers in different diseases, targeted against specific ligands is represented in Table 5.
Table 5.
DNA aptamers and their role in different diseases, targeted against specific ligands
| Disease | DNA aptamer name | Length (mer) | Ligand/target | References |
|---|---|---|---|---|
| Infectious diseases | 2008a | 35 | Plasmodium falciparum LDH (PfLDH) | Cheung et al. (2013) |
| Sequence (2) | 74 | HA protein (H5N1) | Wang et al. (2013) | |
| 4C6 | 45 | Truncated murine prion protein (H-MoPrP90-231) | Xiao et al. (2012) | |
| LmWC-25R and LmHSP-7b/11R | 36 | Leishmania promastigote and hydrophilic surface protein (HSP) | Bruno et al. (2014) | |
| Cancer | 32 | 30 | Glioblastoma multiforme cells (U87Δ) epidermal growth factor receptor variant III (EGFRvIII) | Tan et al. (2013) |
| Aptamer BC15 | 111 | Heterogenous nuclear ribonucleoprotein A1 (hnRNP A1) | Li et al. (2009) | |
| DNA aptamer HB5 | 19 | Her 2 (human EGFR 2) | Liu et al. (2012) | |
| DNA aptamer A and C | 32 | TTF1 (a member of the NK homeodomain transcription factor) | Murphy et al. (2003) | |
| SYL3 DNA aptamer | 38 | EpCAM (epithelial cell adhesion molecule) | Song et al. (2013) | |
| MUC1 DNA aptamer | 33 | MUC1 peptide | Ferreira et al. (2006) | |
| AGC 03 | 40 | Gastric cancer cells (HGC-27) | Zhang et al. (2014) | |
| C-2 | 50 | Liver cancer cells (HepG2) | Ninomiya et al. (2013) | |
| Cy-apt20 | 52 | Gastric carcinoma cells (AGS) | Cao et al. (2014) | |
| JHIT2 | 25 | Liver cancer cells (HepG2) | Xu et al. (2015) | |
| GMB128 | 45 | Glioblastoma cells (U118-MG) | Li et al. (2014a, b) | |
| GMT3 | 42 | Glioblastoma multiforme cells (A172) | Bayrac et al. (2011) | |
| LXL-1 | 45 | Metastatic breast cancer cells (MDA-MB-231) | Urata et al. (1992) | |
| SYL3-C | 40 | Solid cancer | Song et al. (2013) | |
| Vea5 (SL2-B) | 30 | Cancer cells biomarker: VEGF165) | Kaur and Yung (2012); Hasegawa et al. (2008) | |
| Wy-5a | 45 | Prostate cancer cells (PC-3, metastatic) | Wang et al. (2014a, b, c, d) | |
| XL-33 | 45 | Metastatic colon cancer cells (SW620) | Li et al. (2015) | |
| XQ-2d | 42 | Pancreatic ductal adenocarcinoma (PDAC) cells (PL45) | Wu et al. (2015) | |
| y119 | 40 | Cholangiocarcinoma cells (QBC-939) | Wan et al. (2015) | |
| Cardiovascular disease | Hcy8 | 60 | L-homocysteine | McKeague et al. (2013) |
| Myo040-7-27 | 40 | Myoglobin | Wang et al. (2014a, b, c, d) | |
| Food borne disease | DNA aptamer | 20 | OmpC | Han et al. (2013) |
Applications of RNA aptamers
Recently, a good momentum has been found in RNA aptamer technology because of approval by food and drug administration (FDA) (Stein and Castanotto 2017). Different RNA aptamers have been synthesized against intra- and extracellular components of signaling pathways, in addition to cell surface markers. The therapeutic applications of different RNAs include, short hairpin RNA (shRNA), antisense oligonucleotides (AS OGNs), siRNA, micro RNA, ribozyme and RNA aptamers (Sundaram et al. 2013).
RNA aptamers possess unique advantages as compared to other types of RNAs, as they can directly bind to extracellular targets, to either activate or inhibit their functions. RNA aptamers are efficient therapeutic agents in the blood stream, cell surface proteins and ocular divisions (Shigdar et al. 2011; Ng et al. 2006). In addition to binding with cell surface proteins, RNA aptamers are used as aptamosomes to deliver different therapeutic items including small molecules, antioxidants or peptides.
In 2004, Pegaptanib, was the first RNA aptamer synthesized against VEGF, for the therapeutic use against age-related macular degeneration. As we now clearly know that VEGF plays a major role in angiogenesis, through binding with its receptor, VEGF-R1 and -R2. By some alternate splicing, the VEGF gene expresses four main isoforms at different amino acid residues, like 121, 165, 189 and 206. During diabetic macular edema and age-related macular degeneration, an unusual neovascularization occurs due to overexpression of VEGF165 isoform. The RNA aptamers have been synthesized precisely, targeted against VEGF165, with enhanced affinity and efficient specificity. The pre-clinical trials have clearly shown that these aptamers possess great inhibitory activity against VEGF165 functions for vascular permeability and endothelial mitogen promotion.
Recently, an innovative antitumor drug-encapsulated liposome was engineered against prostate-specific membrane antigen and conjugated with RNA aptamer. This RNA–aptamosome was synthesized by the post-insertion method, involving the attachment of RNA–aptamer-conjugated micelles. These nanosomes (∼100 nm) aptamer-liganded liposomes possessed specificity for LNCaP prostate epithelial cells expressing PMSA. The expression of PMSA significantly increased the possibility of such targeted RNA–aptamosomes binding and uptake as compared to liposomes lacking PMSA aptamer.
The RNA–aptamosomes carrying the anticancer drug doxorubicin were notably more cyto-toxic against LNCaP cells as compared to non-targeted tumor cells. To further clarify the efficiency of the anticancer activity of doxorubicin and the role of PMSA aptamosome, LNCaP administered xenograft nude mice were selected. It has been demonstrated that upon the administration of such liposomes, there occurs regression of tumor size in LNCaP xenograft mice. These experiments clearly reveal the importance of such aptamosomes in clinical practice and the minimal toxicity of such chemotherapeutics. At present, there are many RNA aptamers under preclinical and clinical trials for the treatment of different diseases like diabetes and cancer. The role of some special RNA aptamers with varied size in different diseases, targeted against different ligands is represented in Table 6.
Table 6.
RNA aptamers targeted against different ligands and their role in different diseases
| Disease | RNA aptamer name | Length (mer) | Target | References |
|---|---|---|---|---|
| Oncogenic protein | RNA aptamer G4 | 139 | K-Ras-derived protein | Gilbert et al. (1997) |
| Hepatocellular carcinoma | AFP aptamer | 33 | AFP (fetal protein) | Lee and Lee (2012) |
| Breast cancer (human breast adenocarcinoma cell line, MCF-7) | Class III GSH aptamer 8.17 | 41.8 | Glutathione | Bala et al. (2011) |
| Breast and lung cancer | Anti-EGFR aptamer (E07) | 2.4 | Epidermal growth factor receptor | Li et al. (2011) |
| Cancer | 2′F RNA and 2′NH2 aptamer | na | Human keratinocyte growth factor | Pagratis et al. (1997) |
| Cancer and inflammation | RNA aptamer | 5.4 | NF-κB p50 homodimer | Huang et al. (2003) |
| Prostate cancer | Anti-PMSA chimera | na | Prostate-specific membrane antigen | McNamara et al. (2006) |
| Colon cancer | Aptamer number 5 | 100 | T cell factor 1 | Lee et al. (2005a, b) |
| Glioblastoma breast cancer | TTA1 | 5 | Tenascin-C | Hicke et al. (2001, 2006) |
| Ovarian cancer | RNA aptamer | 1 | TGFβ type III receptor | Ohuchi et al. (2006) |
| Wilm’s tumor | RNA aptamer 22 | 700 | WT1 | Bardeesy and Pelletier (1998) |
| Colon cancer | RNA aptamer | 5 | β-Catenin | Lee et al. (2006) |
| Carcinogenic and DNA damaging agent | MDA aptamer M1 | 450 | MDA(4,4′methylene-dianiline) | Brockstedt et al. (2004) |
| HIV | RNA aptamer | 19–36 | Rev peptide | Xu and Ellington et al. (1996) |
| RNA aptamer S66A-C6, | 406 | V3 loop of gp120 | Gronewold et al. (2009) | |
| RNA aptamer S69A-C15 | 637 | HIV-1RT | Gronewold et al. (2009) | |
| Anti-gp120 aptamer chimera | na | Glycoprotein 120 (gp120) | Zhou et al. (2008) | |
| Neuromuscular disorder | Class 1 and Class II AChR aptamer | 2 and 12 | Acetylcholine receptor | Ulrich et al. (1998) |
| Antiviral activity via modulating IFN ⍺/β production | RNA aptamer | 37 | RIG1 (1-925aa) | Hwang et al. (2012) |
| Malaria | DBL1⍺-specific RNA aptamer | 33 | Erythrocyte membrane protein 1 (PfEMP1) | Barfod et al. (2009) |
| Inflammation | RNA aptamer | 17 | L-selectin | O'Codnnell et al, (1996) |
| Age-related macular degeneration | Pegaptanib sodium, NX 1838 | 0.05 | VEGF 165 | Lee et al. (2005a, b) |
| Alzheimer’s disease | RNA aptamer | 29–48 | Amyloid beta-peptide A4 (I-40) | Ylera et al. (2002) |
| Alzheimer’s disease | RNA apt TH14 | 280 | Beta-Secretase BACE1 (B1-CT) | Rentmeister et al. (2006) |
Limitations and future prospects of aptamers
The use and selection of an aptamer is relatively a challenging task, as there are several factors that need to be addressed before a fruitful selection of an experiment can be managed. First, the fine purification of a target protein is a must, and in addition to this, the library design and its amplification are also keys to get a satisfactory result. The nucleic acid aptamers have certain limitations as compared to peptide aptamers, as the limited set of nucleotides as building blocks of nucleic acid library. The four nucleotides, either in RNA or DNA in conjunction with ribose or deoxyribose, have a limited chemical diversity in comparison with 21 proteogenic amino acids which facilitate different types of molecular interactions and chemical bonds. Furthermore, the in vitro experiments for the selection of a specific aptamer have uncertain predictions of success as a specific combination of the nucleic acid library with a target protein. However, these limitations of aptamer use have been overcome recently by the introduction of at least an additional chemical entity by one nucleotide modification at the C5 position of uridine. The new aptamer variants are known as clickmers and SOMAmers (Tolle and Mayer 2013; Rohloff et al. 2014; Tolle et al. 2015).
One more limitation in applicability of the aptamers is their inability to cross the plasma membrane by passive diffusion. However, several strategies are applied to overcome this challenge like the use of nanoparticles (Ryou et al. 2011), transfection with liposomes (Hwang et al. 2012), or by viral or plasmid delivery (Choi et al. 2006). In addition to this, the patch-clamp technique is a sophisticated and straightforward method, especially, for the investigation of neuronal the cells (Lennarz et al. 2015).
While the initial stages in any research and technology have limitations and doubts, and are hardly green-signaled, in contrast, the aptamer science has made substantial advances over the last two decades. The limitations and challenges of aptamers are based on issues of cost and manufacture, route of administration, efficiency, and applicability (Bruno 2015). Nevertheless, the overall advantages and vivid future outlooks of aptamers outweighs their most of the limitations. Aptamers have extraordinary target sensitivity and specificity, opportunities for substitute formulations, the scale of preparation, compatible biophysical and pharmacokinetic properties, program of administration, and laboratory economics. So aptamers have professionally found themselves a commanding niche, and are well known as a promising new class of super medication.
The proof-of-concept trials, which illustrate that aptamers can precisely bind and control the functioning of different biomedically important proteins, promise us very well for upcoming aptamer-based diagnostics and drugs development. Our prescient expectation is that the next phase of aptamer development and their clinical applications will revolutionize the proper drug discovery and efficiently targeted delivery processes, as well as the way of diagnosis, treatment and prevention of diseases (Rozenblum et al. 2016).
Conclusion
The advancement of nucleic acid aptamers and their broad range applications in diverse human disease diagnosis and therapy, have led these compounds a mighty potential to be employed as an alternative to antibodies. Even though antibodies are magnificently and comprehensively used in the treatment of different human diseases, these antibodies are still expensive in all parameters. Nucleic acid aptamers are anticipated to reach a parallel success to antibodies in near future. Because of their good stability, lower cost of production and easy manipulation, nucleic acid aptamers are very promising to be comprehensively reviewed and applied in human disease management.
Although, the modified nucleic acid aptamers have improved stability, the higher expenditure of chemically modified RNA production might limit their study and applications to some extent. However, these RNA aptamers have a good advantage in presenting additional complex and varied 3D structures, which is very advantageous for selecting such aptamers with greater affinity for highly complex targets required for specific disease therapy. In this aspect, DNA aptamers may possess extra promising clinical applications in disease diagnosis and in vivo imaging, while the extra modified RNA aptamers may retain more assuring applications in disease therapy. In this period of personalized medicine, nucleic acid aptamer-based diagnostics and therapeutics is assumed to have a greater potential for broad range applications, as the aptamers possess a good flexibility to bind specifically to broad range of molecular targets and generally cancer cells. Some of the limitations of aptamers like nuclease breakdown, fast renal elimination and cross-reactivity of some aptamers needs are analyzed in future research. Even though aptamers have wide applications, there are still so many limitations that continue to be tackled.
Author contributions
KSA, AAK, and AA have contributed to the concept, design, diagrams and information collection of the work. MAA and GTB have reviewed it thoroughly. FA and AHR have contributed in reviewing and final approval of the work.
Compliance with ethical standards
Conflict of interest
All the authors state that there is no conflict of interest.
References
- Ahirwar R, Nahar P. Development of an aptamer-affinity chromatography for efficient single step purification of Concanavalin A from Canavalia ensiformis. J Chrom B. 2015;997:105–109. doi: 10.1016/j.jchromb.2015.06.003. [DOI] [PubMed] [Google Scholar]
- Ai J, Xu Y, Lou B, Li D, Wang E. Multifunctional AS1411-functionalized fluorescent gold nanoparticles for targeted cancer cell imaging and efficient photodynamic. Therapy Talanta. 2014;118:54–60. doi: 10.1016/j.talanta.2013.09.062. [DOI] [PubMed] [Google Scholar]
- Alibolandi M, Ramezani M, Sadeghi F, Abnous K, Hadizadeh F. Epithelial cell adhesion molecule aptamer conjugated PEG–PLGA nanopolymersomes for targeted delivery of doxorubicin to human breast adenocarcinoma cell line in vitro. Int J Pharmaceut. 2015;479:241–251. doi: 10.1016/j.ijpharm.2014.12.035. [DOI] [PubMed] [Google Scholar]
- Alizadeh L, Alizadeh E, Zarebkohan A, Ahmadi E, Rahmati-Yamchi M, Salehi R. AS1411 aptamer-functionalized chitosan-silica nanoparticles for targeted delivery of epigallocatechin gallate to the SKOV-3 ovarian cancer cell lines. J Nanoparticle Res. 2020;22:1–14. [Google Scholar]
- Alshaer W, Hillaireau H, Vergnaud J, Ismail S, Fattal E. Functionalizing liposomes with anti-CD44 aptamer for selective targeting of cancer cells. Bioconj Chem. 2015;26:1307–1313. doi: 10.1021/bc5004313. [DOI] [PubMed] [Google Scholar]
- Ara MN, Hyodo M, Ohga N, Hida K, Harashima H. Development of a novel DNA aptamer ligand targeting to primary cultured tumor endothelial cells by a cell-based SELEX method. PLoS ONE. 2012;7:e50174. doi: 10.1371/journal.pone.0050174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek SE, Lee KH, Park YS, Oh D-K, Oh S, Kim K-S, Kim D-E. RNA aptamer-conjugated liposome as an efficient anticancer drug delivery vehicle targeting cancer cells in vivo. J Control Release. 2014;196:234–242. doi: 10.1016/j.jconrel.2014.10.018. [DOI] [PubMed] [Google Scholar]
- Bagalkot V, Farokhzad OC, Langer R, Jon S. An aptamer-doxorubicin physical conjugate as a novel targeted drug-delivery platform. Angew Chem Int Ed. 2006;45:8149–8152. doi: 10.1002/anie.200602251. [DOI] [PubMed] [Google Scholar]
- Bala J, Bhaskar A, Varshney A, Singh AK, Dey S, Yadava P. In vitro selected RNA aptamer recognizing glutathione induces ROS-mediated apoptosis in the human breast cancer cell line MCF 7. RNA Biol. 2011;8:101–111. doi: 10.4161/rna.8.1.14116. [DOI] [PubMed] [Google Scholar]
- Bardeesy N, Pelletier J. Overlapping RNA and DNA binding domains of the wt1 tumor suppressor gene product. Nuc Acid Res. 1998;26:1784–1792. doi: 10.1093/nar/26.7.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barfod A, Persson T, Lindh J. In vitro selection of RNA aptamers against a conserved region of the Plasmodium falciparum erythrocyte membrane protein 1. Parasitol Res. 2009;105:1557–1566. doi: 10.1007/s00436-009-1583-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barman J. Targeting cancer cells using aptamers: cell-SELEX approach and recent advancements. RSC Adv. 2015;5:11724–11732. [Google Scholar]
- Bayrac AT, Sefah K, Parekh P, Bayrac C, Gulbakan B, Oktem HA, Tan WH. In vitro selection of DNA aptamers to glioblastoma multiforme. ACS Chem Neurosci. 2011;2:175–181. doi: 10.1021/cn100114k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beloqui A, des Rieux A, Preat V, Mechanisms of transport of polymeric and lipidic nanoparticles across the intestinal barrier. Adv Drug Deliv Rev. 2016;106:242–255. doi: 10.1016/j.addr.2016.04.014. [DOI] [PubMed] [Google Scholar]
- Berezovski M, Musheev M, Drabovich A, Krylov SN. Non-SELEX selection of aptamers. J Am Chem Soc. 2006;128:1410–1411. doi: 10.1021/ja056943j. [DOI] [PubMed] [Google Scholar]
- Bianchini M, Radrizzani M, Brocardo MG, Reyes GB, Solveyra CG, Santa-Coloma TA. Specific oligobodies against ERK-2 that recognize both the native and the denatured state of the protein. J Immunol Methods. 2001;252:191–197. doi: 10.1016/s0022-1759(01)00350-7. [DOI] [PubMed] [Google Scholar]
- Brandl M. Liposomes as drug carriers: a technological approach. Biotechnol Ann Rev. 2001;7:59–85. doi: 10.1016/s1387-2656(01)07033-8. [DOI] [PubMed] [Google Scholar]
- Brockstedt U, Uzarowska A, Montpetit A, Pfau W, Labuda D. In vitro evolution of RNA aptamers recognizing carcinogenic aromatic amines. Biochem Biophy Res Commun. 2004;313:1004–1008. doi: 10.1016/j.bbrc.2003.12.030. [DOI] [PubMed] [Google Scholar]
- Bruno JG. Predicting the uncertain future of aptamer-based diagnostics and therapeutics. Molecules. 2015;20:6866–6887. doi: 10.3390/molecules20046866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno JG, Richarte AM, Phillips T, Savage AA, Sivils JC, Greis A, Mayo MW. Development of a fluorescent enzyme-linked DNA aptamer-magnetic bead sandwich assay and portable fluorometer for sensitive and rapid leishmania detection in sandflies. J Fluoresc. 2014;24:267–277. doi: 10.1007/s10895-013-1315-6. [DOI] [PubMed] [Google Scholar]
- Bugaut A, Toulme JJ, Rayner B. SELEX and dynamic combinatorial chemistry interplay for the selection of conjugated RNA aptamers. Org Biomol Chem. 2006;4:4082–4088. doi: 10.1039/b610890c. [DOI] [PubMed] [Google Scholar]
- Burke DH, Willis JH. Recombination, RNA evolution, and bifunctional RNA molecules isolated through chimeric SELEX. RNA. 1998;4(9):1165–1175. doi: 10.1017/s1355838298980542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z, Tong R, Mishra A, Xu W, Wong GC, Cheng J, Lu Y. Reversible cell-specific drug delivery with aptamer-functionalized liposomes. Angew Chem Int Ed. 2009;48:6494–6498. doi: 10.1002/anie.200901452. [DOI] [PubMed] [Google Scholar]
- Cao HY, Yuan AH, Chen W, Shi XS, Miao Y. A DNA aptamer with high affinity and specificity for molecular recognition and targeting therapy of gastric cancer. BMC Cancer. 2014;14(1):1–9. doi: 10.1186/1471-2407-14-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Yang S. Replacing antibodies with aptamers in lateral flow immunoassay. Biosens Bioelectron. 2015;71:230–242. doi: 10.1016/j.bios.2015.04.041. [DOI] [PubMed] [Google Scholar]
- Cheung YW, Kwok J, Law AW, Watt RM, Kotaka M, Tanner JA. Structural basis for discriminatory recognition of Plasmodium lactate dehydrogenase by a DNA aptamer. Proc Natl Acad Sci USA. 2013;110:15967–15972. doi: 10.1073/pnas.1309538110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho M, Xiao Y, Nie J, Stewart R, Csordas AT, Oh SS, Thomson JA, Soh HT. Quantitative selection of DNA aptamers through microfluidic selection and high-throughput sequencing. Proc Natl Acad Sci USA. 2010;107:15373–15378. doi: 10.1073/pnas.1009331107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KH, Park MW, Lee SY, Jeon MY, Kim MY, Lee HK, Yu J, Kim HJ, Han K, Lee H, Park K. Intracellular expression of the T cell factor-1 RNA aptamer as an intramer. Mol Can Therap. 2006;5:2428–2434. doi: 10.1158/1535-7163.MCT-05-0204. [DOI] [PubMed] [Google Scholar]
- Coulter LR, Landree MA, Cooper TA. Identification of a new class of exonic splicing enhancers by in vivo selection. Mol Cell Biol. 1997;17:2143–2150. doi: 10.1128/mcb.17.4.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dam DH, Culver KS, Odom TW. Grafting aptamers onto gold nanostars increases in vitro efficacy in a wide range of cancer cell types. Mol Pharmaceut. 2014;11:580–587. doi: 10.1021/mp4005657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Avino C, Palmieri D, Braddom A, Zanesi N, James C, Cole S, Salvatore F, Croce CM, De Lorenzo C. A novel fully human anti-NCL immunoRNase for triple-negative breast cancer therapy. Oncotarget. 2016;7:87016. doi: 10.18632/oncotarget.13522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deissler HL, Lang GE. Effect of VEGF165 and the VEGF aptamer pegaptanib (Macugen) on the protein composition of tight junctions in microvascular endothelial cells of the retina. Klin Monatsbl Augenh. 2008;225:863–867. doi: 10.1055/s-2008-1027767. [DOI] [PubMed] [Google Scholar]
- Du F, Guo L, Qin Q, Zheng X, Ruan G, Li J, Li G. Recent advances in aptamer-functionalized materials in sample preparation TrAC. Trends Anal Chem. 2015;67:134–146. [Google Scholar]
- Ellington AD, Szostak JW. vitro selection of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature. 1992;355:850–852. doi: 10.1038/355850a0. [DOI] [PubMed] [Google Scholar]
- El-Say KM, El-Sawy HS. Polymeric nanoparticles: promising platform for drug delivery. Int J Pharm. 2017;528:675–691. doi: 10.1016/j.ijpharm.2017.06.052. [DOI] [PubMed] [Google Scholar]
- Eremeeva E, Fikatas A, Margamuljana L, Abramov M, Schols D, Groaz E, Herdewijn P. Highly stable hexitol based XNA aptamers targeting the vascular endothelial growth factor. Nucleic Acid Res. 2019;47:4927–4939. doi: 10.1093/nar/gkz252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Tan W. Aptamers generated from cell-SELEX for molecular medicine: a chemical biology approach. Acc Chem Res. 2010;43:48–57. doi: 10.1021/ar900101s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira CSM, Matthews CS, Missailidis S. DNA aptamers that bind to MUC1 tumour marker: design and characterization of MUC1-binding single-stranded DNA aptamers. Tumor Biol. 2006;27:289–301. doi: 10.1159/000096085. [DOI] [PubMed] [Google Scholar]
- Ferreira-Bravo AI, Cozens C, Holliger P, DeStefano JJ. Selection of 2′-deoxy-2′-fluoroarabinonucleotide (FANA) aptamers that bind HIV-1 reverse transcriptase with picomolar affinity. Nucleic Acid Res. 2015;43:9587–9599. doi: 10.1093/nar/gkv1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleige E, Quadir MA, Haag R. Stimuli-responsive polymeric nanocarriers for the controlled transport of active compounds: concepts and applications. Adv Drug Deliv Rev. 2012;64:866–884. doi: 10.1016/j.addr.2012.01.020. [DOI] [PubMed] [Google Scholar]
- Gilbert BA, Sha M, Wathen ST, Rando RR. RNA aptamers that specifically bind to a K ras-derived farnesylated peptide. Bioorg Med Chem. 1997;5:1115–1122. doi: 10.1016/s0968-0896(97)00047-3. [DOI] [PubMed] [Google Scholar]
- Golden MC, Collins BD, Willis MC, Koch TH. Diagnostic potential of PhotoSELEX-evolved ssDNA aptamers. J biotechnol. 2000;81:167–178. doi: 10.1016/s0168-1656(00)00290-x. [DOI] [PubMed] [Google Scholar]
- Gopinath SCB. Methods developed for SELEX. Anal Bioanal Chem. 2007;387:171–182. doi: 10.1007/s00216-006-0826-2. [DOI] [PubMed] [Google Scholar]
- Goringer HU, Adler A, Forster N, Homann M. Post-SELEX chemical optimization of a trypanosome-specific RNA aptamer. Comb Chem High Throughput Screen. 2008;11:16–23. doi: 10.2174/138620708783398331. [DOI] [PubMed] [Google Scholar]
- Gronewold TMA, Baumgartner A, Hierer J, Baumgartner A, Hierer J, Sierra S, Blind M, Schäfer F, Blümer J, Tillmann T, Kiwitz A, Kaiser R, Zabe-Kühn M. Kinetic binding analysis of aptamers targeting HIV-1 proteins by a combination of a microbalance array and mass spectrometry (MAMS) J Prot Res. 2009;8:3568–3577. doi: 10.1021/pr900265r. [DOI] [PubMed] [Google Scholar]
- Han SR, Lee S-W. In vitro selection of RNA aptamer specific to salmonella typhimurium. J Microbiol Biotechnol. 2013;23:878–884. doi: 10.4014/jmb.1212.12033. [DOI] [PubMed] [Google Scholar]
- Hasegawa H, Sode K, Ikebukuro K. Selection of DNA aptamers against VEGF(165) using a protein competitor and the aptamer blotting method. Biotechnol Lett. 2008;30:829–834. doi: 10.1007/s10529-007-9629-6. [DOI] [PubMed] [Google Scholar]
- Hermann T, Patel DJ. Adaptive recognition by nucleic acid aptamers. Science. 2000;287:820–825. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- Hianik T, Ostatna V, Sonlajtnerova M, Grman I. Influence of ionic strength, pH and aptamer configuration for binding affinity to thrombin. Bio-electrochemistry. 2007;70:127–133. doi: 10.1016/j.bioelechem.2006.03.012. [DOI] [PubMed] [Google Scholar]
- Hicke BJ, Marion C, Chang YF, Gould T, Lynott CK, Parma D, Schmidt PG, Warren S. Tenascin-C aptamers are generated using tumor cells and purified protein. J Biol Chem. 2001;276:48644–48654. doi: 10.1074/jbc.M104651200. [DOI] [PubMed] [Google Scholar]
- Hicke BJ, Stephens AW, Gould T, Chang YF, Lynott CK, Heil J, Borkowski S, Hilger CS, Cook G, Warren S, Schmidt PG. Tumor targeting by an aptamer. J Nuc Med. 2006;47:668–678. [PubMed] [Google Scholar]
- Ho LC, Wu WC, Chang CY, Hsieh HH, Lee CH, Chang HT. Aptamer-conjugated polymeric nanoparticles for the detection of cancer cells through “turn-on” retro-self-quenched fluorescence. Anal Chem. 2015;87:4925–4932. doi: 10.1021/acs.analchem.5b00569. [DOI] [PubMed] [Google Scholar]
- Homann M, Goringer HU. Combinatorial selection of high affinity RNA ligands to live African trypanosomes. Nucleic Acids Res. 1999;27:2006–2014. doi: 10.1093/nar/27.9.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoon S, Zhou B, Janda KD, Brenner S, Scolnick J. Aptamer selection by high-throughput sequencing and informatic analysis. Biotechniques. 2011;51:413–416. doi: 10.2144/000113786. [DOI] [PubMed] [Google Scholar]
- Hoppe-Seyler F, Crnkovic-Mertens I, Tomai E, Butz K. Peptide aptamers: specific inhibitors of protein function. Curr Mol Med. 2004;4:529–538. doi: 10.2174/1566524043360519. [DOI] [PubMed] [Google Scholar]
- Hua X, Zhou Z, Yuan L, Liu S. Selective collection and detection of MCF-7 breast cancer cells using aptamer-functionalized magnetic beads and quantum dots based nano-bio-probes. Anal Chim Acta. 2013;788:135–140. doi: 10.1016/j.aca.2013.06.001. [DOI] [PubMed] [Google Scholar]
- Huang D-B, Vu D, Cassiday LA, Zimmerman JM, Maher LJ, III, Ghosh G. Crystal structure of NF-κB (p50)2 complexed to a high-affinity RNA aptamer. Proc Nat Acad Sci (USA) 2003;100:9268–9273. doi: 10.1073/pnas.1632011100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y-F, Sefah K, Bamrungsap S, Chang H-T, Tan W. Selective photothermal therapy for mixed cancer cells using aptamer-conjugated nanorods. Langmuir. 2008;24:11860–11865. doi: 10.1021/la801969c. [DOI] [PubMed] [Google Scholar]
- Huang CJ, Lin HI, Shiesh SC, Lee GB. Integrated microfluidic system for rapid screening of CRP aptamers utilizing systematic evolution of ligands by exponential enrichment (SELEX) Biosens Bioelectron. 2010;25(7):1761–1766. doi: 10.1016/j.bios.2009.12.029. [DOI] [PubMed] [Google Scholar]
- Hwang SY, Sun HY, Lee KH, Oh BH, Cha YJ, Kim BH, Yoo JY. 5′-Triphosphate-RNA-independent activation of RIG-I via RNA aptamer with enhanced antiviral activity. Nucleic Acid Res. 2012;40:2724–2733. doi: 10.1093/nar/gkr1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenison RD. High-resolution molecular discrimination by RNA. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- Kang KN, Lee YS. Trends in biotechnology. Berlin, Heidelberg: Springer; 2012. RNA aptamers: a review of recent trends and applications in future; pp. 153–169. [DOI] [PubMed] [Google Scholar]
- Kang H, O'Donoghue MB, Liu H, Tan W. A liposome-based nanostructure for aptamer directed delivery. Chem Commun (Camb Eng) 2010;46:249–251. doi: 10.1039/b916911c. [DOI] [PubMed] [Google Scholar]
- Kaur H, Yung LY. Probing high affinity sequences of DNA aptamer against VEGF165. PLoS ONE. 2012;7:e31196. doi: 10.1371/journal.pone.0031196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami J, Imanaka H, Yokota Y, Sugimoto N. In vitro selection of aptamers that act with Zn2+ J Inorg Biochem. 2000;82:197–206. doi: 10.1016/s0162-0134(00)00158-6. [DOI] [PubMed] [Google Scholar]
- Khan AA, Allemailem KS, Almatroodi SA, Almatroudi A, Rahmani AH. Recent strategies towards the surface modification of liposomes: an innovative approach for different clinical applications. 3 Biotech. 2020;10:1–15. doi: 10.1007/s13205-020-2144-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratschmer C, Levy M. Targeted delivery of auristatin-modified toxins to pancreatic cancer using aptamers. Mol Ther Nucleic Acids. 2018;10:227–236. doi: 10.1016/j.omtn.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruspe S, Hahn U. An aptamer intrinsically comprising 5-fluoro-2’-deoxyuridine for targeted chemotherapy. Angew Chem Int Ed. 2014;53:10541–10544. doi: 10.1002/anie.201405778. [DOI] [PubMed] [Google Scholar]
- Lee YJ, Lee S-W. Regression of hepatocarcinoma cells using RNA aptamer specific to alpha-fetoprotein. Biochem Biophy Res Commun. 2012;417:521–527. doi: 10.1016/j.bbrc.2011.11.153. [DOI] [PubMed] [Google Scholar]
- Lee J-H, Canny MD, De Erkenez A, Krilleke D, Ng YS, Shima DT, Pardi A, Jucker F. A therapeutic aptamer inhibits angiogenesis by specifically targeting the heparin binding domain of VEGF165. Proc Nat Acad Sci (USA) 2005;102:18902–18907. doi: 10.1073/pnas.0509069102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Park MW, Yang EG, Yu J, Jeong S. An RNA aptamer that binds to the β-catenin interaction domain of TCF-1 protein. Biochem Biophys Res Commun. 2005;327:294–299. doi: 10.1016/j.bbrc.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Lee HK, Choi YS, Park YA, Jeong S. Modulation of oncogenic transcription and alternative splicing by β-catenin and an RNA aptamer in colon cancer cells. Can Res. 2006;66:10560–10566. doi: 10.1158/0008-5472.CAN-06-2526. [DOI] [PubMed] [Google Scholar]
- Lee JH, Jucker F, Pardi A. Imino proton exchange rates imply an induced-fit binding mechanism for the VEGF165-targeting aptamer, Macugen. FEBS lett. 2008;582:1835–1839. doi: 10.1016/j.febslet.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner M, Poturnayova A, Lamprecht C, Weich S, Snejdarkova M, Karpisova I, Hianik T, Ebner A. Characterization of the specific interaction between the DNA aptamer sgc8c and protein tyrosine kinase-7 receptors at the surface of T cells by biosensing AFM. Anal Bioanal Chem. 2017;409:2767–2776. doi: 10.1007/s00216-017-0238-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennarz S, Alich TC, Kelly T, Blind M, Beck H, Mayer G. Selective aptamer-based control of intraneuronal signaling. Angew Chem Int Ed. 2015;54:5369–5373. doi: 10.1002/anie.201409597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhard V, Alasino RV, Bianco ID, Garro AG, Heredia V, Beltramo DM. Self-assembled micelles of monosialogangliosides as nanodelivery vehicles for taxanes. J Controlled Release. 2012;162:619–627. doi: 10.1016/j.jconrel.2012.07.031. [DOI] [PubMed] [Google Scholar]
- Levy-Nissenbaum E, Radovic-Moreno AF, Wang AZ, Langer R, Farokhzad OC. Nanotechnology and aptamers: applications in drug delivery. Trends Biotechnol. 2008;26:442–449. doi: 10.1016/j.tibtech.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Li S, Xu H, Ding H, Huang Y, Cao X, Yang G, Li J, Xie Z, Meng Y, Li X, Zhao Q. Identification of an aptamer targeting hnRNP A1 by tissue slide-based SELEX. J Pathol. 2009;218:327–336. doi: 10.1002/path.2543. [DOI] [PubMed] [Google Scholar]
- Li N, Nguyen HH, Byrom M, Ellington AD. Inhibition of cell proliferation by an anti-EGFR aptamer. PLoS ONE. 2011;6:e20299. doi: 10.1371/journal.pone.0020299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Hou J, Liu X, Guo Y, Wu Y, Zhang L, Yang Z. Nucleolin-targeting liposomes guided by aptamer AS1411 for the delivery of siRNA for the treatment of malignant melanomas. Biomaterials. 2014;35:3840–3850. doi: 10.1016/j.biomaterials.2014.01.019. [DOI] [PubMed] [Google Scholar]
- Li XL, Zhang WY, Liu L, Zhu Z, Ouyang GL, An Y, Zhao CY, Yang CJ. In vitro selection of DNA aptamers for metastatic breast cancer cell recognition and tissue imaging. Anal Chem. 2014;86:6596–6603. doi: 10.1021/ac501205q. [DOI] [PubMed] [Google Scholar]
- Li X, An Y, Jin J, Zhu Z, Hao L, Liu L, Shi Y, Fan D, Ji T, Yang CJ. Evolution of DNA aptamers through in vitro metastatic-cell-based systematic evolution of ligands by exponential enrichment for metastatic cancer recognition and imaging. Anal Chem. 2015;87:4941–4948. doi: 10.1021/acs.analchem.5b00637. [DOI] [PubMed] [Google Scholar]
- Li FF, Lu J, Liu J, Liang C, Wang ML, Wang LY, Li DF, Yao HZ, Zhang QL, Wen J, Zhang ZK. A water-soluble nucleolin aptamer-paclitaxel conjugate for tumor-specific targeting in ovarian cancer. Nat Commun. 2017;8:1390–1403. doi: 10.1038/s41467-017-01565-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao ZX, Chuang EY, Lin CC, Ho YC, Lin KJ, Cheng PY, Chen KJ, Wei HJ, Sung HW. An AS1411 aptamer-conjugated liposomal system containing a bubble-generating agent for tumor-specific chemotherapy that overcomes multidrug resistance. J Control Release. 2015;208:42–51. doi: 10.1016/j.jconrel.2015.01.032. [DOI] [PubMed] [Google Scholar]
- Lipi F, Chen S, Chakravarthy M, Rakesh S, Veedu RN. In vitro evolution of chemically-modified nucleic acid aptamers: pros and cons, and comprehensive selection strategies. RNA Biol. 2016;13:1232–1245. doi: 10.1080/15476286.2016.1236173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Liu D, Wang L, Zhang J, Zhang N. Docetaxel-loaded pluronic p123 polymeric micelles: in vitro and in vivo evaluation. Int J Mol Sci. 2011;12:1684–1696. doi: 10.3390/ijms12031684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Duan J-H, Song Y-M, Ma J, Wang FD, Lu X, Yang XD. Novel HER2 aptamer selectively delivers cytotoxic drug to HER2-positive breast cancer cells in vitro. J Transl Med. 2012;10:148. doi: 10.1186/1479-5876-10-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long SB, Long MB, White RR, Sullenger BA. Crystal structure of an RNA aptamer bound to thrombin. RNA. 2008;14:2504–2512. doi: 10.1261/rna.1239308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Liu J, Ali MM, Mahmood MAI, Labanieh L, Lu M, Iqbal SM, Zhang Q, Zhao W, Wan Y. Nucleic acid aptamers in cancer research, diagnosis and therapy. Chem Soc Rev. 2015;44:1240–1256. doi: 10.1039/c4cs00357h. [DOI] [PubMed] [Google Scholar]
- Ma Q, Lee D, Tan YQ, Wong G, Gao Z. Synthetic genetic polymers: advances and applications. Polym Chem. 2016;7(33):5199–5216. [Google Scholar]
- Mascini M. Aptamers in bioanalysis. Amsterdam: Wiley; 2009. [Google Scholar]
- Mastico RA, Talbot SJ, Stockley PG. Multiple presentation of foreign peptides on the surface of an RNA-free spherical bacteriophage capsid. J Gen Virol. 1993;74:541–548. doi: 10.1099/0022-1317-74-4-541. [DOI] [PubMed] [Google Scholar]
- McKeague M, Foster A, Miguel Y, Giamberardino A, Verdin C, Chan JY, DeRosa MC. Development of a DNA aptamer for direct and selective homocysteine detection in human serum. RSC Adv. 2013;3:24415–24422. [Google Scholar]
- McNamara JO, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E, Sullenger BA, Giangrande PH. Cell type specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol. 2006;24:1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- Mei H, Liao JY, Jimenez RM, Wang Y, Bala S, McCloskey C, Switzer C, Chaput JC. Synthesis and evolution of a threose nucleic acid aptamer bearing 7-deaza-7-substituted guanosine residues. J Am Chem Soc. 2018;140:5706–5713. doi: 10.1021/jacs.7b13031. [DOI] [PubMed] [Google Scholar]
- Mendonsa SD, Bowser MT. In vitro evolution of functional DNA using capillary electrophoresis. J Am Chem Soc. 2004;126:20–21. doi: 10.1021/ja037832s. [DOI] [PubMed] [Google Scholar]
- Meng L, Sefah K, O’Donoghue MB, Zhu GZ, Shangguan DH, Noorali A, Chen Y, Zhou L, Tan WH. Silencing of PTK7 in colon cancer cells: caspase-10-dependent apoptosis via mitochondrial pathway. PLoS ONE. 2010;5:e14018–e14029. doi: 10.1371/journal.pone.0014018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Z, Westhof E. RNA structure: advances and assessment of 3D structure prediction. Annu Rev Biophys. 2017;46:483–503. doi: 10.1146/annurev-biophys-070816-034125. [DOI] [PubMed] [Google Scholar]
- Min K, Cho M, Han SY, Shim YB, Ku J, Ban C. A simple and direct electrochemical detection of interferon-γ using its RNA and DNA aptamers. Biosens Bioelectron. 2008;23:1819–1824. doi: 10.1016/j.bios.2008.02.021. [DOI] [PubMed] [Google Scholar]
- Mody VV, Siwale R, Singh A, Mody HR. Introduction to metallic nanoparticles. J Pharm Bioallied Sci. 2010;2:282–289. doi: 10.4103/0975-7406.72127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy MB, Fuller ST, Richardson PM, Doyle SA. An improved method for the in vitro evolution of aptamers and applications in protein detection and purification. Nuc Acid Res. 2003;31:e110. doi: 10.1093/nar/gng110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng EW, Shima DT, Calias P, Cunningham ET, Guyer DR, Adamis AP. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat Rev Drug Discov. 2006;5:123–132. doi: 10.1038/nrd1955. [DOI] [PubMed] [Google Scholar]
- Ninomiya K, Kaneda K, Kawashima S, Miyachi Y, Ogino C, Shimizu N. Cell-SELEX based selection and characterization of DNA aptamer recognizing human hepatocarcinoma. Bioorg Med Chem Lett. 2013;23:1797–1802. doi: 10.1016/j.bmcl.2013.01.040. [DOI] [PubMed] [Google Scholar]
- Nitsche A, Kurth A, Dunkhorst A, Pänke O, Sielaff H, Junge W, Muth D, Scheller F, Stöcklein W, Dahmen C, Pauli G. One-step selection of Vaccinia virus-binding DNA aptamers by MonoLEX. BMC Biotechnol. 2007;7:48. doi: 10.1186/1472-6750-7-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu WJ, Chen XG, Tan WH, Veige AS. N-heterocyclic carbene-Gold(I) complexes conjugated to a leukemia-specific DNA aptamer for targeted drug delivery. Angew Chem Int Ed. 2016;55:8889–8893. doi: 10.1002/anie.201602702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Codnnell D, Koenig A, Jennings S, Hicke B, Han HL, Fitzwater T, Chang YF, Varki N, Parma D, Varki A. Calcium dependent oligonucleotide antagonists specific for l-selectin. Proc Nat Acad Sci (USA) 1996;93:5883–5887. doi: 10.1073/pnas.93.12.5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donoghue MB. A liposome-based nanostructure for aptamer directed delivery. Chem Commun. 2010;46:249–251. doi: 10.1039/b916911c. [DOI] [PubMed] [Google Scholar]
- Ohuchi SP, Ohtsu T, Nakamura Y. Selection of RNA aptamers against recombinant transforming growth factor-β type III receptor displayed on cell surface. Biochimie. 2006;88:897–904. doi: 10.1016/j.biochi.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Orava EW, Cicmil N, Gariepy J. Delivering cargoes into cancer cells using DNA aptamers targeting internalized surface portals. BBA-Biomembr. 2010;1798:2190–2200. doi: 10.1016/j.bbamem.2010.02.004. [DOI] [PubMed] [Google Scholar]
- Osborne SE, Matsumura I, Ellington AD. Aptamers as therapeutic and diagnostic reagents: problems and prospects. Curr Opin Chem Biol. 1997;1:5–9. doi: 10.1016/s1367-5931(97)80102-0. [DOI] [PubMed] [Google Scholar]
- Ouellet E, Foley JH, Conway EM, Haynes C. Hi-Fi SELEX: a high-fidelity digital-PCR based therapeutic aptamer discovery platform. Biotechnol Bioeng. 2015;112(8):1506–1522. doi: 10.1002/bit.25581. [DOI] [PubMed] [Google Scholar]
- Pagratis NC, Bell C, Chang YF, Jennings S, Fitzwater T, Jellinek D, Dang C. Potent 2ʹ-amino- and 2-fluoro-2-deoxyribonucleotide RNA inhibitors of keratinocyte growth factor. Nat Biotechnol. 1997;15:68–73. doi: 10.1038/nbt0197-68. [DOI] [PubMed] [Google Scholar]
- Peng L, Stephens BJ, Bonin K, Cubicciotti R, Guthold M. A combined atomic force/fluorescence microscopy technique to select aptamers in a single cycle from a small pool of random oligonucleotides. Micros Res Tech. 2007;70:372–381. doi: 10.1002/jemt.20421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JA, Lopez-Colon D, Zhu Z, Xu Y, Tan W. Applications of aptamers in cancer cell biology. Anal Chim Acta. 2008;621:101–108. doi: 10.1016/j.aca.2008.05.031. [DOI] [PubMed] [Google Scholar]
- Pinheiro VB, Holliger P. The XNA world: progress towards replication and evolution of synthetic genetic polymers. Curr Opin Chem Biol. 2012;16:245–252. doi: 10.1016/j.cbpa.2012.05.198. [DOI] [PubMed] [Google Scholar]
- Pinheiro VB, Holliger P. Towards XNA nanotechnology: new materials from synthetic genetic polymers. Trends Biotechnol. 2014;32(6):321–328. doi: 10.1016/j.tibtech.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plapied L, Duhem N, des Rieux A, Preat V. Fate of polymeric nanocarriers for oral drug delivery. Curr Opin Colloid Interface Sci. 2011;16:228–237. [Google Scholar]
- Prakash TP. An overview of sugar-modified oligonucleotides for antisense therapeutics. Chem Biodiver. 2011;8:1616–1641. doi: 10.1002/cbdv.201100081. [DOI] [PubMed] [Google Scholar]
- Radom F, Jurek PM, Mazurek MP, Otlewski J, Jelen F. Aptamers: molecules of great potential. Biotechnol Adv. 2013;31:1260–1274. doi: 10.1016/j.biotechadv.2013.04.007. [DOI] [PubMed] [Google Scholar]
- Rangel AE, Chen Z, Ayele TM, Heemstra JM. In vitro selection of an XNA aptamer capable of small-molecule recognition. Nucleic Acid Res. 2018;46:8057–8068. doi: 10.1093/nar/gky667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravelet C, Grosset C, Peyrin E. Liquid chromatography, electrochromatography and capillary electrophoresis applications of DNA and RNA aptamers. J Chromat A. 2006;1117:1–10. doi: 10.1016/j.chroma.2006.03.101. [DOI] [PubMed] [Google Scholar]
- Rentmeister A, Bill A, Wahle T, Walter J, Famulok M. RNA aptamers selectively modulate protein recruitment to the cytoplasmic domain of β-secretase BACE1 in vitro. RNA. 2006;12:1650–1660. doi: 10.1261/rna.126306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohloff JC, Gelinas AD, Jarvis TC, Ochsner UA, Schneider DJ, Gold L, Janjic N. Nucleic acid ligands with protein-like side chains: modified aptamers and their use as diagnostic and therapeutic agents. Mol Ther Nucleic Acids. 2014;3:201. doi: 10.1038/mtna.2014.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenblum GT, Lopez VG, Vitullo AD, Radrizzani M. Aptamers: current challenges and future prospects. Expert Opin Drug Dis. 2016;11:127–135. doi: 10.1517/17460441.2016.1126244. [DOI] [PubMed] [Google Scholar]
- Rui M, Xin Y, Li R, Ge Y, Feng C, Xu X. Targeted biomimetic nanoparticles for synergistic combination chemotherapy of paclitaxel and doxorubicin. Mol Pharmaceut. 2017;14:107–123. doi: 10.1021/acs.molpharmaceut.6b00732. [DOI] [PubMed] [Google Scholar]
- Ryou SM, Kim JM, Yeom JH, Hyun S, Kim S, Han MS, Kim SW, Bae J, Rhee S, Lee K. Gold nanoparticle-assisted delivery of small, highly structured RNA into the nuclei of human cells. Biochem Biophy Res Commun. 2011;416:178–183. doi: 10.1016/j.bbrc.2011.11.020. [DOI] [PubMed] [Google Scholar]
- Rytting E, Nguyen J, Wang X, Kissel T. Biodegradable polymeric nanocarriers for pulmonary drug delivery. Expert Opin Drug Del. 2008;5:629–639. doi: 10.1517/17425247.5.6.629. [DOI] [PubMed] [Google Scholar]
- Sefah K, Shangguan D, Xiong X, O’donoghue MB, Tan W. Development of DNA aptamers using cell-SELEX. Nat Protoc. 2010;5:1169. doi: 10.1038/nprot.2010.66. [DOI] [PubMed] [Google Scholar]
- Shigdar S, Lin J, Yu Y, Pastuovic M, Wei M, Duan W. RNA aptamer against a cancer stem cell marker epithelial cell adhesion molecule. Cancer sci. 2011;102:991–998. doi: 10.1111/j.1349-7006.2011.01897.x. [DOI] [PubMed] [Google Scholar]
- Shoji A, Kuwahara M, Ozaki H, Sawai H. Modified DNA aptamer that binds the (R)-isomer of a thalidomide derivative with high enantioselectivity. J Am Chem Soc. 2007;129:1456–1464. doi: 10.1021/ja067098n. [DOI] [PubMed] [Google Scholar]
- Singer BS, Shtatland T, Brown D, Gold L. Libraries for genomic SELEX. Nucleic Acids Res. 1997;25(4):781–786. doi: 10.1093/nar/25.4.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Koshkin AA, Wengel J, Nielsen P. LNA (locked nucleic acids): synthesis and high-affinity nucleic acid recognition. Chem Commun. 1998;4:455–456. [Google Scholar]
- Song YL, Zhu Z, An Y, Zhang WT, Zhang HM, Liu D, Yu CD, Duan W, Yang CJ. Selection of DNA aptamers against epithelial cell adhesion molecule for cancer cell imaging and circulating tumor cell capture. Anal Chem. 2013;85:4141–4149. doi: 10.1021/ac400366b. [DOI] [PubMed] [Google Scholar]
- Soukup GA, Emilsson GA, Breaker RR. Altering molecular recognition of RNA aptamers by allosteric selection. J Mol Biol. 2000;298:623–632. doi: 10.1006/jmbi.2000.3704. [DOI] [PubMed] [Google Scholar]
- Soundararajan S, Chen W, Spicer EK, Courtenay-Luck N, Fernandes DJ. The nucleolin targeting aptamer AS1411 destabilizes Bcl-2 messenger RNA in human breast cancer cells. Cancer Res. 2008;68:2358–2365. doi: 10.1158/0008-5472.CAN-07-5723. [DOI] [PubMed] [Google Scholar]
- Soundararajan S, Wang L, Sridharan V, Chen W, Courtenay-Luck N, Jones D, Spicer EK, Fernandes DJ. Plasma membrane nucleolin is a receptor for the anticancer aptamer AS1411 in MV4-11 leukemia cells. Mol Pharm. 2009;76:984–991. doi: 10.1124/mol.109.055947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein CA, Castanotto D. FDA-approved oligonucleotide therapies in 2017. Mol Ther. 2017;25:1069–1075. doi: 10.1016/j.ymthe.2017.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltenburg R, Reinemann C, Strehlitz B. FluMag-SELEX as an advantageous method for DNA aptamer selection. Anal Bioanal Chem. 2005;383:83–91. doi: 10.1007/s00216-005-3388-9. [DOI] [PubMed] [Google Scholar]
- Stoltenburg R, Nikolaus N, Strehlitz B. Capture-SELEX: selection of DNA aptamers for aminoglycoside antibiotics. J Anal Methods Chem. 2012;2012:1–14. doi: 10.1155/2012/415697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Su J, Meng Q, Yin Q, Chen L, Gu W, Zhang P, Zhang Z, Yu H, Wang S, Li Y. Cancer-cell-biomimetic nanoparticles for targeted therapy of homotypic tumors. Adv Mater. 2016;28:9581–9588. doi: 10.1002/adma.201602173. [DOI] [PubMed] [Google Scholar]
- Sundaram P, Kurniawan H, Byrne ME, Wower J. Therapeutic RNA aptamers in clinical trials. Eur J Pharmaceut Sci. 2013;48:259–271. doi: 10.1016/j.ejps.2012.10.014. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Sakota E, Nakamura Y. The efficient cell-SELEX strategy, Icell-SELEX, using isogenic cell lines for selection and counter-selection to generate RNA aptamers to cell surface proteins. Biochimie. 2016;131:77–84. doi: 10.1016/j.biochi.2016.09.018. [DOI] [PubMed] [Google Scholar]
- Tan Y, Shi YS, Wu XD, Liang HY, Gao YB, Li SJ, Zhang XM, Wang F, Gao TM. DNA aptamers that target human glioblastoma multiforme cells overexpressing epidermal growth factor receptor variant III in vitro. Acta Pharmacol Sin. 2013;34:1491–1498. doi: 10.1038/aps.2013.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao W, Zeng X, Wu J, Zhu X, Yu X, Zhang X, Zhang J, Liu G, Mei L. Polydopamine-based surface modification of novel nanoparticle-aptamer bioconjugates for in vivo breast cancer targeting and enhanced therapeutic effects. Theranostics. 2016;6:470. doi: 10.7150/thno.14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolle F, Mayer G. Dressed for success–applying chemistry to modulate aptamer functionality. Chem Sci. 2013;4:60–67. [Google Scholar]
- Tolle F, Brandle GM, Matzner D, Mayer G. A versatile approach towards nucleobase-modified aptamers. Angew Chem Int Ed. 2015;54:10971–10974. doi: 10.1002/anie.201503652. [DOI] [PubMed] [Google Scholar]
- Torres LL, Pinheiro VB. Xenobiotic nucleic acid (XNA) synthesis by Phi29 DNA polymerase. Curr Protoc Chem Biol. 2018;10(2):e41. doi: 10.1002/cpch.41. [DOI] [PubMed] [Google Scholar]
- Tsukioka Y, Matsumura Y, Hamaguchi T, Koike H, Moriyasu F, Kakizoe T. Pharmaceutical and biomedical differences between micellar doxorubicin (NK911) and liposomal doxorubicin (Doxil) Jpn J Cancer Res. 2002;93:1145–1153. doi: 10.1111/j.1349-7006.2002.tb01217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker WO, Shum KT, Tanner JA. G-quadruplex DNA aptamers and their ligands: structure, function and application. Curr Pharmaceut Design. 2012;18:2014–2026. doi: 10.2174/138161212799958477. [DOI] [PubMed] [Google Scholar]
- Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249(4968):505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Ulrich H. RNA towards medicine. Berlin, Heidelberg: Springer; 2006. RNA aptamers: from basic science towards therapy; pp. 305–326. [DOI] [PubMed] [Google Scholar]
- Ulrich H, Ippolito JE, Pagan OR, Eterovic VA, Hann RM, Shi H, Lis JT, Eldefrawi ME, Hess GP. In vitro selection of RNA molecules that displace cocaine from the membrane bound nicotinic acetylcholine receptor. Proc Nat Acad Sci (USA) 1998;95:14051–14056. doi: 10.1073/pnas.95.24.14051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich H, Martins AHB, Pesquero JB. RNA and DNA aptamers in cytomics analysis. Cytometry Part A. 2004;59:220–231. doi: 10.1002/cyto.a.20056. [DOI] [PubMed] [Google Scholar]
- Urata H, Ogura E, Shinohara K, Ueda Y, Akagi M. Synthesis and properties of mirror-image DNA. Nucleic Acids Res. 1992;20:3325–3332. doi: 10.1093/nar/20.13.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandghanooni S, Eskandani M, Barar J, Omidi Y. AS1411 aptamer-decorated cisplatin-loaded poly (lactic-co-glycolic acid) nanoparticles for targeted therapy of miR-21-inhibited ovarian cancer cells. Nanomedicine. 2018;13:2729–2758. doi: 10.2217/nnm-2018-0205. [DOI] [PubMed] [Google Scholar]
- Vater A, Jarosch F, Buchner K, Klussmann S. Short bioactive Spiegelmers to migraine-associated calcitonin gene-related peptide rapidly identified by a novel approach: tailored-SELEX. Nucleic Acids Res. 2003;31:130. doi: 10.1093/nar/gng130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Ye L, Yang XH, Guo QP, Wang KM, Huang ZX, Tan YY, Yuan BY, Xie Q. Cell-SELEX based selection and optimization of DNA aptamers for specific recognition of human cholangiocarcinoma QBC-939 cells. Analyst. 2015;140:5992–5997. doi: 10.1039/c5an01055a. [DOI] [PubMed] [Google Scholar]
- Wang E, Wu R, Niu HY, Cai J. Improving the stability of aptamers by chemical modification. Curr Med Chem. 2011;18:4126–4138. doi: 10.2174/092986711797189565. [DOI] [PubMed] [Google Scholar]
- Wang P, Yang Y, Hong H, Zhang Y, Cai W, Fang D. Aptamers as therapeutics in cardiovascular diseases. Curr Med Chem. 2011;18:4169–4174. doi: 10.2174/092986711797189673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Zhao J, Jiang T, Kwon YM, Lu H, Jiao P, Liao M, Li Y. Selection and characterization of DNA aptamers for use in detection of avian influenza virus H5N1. J Virol Methods. 2013;189:362–369. doi: 10.1016/j.jviromet.2013.03.006. [DOI] [PubMed] [Google Scholar]
- Wang Q, Liu W, Xing Y, Yang X, Wang K, Jiang R, Wang P, Zhao Q. Screening of DNA aptamers against myoglobin using a positive and negative selection units integrated microfluidic chip and its biosensing application. Anal Chem. 2014;86:6572–6579. doi: 10.1021/ac501088q. [DOI] [PubMed] [Google Scholar]
- Wang R, Zhu G, Mei L, Xie Y, Ma H, Ye M, Qing F-L, Tan W. Automated modular synthesis of aptamer-drug conjugates for targeted drug delivery. J Am Chem Soc. 2014;136:2731–2734. doi: 10.1021/ja4117395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Luo Y, Bing T, Chen Z, Lu M, Zhang N, Shangguan D, Gao X. DNA aptamer evolved by cell-SELEX for recognition of prostate cancer. PLoS ONE. 2014;9:e100243. doi: 10.1371/journal.pone.0100243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GQ, Maheshwari N, Eisenstein M, Arcila ML, Kosik KS, Soh HT. Particle display: a quantitative screening method for generating high-affinity aptamers. Angew Chem Int Ed Engl. 2014;126:4896–4901. doi: 10.1002/anie.201309334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Yao H, Meng Y, Wang Y, Yan X, Huang R. Specific aptamer- conjugated mesoporous silica–carbon nanoparticles for HER2-targeted chemo- photothermal combined therapy. Acta Biomat. 2015;16:196–205. doi: 10.1016/j.actbio.2015.01.002. [DOI] [PubMed] [Google Scholar]
- Wang X, Zhou X, Wang J, Cao Z, Zhang L, Tang R. Acid-labile copolymer micelles cross-linked by a twin ortho ester cross-linking agent: synthesis, characterization, and evaluation. Macromol Chem Phys. 2016;217:2182–2190. [Google Scholar]
- White RR, Sullenger BA, Rusconi CP. Developing aptamers into therapeutics. J Clin Invest. 2000;106:929–934. doi: 10.1172/JCI11325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R, Rusconi C, Scardino E, Wolberg A, Lawson J, Hoffman M, Sullenger B. Generation of species cross-reactive aptamers using "toggle" SELEX. Mol Ther. 2001;4(6):567–573. doi: 10.1006/mthe.2001.0495. [DOI] [PubMed] [Google Scholar]
- Wilner SE, Wengerter B, Maier K, de Lourdes B, Magalhaes M, Del Amo DS, Pai S, Opazo F, Rizzoli SO, Yan A, Levy M. An RNA alternative to human transferrin: a new tool for targeting human cells. Mol Ther Nucleic Acids. 2012;1:e21–e21. doi: 10.1038/mtna.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolter O, Mayer G. Aptamers as valuable molecular tools in neurosciences. J Neurosci. 2017;37:2517–2523. doi: 10.1523/JNEUROSCI.1969-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Curran JF. An allosteric synthetic DNA. Nucleic Acids Res. 1999;27:1512–1516. doi: 10.1093/nar/27.6.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Sefah K, Liu H, Wang R, Tan W. DNA aptamer-micelle as an efficient detection/delivery vehicle toward cancer cells. Proc Natl Acad Sci USA. 2010;107:5–10. doi: 10.1073/pnas.0909611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Zhao Z, Bai H, Fu T, Yang C, Hu X, Liu Q, Champanhac C, Teng IT, Ye M, Tan W. DNA aptamer selected against pancreatic ductal adenocarcinoma for in vivo imaging and clinical tissue recognition. Theranostics. 2015;5:985–994. doi: 10.7150/thno.11938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao SJ, Hu PP, Xiao GF, Wang Y, Liu Y, Huang CZ. Label-free detection of prion protein with its DNA aptamer through the formation of T-Hg2+-T configuration. J Phys Chem B. 2012;116:9565–9569. doi: 10.1021/jp302522b. [DOI] [PubMed] [Google Scholar]
- Xing H, Hwang K, Li J, Torabi SF, Lu Y. DNA aptamer technology for personalized medicine. Curr Opin Chem Eng. 2014;4:79–87. doi: 10.1016/j.coche.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Ellington AD. Anti-peptide aptamers recognize amino acid sequence and bind a protein epitope. Proc Nat Acad Sci (USA) 1996;93:7475–7480. doi: 10.1073/pnas.93.15.7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu JH, Teng IT, Zhang LQ, Delgado S, Champanhac C, Cansiz S, Wu CC, Shan H, Tan WH. Molecular recognition of human liver cancer cells using DNA aptamers generated via cell-SELEX. PLoS ONE. 2015;10:e0125863. doi: 10.1371/journal.pone.0125863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Gao X, Zhang Z. Isolation and characterization of 2′-amino-modified RNA aptamers for human TNFα. Genom Proteom Bioinform. 2004;2:32–42. doi: 10.1016/S1672-0229(04)02005-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Wei T, Goldberg H, Wang W, Cullion K, Kohane DS. Getting drugs across biological barriers. Adv Mater. 2017;2017:29. doi: 10.1002/adma.201606596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, He X, Wang K, Qing Z, Wu X, Shi H, Yang X. One-step engineering of silver nanoclusters–aptamer assemblies as luminescent labels to target tumor cells. Nanoscale. 2012;4:110–112. doi: 10.1039/c1nr11265a. [DOI] [PubMed] [Google Scholar]
- Ylera F, Lurz R, Erdmann VA, Furste JP. Selection of RNA aptamers to the Alzheimer’s disease amyloid peptide. Biochem Biophy Res Commun. 2002;290:1583–1588. doi: 10.1006/bbrc.2002.6354. [DOI] [PubMed] [Google Scholar]
- Yoo S, Huang KW, Reebye V, Spalding D, Przytycka TM, Wang YJ, Swiderski P, Li L, Armstrong B, Reccia I, Zacharoulis D. Aptamer-drug conjugates of active metabolites of nucleoside analogs and cytotoxic agents inhibit pancreatic tumor cell growth. Mol Ther Nucleic Acids. 2017;17:80–88. doi: 10.1016/j.omtn.2016.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XJ, Zhang J, Ma YY, Pei XY, Liu QM, Lu B, Jin L, Wang JC, Liu J. A cell-based single-stranded DNA aptamer specifically targets gastric cancer. Int J Biochem Cell Biol. 2014;46:1–8. [PubMed] [Google Scholar]
- Zhang J, Chen R, Fang X, Chen F, Wang Y, Chen M. Nucleolin targeting AS1411 aptamer modified pH-sensitive micelles for enhanced delivery and antitumor efficacy of paclitaxel. Nano Res. 2015;8:201–218. [Google Scholar]
- Zhang K, Liu M, Tong X, Sun N, Zhou L, Cao Y, Wang J, Zhang H, Pei R. Aptamer-modified temperature-sensitive liposomal contrast agent for magnetic resonance imaging. Biomacromol. 2015;16:2618–2623. doi: 10.1021/acs.biomac.5b00250. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yu Z, Jiang F, Fu P, Shen J, Wu W, Li J. Two DNA aptamers against avian influenza H9N2 virus prevent viral infection in cells. PLoS ONE. 2015;10:e0123060. doi: 10.1371/journal.pone.0123060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Wang SB, Wu WG, Kankala RK, Chen AZ, Liu YG, Fan JQ. Co-delivery of doxorubicin and AS1411 aptamer by poly (ethylene glycol)-poly (β-amino esters) polymeric micelles for targeted cancer therapy. J Nanoparticle Res. 2017;19:224. [Google Scholar]
- Zhao F, Zhou J, Su X, Wang Y, Yan X, Jia S, Du B. A smart responsive dual aptamers-targeted bubble-generating nanosystem for cancer triplex therapy and ultrasound imaging. Small. 2017;13:20. doi: 10.1002/smll.201603990. [DOI] [PubMed] [Google Scholar]
- Zhou J, Rossi JJ. The therapeutic potential of cell-internalizing aptamers. Curr Top Med Chem. 2009;9:1144–1157. doi: 10.2174/156802609789630893. [DOI] [PubMed] [Google Scholar]
- Zhou J, Rossi J. Aptamers as targeted therapeutics: current potential and challenges. Nat Rev Drug Discov. 2017;16:181. doi: 10.1038/nrd.2016.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Han WF, Landree LE, Thupari JN, Pinn ML, Bililign T, Kim EK, Vadlamudi A, Medghalchi SM, El Meskini R, Ronnett GV. Fatty acid synthase inhibition activates AMP-activated protein kinase in SKOV3 human ovarian cancer cells. Cancer Res. 2007;67:2964–2971. doi: 10.1158/0008-5472.CAN-06-3439. [DOI] [PubMed] [Google Scholar]
- Zhou J, Li H, Li S, Zaia J, Rossi JJ. Novel dual inhibitory function aptamer-siRNA delivery system for HIV-1 therapy. Mol Ther. 2008;16:1481–1489. doi: 10.1038/mt.2008.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Meng L, Ye M, Yang L, Sefah K, O’Donoghue MB, Chen Y, Xiong X, Huang J, Song E, Tan W. Self-assembled aptamer-based drug carriers for bispecific cytotoxicity to cancer cells. Chem Asian J. 2012;7:1630–1636. doi: 10.1002/asia.201101060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Ye M, Donovan MJ, Song E, Zhao Z, Tan W. Nucleic acid aptamers: an emerging frontier in cancer therapy. Chem Commun. 2012;48:10472–10480. doi: 10.1039/c2cc35042d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu GZ, Zheng J, Song EQ, Donovan M, Zhang KJ, Liu C, Tan WH. Self-assembled, aptamer tethered DNA nanotrains for targeted transport of molecular drugs in cancer theranostics. Proc Natl Acad Sci USA. 2013;110:7998–8003. doi: 10.1073/pnas.1220817110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Huang H, Dong S, Ge L, Zhang Y. Progress in aptamer-mediated drug delivery vehicles for cancer targeting and its implications in addressing chemotherapeutic challenges. Theranostics. 2014;4:931–944. doi: 10.7150/thno.9663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Liu G, Kai M. DNA aptamers in the diagnosis and treatment of human diseases. Molecules. 2015;20(12):20979–20997. doi: 10.3390/molecules201219739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo Z, Yu Y, Wang M, Li J, Zhang Z, Liu J, Wu X, Lu A, Zhang G, Zhang B. Recent advances in SELEX technology and aptamer applications in biomedicine. Int J Mol Sci. 2017;18(10):2142. doi: 10.3390/ijms18102142. [DOI] [PMC free article] [PubMed] [Google Scholar]