Abstract
Purpose
Reducing toxicity, immunogenicity, and costs of small interfering RNAs (siRNA) carrier materials are key goals for RNA interference (RNAi) technology transition from bench to bed. Recently, calcium ions (Ca2+) have garnered attention as a novel, alternative material for delivering siRNA to cells. However, the tolerance for Ca2+ concentration varies in different cell types, which has limited its applications in vivo. Bovine serum albumin (BSA) can bind to Ca2+ through chelation. Moreover, BSA is a favorable coating material for nanoparticles owing to its excellent biocompatibility. Therefore, we hypothesized that coating Ca2+-siRNA with BSA helps buffer Ca2+ toxicity in vivo.
Methods
BSA-Ca2+-siRNA nanoparticles were prepared, and the size, shape, encapsulation, and release efficiency were characterized using atomic force microscopy, scanning electronic microcopy, and gel electrophoresis. Binding nanoparticles were evaluated using attenuated total reflection-Fourier-transform infrared spectroscopy. The cellular uptake, intracellular release, cytotoxicity, and gene knockdown of nanoparticles were evaluated in periodontal ligament stem cells (PDLSCs) using laser-scanning confocal microscope, flow cytometry, and real-time quantitative polymerase chain reaction.
Results
BSA and Ca2+-siRNA could form a stable nano-scale complex (~140 nm in diameter). The nanocomplexes could maintain siRNA release for more than 1 week in neutral phosphate-buffered saline (PBS) and could induce accelerated degradation in acidic PBS (pH 5.0). The nanoparticles were taken up by the cells, primarily through macropinocytosis, and were then released intracellularly through the acidification of endosomes/lysosomes. Importantly, the BSA-Ca2+ carrier had high transfection efficiency and biocompatibility both in vitro and in vivo. To demonstrate the therapeutic potential of our BSA coating-optimized Ca2+-siRNA technology, we showed that BSA-Ca2+-siWWP1 complexes strongly enhanced the osteogenic differentiation of inflammatory PDLSCs.
Conclusion
BSA-Ca2+ could potentially be used for siRNA delivery, which is not only highly efficient and cost-effective but also biocompatible to host tissues owing to the BSA coating.
Keywords: bovine serum albumin, calcium ions, osteogenic differentiation, small interfering RNA
Introduction
Owing to its high specificity and effectiveness, RNA interference (RNAi) is used in biomedicine to study target genes, and the potential therapeutic applications of this technique are emerging.1 However, the clinical implementation of RNAi therapy is hampered by factors such as enzymatic degradation, cell targeting, uptake, and endosomal escape under physiological environments.2 To overcome these limitations, research has focused on the development of improved small interfering RNAs (siRNA) carrier materials.3 Most carrier materials are cationic lipids or polymers synthesized by exogenous chemical methods and are commonly toxic and immunogenic.4–6 Moreover, the preparation of such nanocarriers often requires complex technology and is associated with a high cost.7 Therefore, developing convenient and biocompatible siRNA vectors remains a challenge. Reportedly, calcium ions (Ca2+) are adsorbed on siRNA surface and spontaneously form uniform-sized Ca2+-siRNA nanoparticles with an average diameter of 100 nm.2,5,7 These nanoparticles can be taken up by mesenchymal stem cells (MSCs) or some cancer cells, resulting in efficient, convenient, and low-cost siRNA delivery.2,7 However, high Ca2+ levels are toxic in vivo and can lead to blood coagulation and acute inflammation.8,9 In addition, we found that in vitro transfection using medium containing high Ca2+ concentrations and without fetal bovine serum (FBS) can induce cell death. Therefore, we hypothesized that FBS, particularly the protein contained within it, plays a crucial role in alleviating Ca2+ toxicity.
Bovine serum albumin (BSA) is the primary component of FBS and can be combined with a variety of cations, anions, and other small molecule substances.10 BSA addition to in vitro cell cultures without FBS can confer physiological and mechanical protection.11 Furthermore, BSA can also be used as a carrier for various drug-delivery applications due to its favorable properties such as high biodegradability, low toxicity, low antigenicity, high stability, ability for long-term storage, and ease of production.12,13 In addition, studies have demonstrated that Ca2+ can chelate with BSA to form microspheres.14 Therefore, we hypothesized that coating Ca2+-siRNA with BSA can help buffer Ca2+ toxicity and can be used for in vivo applications. Herein, we fabricated BSA-Ca2+-siRNA nanoparticles, to the best of our knowledge, for the first time and presented a scheme of the fabrication flow: premixing Ca2+ and siRNA to form nanocomplexes prior to BSA coating on the surface of the Ca/siRNA complexes. The cellular uptake, release, and transfection of nanoparticles were evaluated. Transfection and knockout efficiency were optimized by adjusting the BSA or Ca2+ concentrations and the therapeutic potential of the BSA-Ca2+ delivery system was evaluated through cytotoxicity and hemolysis experiments.
To demonstrate the therapeutic potential of this BSA-Ca2+-siRNA transfection technology, we observed the effect of BSA-Ca2+-siWWP1 on the promotion of osteogenesis in inflammatory PDLSCs. At present, available treatment options for periodontitis mostly focus on controlling inflammation; however, it remains difficult to restore the absorbed alveolar bone to normal. Therefore, the difficulty in delaying bone resorption is also a key limitation in the clinical treatment of periodontitis. The WW domain containing E3 ubiquitin protein ligase 1 (WWP1) is a member of the C2-WW-HECT family of E3 ligases.15,16 Chronic exposure to tumor necrosis factor-α (TNF-α) reduces the osteogenic potential of MSCs by increasing WWP1-mediated degradation of Runx2 and JunB proteins.17,18 Targeted WWP1 knockdown can significantly improve bone phenotype in vivo and in vitro through signaling pathways related to bone morphogenetic proteins.17,19 Therefore, we delivered WWP1 siRNA via a BSA-Ca2+ delivery platform for inducing osteogenesis in inflammatory periodontal ligament stem cells (PDLSCs).
Materials and Methods
Preparation of BSA-Ca2+-siRNA Complexes
The sequences of gene-targeting siRNAs (Sangon, China) dissolved in RNase-free water are shown in Table S1. HEPES (10 mM, pH 7.4), BSA (5–5,000 μg/mL), siRNA (20 μM), and CaCl2 (0.5–2 M stock concentrations) solutions were freshly prepared in RNase-free water at a ratio of 1:1:1:1 to obtain the molar ratios. The scheme of the manufacturing process is as follows. First, equal volumes of CaCl2 and siRNA were vortexed for 45 s and incubated for 25 min at 4°C.7 Then, an equal volume of BSA was added, and the mixture was vortexed for 45 s, followed by incubation for 25 min at 4°C. Finally, an equal volume of HEPES was added, and the mixture was vortexed for 45 s, followed by incubation for 25 min at 4°C prior to use. Lipofectamine 2000 (Invitrogen, USA) transfection complex was prepared in Opti-Minimum Essential Medium (Opti-MEM; Gibco, USA) according to the manufacturer’s instructions. Other reagents, unless specified otherwise, were obtained from Sigma-Aldrich (USA).
Cell Culture
PDLSCs were derived from fresh premolars without caries that were removed for orthodontic purposes from healthy individuals aged 19–31 years. All study donors provided informed consent, and all procedures followed the guidelines of the Ethics Committee of the Stomatological Hospital of Air Force Medical University (approval Number IRB-REV-2017012). PDLSCs were cultured using limiting dilution analysis, and third-to-fifth generation polyclonal PDLSCs were selected for use in this study. According to the manufacturer’s instructions, a lentiviral vector expressing green fluorescent protein (GFP: pRLenti-EF1a-EGFP-P2A-Puro-CMV-MCS-3Flag; Obio, China) was used to infect PDLSCs to obtain GFP-PDLSCs. PDLSCs, with and without GFP expression, were cultured in α-Minimum Essential Medium (α-MEM; Hyclone, USA), supplemented with L-glutamine (Lonza, USA) and 10% FBS (Gibco, USA). Under conventional transfection conditions, PDLSCs were seeded in a 24-well plate at a density of 22,000 cells/well. After 24 h, PDLSCs were incubated with transfection complexes for 3.5 h and replaced with fresh medium.
Characterization of Complexes
Three microliters of BSA-Ca2+-siRNA complexes were deposited on the surface of the silicon wafer. After 20 min, the surface was dried under N2 gas. The morphology of samples was observed via the tapping mode of atomic force microscopy (AFM; Agilent, USA). Apparent diameters of the particles were analyzed using the SPM software (Agilent, USA). The samples were characterized using scanning electronic microcopy (SEM; Thermo Fisher, USA) in a low-dose imaging mode of 5 kV. The particle size was measured using the ImageJ software (National Institutes of Health, USA). Zeta potential and dynamic light scattering (DLS)-measured particle size distribution of complexes were obtained using a Zetasizer Nano (Malvern Panalytical, UK). Approximately 100 µL samples were placed in centrifuge tubes, dried in a lyophilizer (Biocool, China) at −80°C for 24 h, and chemically characterized using attenuated total reflection-Fourier-transform infrared spectroscopy (FTIR; Nexus-670, USA).
Cargo Loading and Release
The loading efficiency of the siRNAs was evaluated by gel electrophoresis. siRNA complexes were loaded onto a gel consisting of 3.5% agarose and SYBR Gold, and then electrophoresed in TAE buffer solution at 110 V and 40 mA. The obtained siRNA signals were imaged using a Gel Doc 2000 (Bio-Rad, USA). BSA-Ca2+-siRNA particles were suspended and incubated in neutral (pH 7.4) or acidic (pH 5.0) PBS at 37°C for 24 h. Subsequently, a spectrophotometer (Thermo Fisher, USA) was used to analyze the siRNA content of the supernatant of the centrifuged samples. The centrifuged particles were re-immersed in PBS and allowed to continue degradation. This step was repeated for 7 days.20
Uptake and Release Pathways
PDLSCs (P3) were seeded on glass-bottomed dishes (MatTek, USA) at a density of 42,000 cells/well. After 24 h, PDLSCs were washed with PBS and treated with each of the inhibitors at its selected concentration (Table 1) for 1 h.5,21 Then, BSA-Ca2+-siRNA nanocomposites (5 μM siRNA, 125 μg/mL BSA, and 250 mM CaCl2) that were freshly diluted 1:50 in α-MEM were added, and cells were incubated for a further 3.5 h. Finally, PDLSCs were analyzed using a flow cytometer (Beckman Coulter, USA) and fixed with 4% paraformaldehyde. The cytoskeleton was stained with AF635-phalloidin (Thermo Fisher, USA), lysosomes/endosomes were stained with LysoTracker (Invitrogen, USA), and the nuclei were stained with Hoechst (Invitrogen, USA). Images were obtained using a laser-scanning confocal microscope (LSCM; Nikon, Japan).
Table 1.
Inhibitors: Functions and Concentrations
| Inhibitor Name | Pathway Inhibited | Chosen Concentration |
|---|---|---|
| Chlorpromazine | Clathrin-dependent | 20 μM |
| Genistein | Caveolae-dependent | 50 μM |
| Amiloride | Actin-dependent | 50 μM |
| Bafilomycin A1 | V-ATPase inhibitor | 75 nM |
| YM201636 | Maturation of early endosome | 2 μM |
Abbreviation: V-ATPase, vacuolar-type H+ ATPase.
Knockdown Efficiency Quantification
Knockdown efficiency was confirmed 72 h post-transfection. A fluorescence microscope (Olympus, Japan) was used to determine the fluorescence intensity of GFP-PDLSCs. Additionally, GFP-PDLSCs were collected by trypsinization and analyzed using a flow cytometer. The geometric mean of FL1 fluorescence intensity was calculated to evaluate the knockdown efficiency. The GFP silencing efficiency percentage was calculated using the following formulas:
% GFP = MFI GFP in sample/MFI GFP in negative control × 100, and
% Silencing = 100–%GFP,
where MFI is the mean fluorescence intensity.
Cytotoxicity Assay
PDLSCs were seeded in 96-well plates (2,200 cells/well) and incubated in α-MEM for 24 h. Next, α-MEM was replaced with serum-free medium containing BSA-Ca2+-siRNA complexes (dilution ratio with medium, 1:50). After 24 h, PDLSCs were washed with PBS and incubated in α-MEM for another 48 h. Cell viability was determined using Cell Counting Kit-8 (CCK-8; Yeasen, China).
Hemolysis Assay
BALB/c mice (8 weeks old) were provided by the Air Force Medical University. All animal experiments were approved by the Experimental Animal Management Ethics Committee of Stomatological Hospital at the Air Force Medical University (approval number 2019-LAEI-kq-016) and were conducted in accordance with the Guidelines for Ethical Review of Laboratory Animal Welfare (GB/T 35892–2018) issued by National Technical Committee 281 on Laboratory Animal Science of Standardization Administration of China (SAC/TC 281).
Red blood cells (RBCs) were obtained by centrifuging blood collected from 10 healthy BALB/c mice (0.1 mL per mouse). siRNA nanocomplexes suspended in PBS at a ratio of 1:200 were incubated with RBCs at 37°C for 3 h. The absorbance (X) of the supernatant from centrifuged samples was measured to estimate the amount of hemoglobin released from RBCs. RBCs were also treated with 1% Triton and PBS to provide positive and negative control samples, respectively.22 The formula for calculating the degree of hemolysis is as follows:
(X from complexes-X from PBS)/(X from Triton-X from PBS) × 100%.
In vivo Cytocompatibility Assay
BALB/c mice (three per group) were injected with 100 μL Ca2+-siRNA (5 mM CaCl2, 100 nM siRNA) alone or complexed with BSA (0.025–25 μg/mL) in the hind leg muscles. In addition, Milli-Q water was used as a negative control. For histological evaluation, legs were collected 48 h after injection and fixed in 4% paraformaldehyde. The sections were stained with hematoxylin and eosin (HE staining).
Osteogenesis-Related Gene Expression
Total RNA was extracted with TRIzol and complementary DNA (cDNA) was obtained using a reverse transcription kit (Qiagen, Germany), according to the manufacturer’s instructions. OneStep RT-PCR Kit (Qiagen, Germany) and ABI 7500 system (Applied Biosystems, USA) were used for amplification and detection. Expression was determined by normalizing with the glyceraldehyde phosphate dehydrogenase (GAPDH) gene, which acted as a control gene (encoding). The related primer sequences are listed in Table 2.
Table 2.
Primers Used for Real-Time Quantitative Polymerase Chain Reaction
| Gene | Forward Primer Sequence (5′-3′) | Reverse Primer Sequence (5′-3′) |
|---|---|---|
| WWP1 | AGGCAGAGATCTGCTTGGTTGA | ACGCAGTCCTTTTTGAAGGCCA |
| ITCH | GCAGAGAGTGGACCAGCACG | TCCCATGTTGTCAACCCGCC |
| Smurf1 | ACATCGTGCGGTGGTTCTGG | GCCGCGCCTGTAGAACCTT |
| Smurf2 | CTCAGGGGTGCCTCAGGAGT | GGGGTGGGGATATGGAGCCT |
| GAPDH | TGCACCACCAACTGCTTGC | GGCATGGACTGTGGTCATGAG |
Abbreviations: WWP1, WW domain-containing E3 ubiquitin protein ligase 1; ITCH, itchy E3 ubiquitin protein ligase; Smurf1, SMAD-specific E3 ubiquitin protein ligase 1; Smurf2, SMAD-specific E3 ubiquitin protein ligase 2.
Osteogenic Differentiation
BSA-Ca2+-siWWP1, BSA-Ca2+-siNC, Lipo-siWWP1, and Milli-Q water were prepared. Three days after PDLSCs inoculation and transfection, osteogenic differentiation was performed in osteogenic medium (Biological Industries, Israel) with or without TNF-α (15 ng/mL). After 2 weeks of induction, the medium was removed, and PDLSCs were fixed in 4% paraformaldehyde for 20 min. Then, the cells were stained with alizarin red S solution for 10 min at 25°C. The sample was imaged through a stereo microscope (Olympus, Japan). Subsequently, the stain was dissolved with a cetylpyridinium chloride solution (10 mM), and the absorbance at 580 nm was measured with a microplate reader.
Statistical Analysis
At least three independent experiments were performed per test, and statistical evaluations of data were performed using Student’s t-test. All results were presented as mean ± standard deviation, and p < 0.05 was considered statistically significant.
Results and Discussion
Synthesis and Characterization of BSA-Ca2+-siRNA Nanoparticles
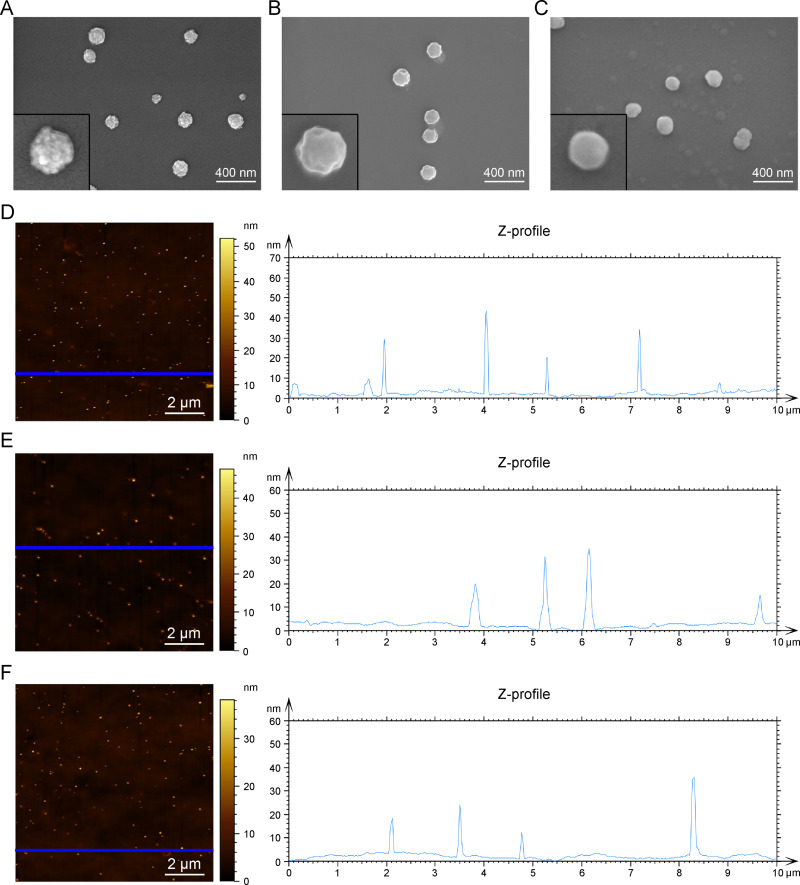
Ca2+ may act as a bridge, connecting the negatively charged siRNA with the protein. SEM observation revealed that BSA, Ca2+, and siRNA formed spherical particles with an average diameter of 139.4 ± 23.9 nm (Figure 1A). Ca2+ is combined with BSA or siRNA to form complexes with diameters of 156.6 ± 28.5 nm (Figure 1B) and 112.1 ± 15.6 nm (Figure 1C), respectively. AFM showed that the surface morphology of BSA-Ca2+-siRNA was rough, unlike that of BSA-Ca2+ and Ca2+-siRNA (smooth surfaces). The formation of BSA-Ca2+-siRNA nanoparticles was also observed by AFM. Notably, the surface morphology of BSA-Ca2+-siRNAs (Figure 1D) was irregular, and the particle sizes were intermediate, between those of BSA-Ca2+ (Figure 1E) and Ca2+-siRNA (Figure 1F). This may be due to the disordered binding of BSA and Ca2+-siRNA.23
Figure 1.
Scanning electron microscope (SEM) images of BSA-Ca2+-siRNA (A), BSA-Ca2+ (B), Ca2+-siRNA (C). Atomic force microscopy (AFM) images and Z-profile (D–F) of BSA-Ca2+-siRNA (D), BSA-Ca2+ (E), Ca2+-siRNA (F). Scale bars = 400 nm and 2 μm (Blue line: a plane scanned in the Z-profile measurement).
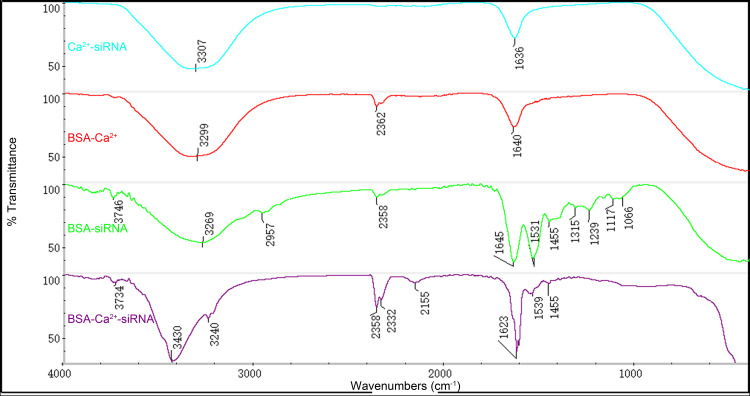
BSA, Ca2+, and siRNA can form nanocomplexes with a surface charge of 4.72 ± 0.37 mV in α-MEM, as observed by DLS (Figure S1). Particles’ diameter was similar to that determined by SEM. The results of FTIR are given in Figure 2. The infrared active vibration of biomolecules is as follows: 3,200–3,650 cm−1 asymmetric and symmetric O-H stretching; 3,250–3,300 cm−1 amide I (N-H stretch); 2,890–2,960 cm−1 C-H stretching of alkenes; 2,100–2,360 cm−1 triple bond (C-C or C-N) or cumulative double bond (C-C-C) stretch; 1,600–1,680 cm−1 C-C stretch (weak peaks) and C-O amide stretch (sharp peaks); 1,480–1,590 cm−1 amide II (N-H bend in plane or C-N stretch) 1,455 cm−1 C-H stretching of alkenes; 1,230–1,330 cm−1 amide III (N-H bend in plane or C-N stretch); and 1,000–1,350 cm−1 amine C-N stretch. The structure of BSA-Ca2+-siRNA contained the maximum numbers of C-C or C-N stretching peaks, and several additional peaks, which were also at a maximum, including 3,430, 2,358, and 2,155 cm−1. However, the structure of BSA-Ca2+-siRNA did not contain peaks specific to BSA-siRNA, such as 1,539, 1,455, and 1,239 cm−1. This suggests that BSA, Ca2+, and siRNA may form a complex rather than exist alone in the form of BSA-siRNA and Ca2+-siRNA (Figure 2).
Figure 2.
Fourier transform infrared spectroscopy (FTIR) spectra analyses of complexes.
Loading and Release of siRNA
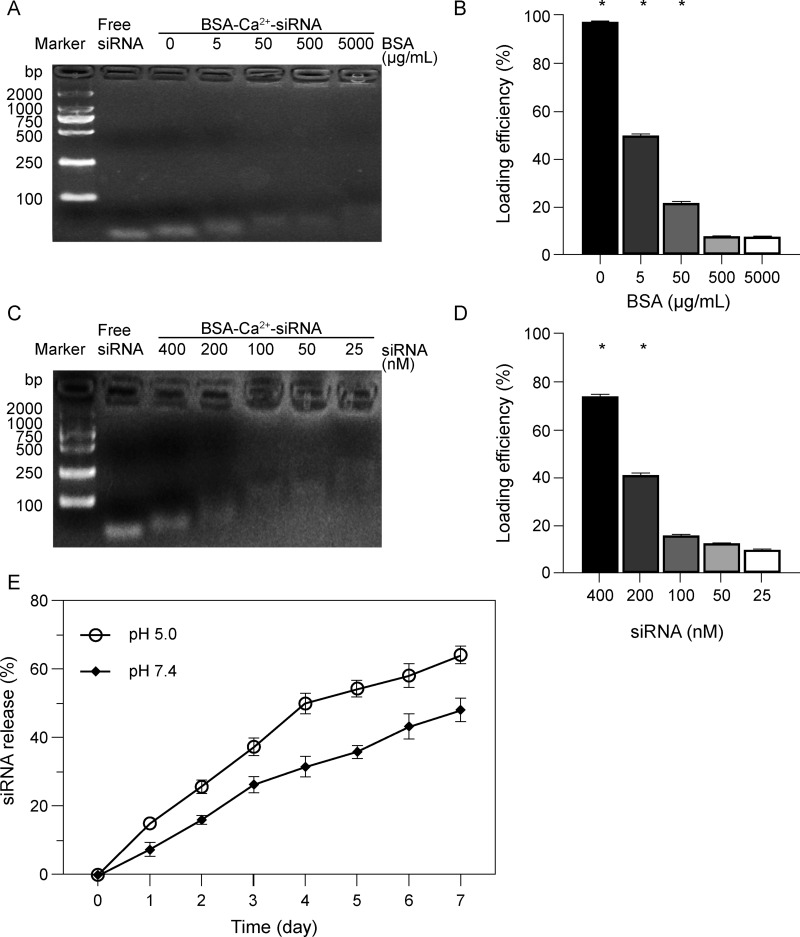
Following BSA and Ca2+ nanoparticle formation, we evaluated the ability of BSA-Ca2+ to directly load siRNA. When the loading concentration of BSA was ≥500 μg/mL, unbound siRNA was only weakly detected in the gel (Figure 3A and B). Similarly, when BSA (500 μg/mL) and CaCl2 (1 M) were loaded with 100 nM siRNA, unbound siRNA remained weakly detected in the gel, indicating that BSA-Ca2+ can load a large amount of siRNA (Figure 3C and D). In addition, a gel blocking test with various concentrations of Ca2+ (0.5–2 M) loaded with siRNA (100 nM) revealed no clear bands (Figure S2). This suggested that the Ca2+-siRNA nanoparticles have poor stability. Conversely, the BSA-Ca2+-siRNA nanoparticles could firmly retain and protect the siRNA cargo, thus confirming the stability of BSA-Ca2+-loaded siRNA.
Figure 3.
Physicochemical characterization of siRNA loading and release. (A and C) Gel electrophoresis assay and (B and D) quantitated by ImageJ of BSA-Ca2+-siRNA processed at various concentrations of bovine serum albumin (BSA) (0–5000 μg/mL) or siRNA (25–400 nM). (E) Release profiles of siRNA from BSA-Ca2+ in PBS (1 mg/mL) at different pH. Significance: *P < 0.05.
We monitored the time/pH-related degradation of BSA-Ca2+-siRNA using a spectrophotometer and found that BSA-Ca2+-siRNA particles could continuously unload siRNA, with release of 48% in neutral PBS by day 7 and 64% in acidic PBS (Figure 3E). This suggests that exposure of BSA-Ca2+-siRNA particles to acidic subcellular compartments (such as endosomes/lysosomes) will enhance their degradability. This phenomenon may be attributed to the pH sensitivity of BSA.24 Under physiological conditions, BSA is adsorbed on the cation core by the negative charge on its surface. As the pH decreases, the electrostatic attraction between the inner core and the BSA gradually weakens.25,26 Therefore, the pH-responsive behavior of BSA-Ca2+ promotes the intracellular release of siRNA.
Cellular Uptake of Nanoparticles
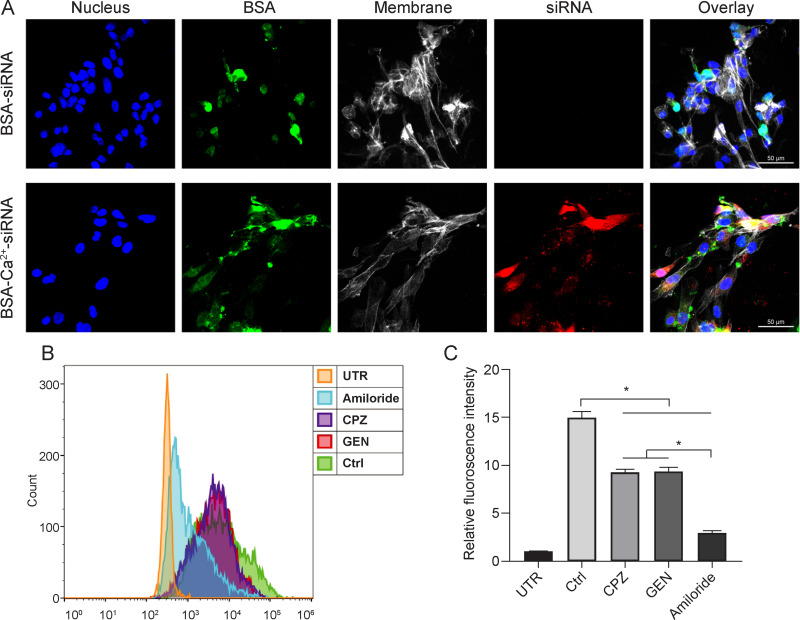
After confirming the effectiveness of siRNA loading/release behavior of BSA-Ca2+, in vitro experiments were conducted to evaluate the intracellular delivery of the loaded siRNA and to confirm the downregulation of target genes. To confirm the efficiency of BSA-Ca2+-siRNA silencing, the GFP gene was selected. As shown in Figure 4A, both Cy3 and FITC fluorescence were observed in cells of the experimental group, while only FITC fluorescence was observed in cells of the control group. This indicated that free siRNA cannot pass into the cell without Ca2+. However, Cy3-siGFP loaded with BSA-Ca2+ can cause cells to emit a bright fluorescent signal, confirming that BSA-Ca2+ can successfully deliver siRNA in cells. Based on flow cytometry analysis, fluorescence could be observed in cells following the uptake of Cy3-siGFP. Compared with untransfected cells, the fluorescence signal of cells treated with BSA-Ca2+-siGFP increased significantly (Figure 4B and C).
Figure 4.
Mechanisms of intracellular uptake and endocytosis of FITC-BSA-Ca2+/Cy3-siRNA complexes in periodontal ligament stem cells (PDLSCs). (A) PDLSCs were stained with AF635-phalloidin and Hoechst and examined under laser scanning confocal microscope (LSCM). (B and C) PDLSCs were pretreated with inhibitors for 1 h and then incubated with transfection complexes for another 3.5 h. Transfection without inhibitors was used as a control (Ctrl). Cellular uptake was analyzed by flow cytometry analysis (B) and quantitated (C). Geometric mean of fluorescence intensity was normalized by untransfected cells (UTR). Significance: *P < 0.05.
To delineate the endocytosis pathway involved in the cellular uptake of BSA-Ca2+-siRNA, three specific inhibitors of endocytosis including chlorpromazine (CPZ), genistein (GEN), and amiloride were used during the transfection process. The intracellular uptake of siRNA was further examined by flow cytometry. Quantitative analysis confirmed that cells pretreated with any inhibitor presented lower levels of siRNA uptake than untreated cells (Figure 4B). The inhibitory effect of amiloride on siRNA uptake was the most notable and exceeded 70% (Figure 4C). This indicated that macropinocytosis is the main uptake mechanism. A recent study showed that consecutive macropinocytosis in macrophages is mediated by calcium-sensing receptors (CaSR).27 Despite the use of different cell types in that study, we speculate that the endocytosis of BSA-Ca2+-siRNA may involve the activation of CaSR. However, further studies are needed to elucidate the exact mechanism underlying the endocytosis of BSA-Ca2+-siRNA and subsequent release from endosomes.
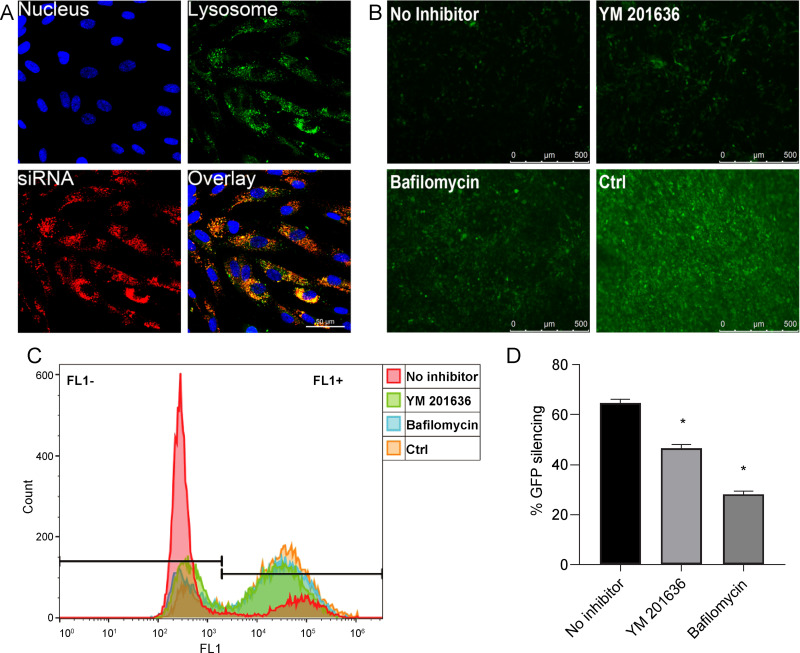
Intracellular Release of Nanoparticles
The intracellular release of Cy3-siRNA was evaluated by LSCM. Following the uptake of BSA-Ca2+-siRNA by cells, a large amount of siRNA was found to be internalized and localized on endosomes/lysosomes (Figure 5A). This indicated that the endosome/lysosome is involved in the release of siRNA. To clarify the escape mechanism of nanoparticles from endosomes, two inhibitors were used: bafilomycin A1, which is a V-ATPase inhibitor that prevents endosome acidification, and YM201636, a PIKfyve inhibitor that prevents early endosomes from maturing into late endosomes. Using fluorescence microscopy, bafilomycin A1 treatment was found to inhibit the silencing effect of siGFP on GFP in PDLSCs, unlike treatment without inhibitor (Figure 5B). The release of siRNA in cells was further evaluated by flow cytometry. Quantitative analysis confirmed that bafilomycin A1 caused a more significant reduction in GFP silencing than YM201636 following 24 h of treatment (Figure 5C and D). This indicated that siRNA release may be primarily mediated by the acidification of endosomes/lysosomes promoting the decomposition of nanocomplexes. After uncomplexing, free Ca2+ increases rapidly and promotes the entry of protons into the endosome/lysosome, causing the rupture of the endosome and the release of siRNA into the cytoplasm. This process of protons entering the endosome/lysosome to replace Ca2+ is termed the “proton sponge” effect.28,29 The rapid de-complexation and siRNA release in the early endosomes are beneficial for maintaining siRNA activity and the high efficiency of gene silencing induced by BSA-Ca2+-siRNA nanocomplexes.
Figure 5.
(A) Fluorescence images of cells treated with BSA-Ca2+-siRNA (Blue: Hoechst-stained nuclei; Green: LysoTracker-stained endosomes/lysosomes; Red: Cy3-siRNA). GFP silencing at 24 h post-transfection in GFP-PDLSCs and the effect of inhibitors on endosomal release were analyzed by fluorescence microscopy (B) and flow cytometry (C) and quantitated (D). The silencing effect was normalized by GFP-PDLSCs with siNC (Ctrl). Significance: *P < 0.05.
Evaluation of the Cytocompatibility of Complexes
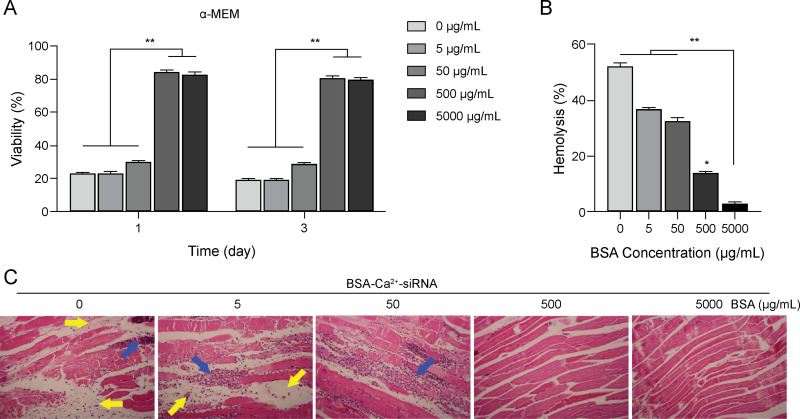
In previous transfection experiments, high Ca2+ concentrations and FBS-free medium were found to induce cell death (Figure S3). Therefore, we optimized Ca2+-siRNA with BSA coating to evaluate the effect of BSA in reducing the cytotoxicity of high Ca2+ concentrations. We found that the carrier with high concentration of BSA (≥500 μg/mL) did not significantly affect cell viability. In contrast, the nanoparticles with low BSA concentration (≤50 μg/mL) were found to have high cytotoxicity (Figure 6A). This indicated that BSA can improve the cellular compatibility of Ca2+-siRNA.
Figure 6.
(A) Cytotoxicity of PDLSCs treated with particles composed of different BSA concentrations (0–5000 ug/mL) after days 1 and 3 was determined by the CCK-8 method. (B) After 6 h of incubation, the hemolysis of BSA-Ca2+ was measured using red blood cells (RBCs) obtained from BALB/c mice (2% hematocrit). The data presents the mean ± SD of the results obtained from four samples. (C) Hematoxylin and eosin (HE) staining of mice muscle tissue (yellow arrows: necrotic muscle cells, blue arrows: infiltrating inflammatory cells). Significance: *P < 0.05, **P < 0.01.
The working solution was diluted with PBS at a ratio of 1:200 to obtain final concentrations for cell hemolysis experiments. No significant hemoglobin leakage was detected in RBCs incubated with the Ca2+-siRNA containing 5,000 μg/mL BSA. In contrast, dose-dependent hemoglobin leakage was observed in RBCs incubated with transfection solution with the BSA concentration of ≤500 μg/mL (Figure 6B). Furthermore, HE staining revealed that decreasing the BSA concentration in the transfection solution resulted in a dose-dependent inflammatory response and necrosis in muscle tissue (Figure 6C). This indicated that BSA can improve the biocompatibility of Ca2+-siRNA and suggested that BSA-Ca2+ may represent a siRNA delivery platform with favorable biological safety. Thus, this carrier may provide a feasible solution for use in the hypodermic, intramuscular, and intravenous injection of siRNA drugs for the purpose of gene therapy.30,31
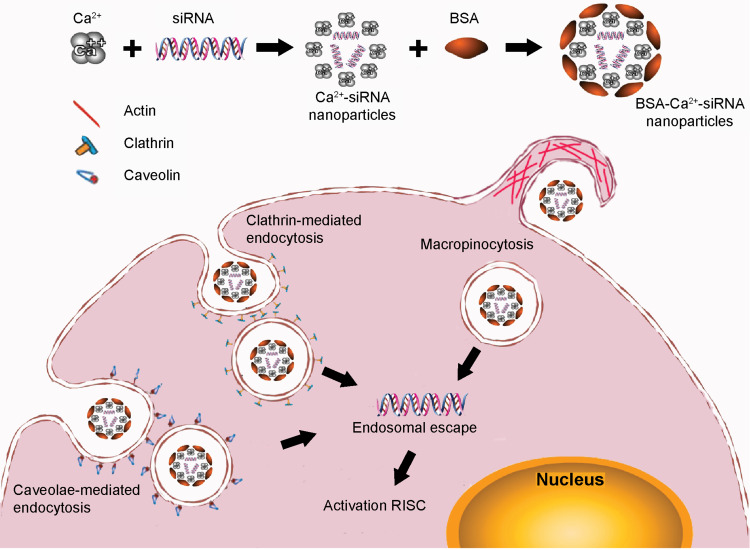
Thus, the fabrication scheme shown in Figure 7 describes our proposed vision of BSA-Ca2+-siRNA nanocomplexes, which comprise a BSA coating on the surface of Ca2+-siRNA nanoparticles. BSA is essential for reducing the toxicity associated with high Ca2+ concentrations and drives the uptake of nanoparticles by cells. The mechanism underlying the internalization of nanoparticles involves a variety of endocytic pathways including clathrin-mediated endocytosis, caveolae-mediated endocytosis, and macropinocytosis. Subsequently, the siRNA released from the endosome reaches the cytosol where the RNA-induced silencing complex is located, resulting in effective siRNA-induced silencing of the target gene. In conclusion, BSA-Ca2+-siRNA transfection is a convenient, safe, and effective method for delivering siRNA/miRNA to PDLSCs.
Figure 7.
Proposed mechanism for BSA-Ca2+-siRNA nanocomplex assembly, cellular uptake, and release. RISC, RNA-induced silencing complex.
Inducing Osteogenic Differentiation of Inflammatory PDLSCs
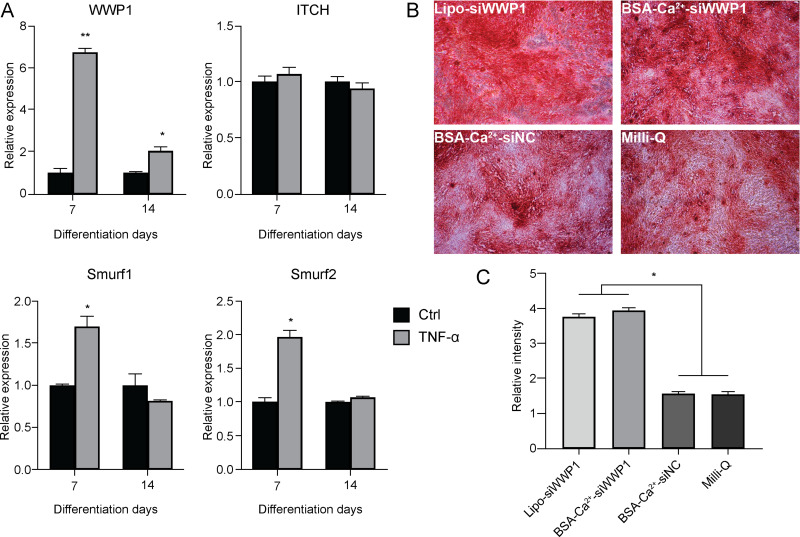
Having established the gene silencing effect through cytosolic siRNA delivery using endosome/lysosome-disruptive BSA-Ca2+ carriers, we further evaluated the method in vitro using a PDLSC osteogenic differentiation assay. Inflammatory cytokines, such as TNF-α, impair the osteogenic differentiation ability of MSCs by mediating the up-regulation of the WWP1 gene17 and downregulation of the ALP, COL-1, and OCN genes.32 This indicates that the ubiquitin-proteasome pathway may negatively influence osteoblast differentiation. Using real-time quantitative polymerase chain reaction (qPCR), we found that the mRNA expression of WWP1, Smurf1, and Smurf2 in PDLSCs was significantly up-regulated 7 days post treatment with TNF-α. Notably, on day 14, WWP1 gene expression remained increased (Figure 8A). This suggested that WWP1 may have a large effect on the inflammatory response of PDLSCs induced by TNF-α.
Figure 8.
Increased expression of WWP1 in PDLSCs stimulated by tumor necrosis factor (TNF)-α. (A) The expression of osteogenesis-related genes (WWP1, ITCH, Smurf1, and Smurf2) was determined by qPCR on days 7 and 14. The expression level of mRNA was normalized to that of GAPDH. Triplicate values are shown, and the data are presented as mean ± SD. PDLSCs were stimulated with TNF-α and transfected with WWP1 siRNA or NC siRNA, and cultured in osteoblast differentiation medium for 2 weeks. (B) Analysis of osteogenic differentiation in vitro by Alizarin Red staining. (C) Evaluation of target gene expression by qPCR. Values are the mean ± SD from three wells. The experiments were repeated three times with similar results. Significance: *P < 0.05, **P < 0.01.
We utilized the novel technology of BSA-Ca2+-siWWP1 particle therapy to inhibit the effect of TNF-α and restore the osteogenic differentiation ability of PDLSCs by blocking the activity of WWP1. Alizarin red staining showed that BSA-Ca2+-siWWP1 significantly promoted the osteogenic differentiation of inflammatory PDLSCs (Figure 8B); there was no significant difference in the transfection efficiency between BSA-Ca2+ and Lipofectamine 2000 (Figure 8C). These results indicated that the revised-BSA optimized Ca2+-siWWP1 nanoparticles have high transfection efficiency and may promote the mineralization of bone defects that occur in periodontitis. Notably, bone resorption inhibition and bone formation promotion are crucial for treating periodontal inflammation. Therefore, siWWP1 can prevent TNF-α from inhibiting bone formation and can enhance the osteogenic effect of PDLSCs; moreover, it may be beneficial for regenerating and repairing bone tissue defects accompanying periodontitis.
There are some limitations to this study. Although we observed the formation of BSA-Ca2+-siRNA nanocomplexes, the binding strength among BSA, Ca2+, and siRNA could not be determined. If such binding is stable, BSA can be modified using antibodies directed against the surface markers of target cells for targeted delivery. In addition, the transfection mechanism of formulated nanoparticles could not be fully elucidated. Moreover, the toxicity and transfection efficiency of BSA-Ca2+-siRNA in other cells, such as tumor cells, are yet to be evaluated. Finally, the in vivo microenvironment is complex, containing numerous phosphates and proteins; therefore, whether these substances affect the stability, biocompatibility, and transfection efficiency of BSA-Ca2+-siRNA remains to be determined.
Conclusion
BSA incorporation does not affect Ca2+-siRNA uptake or knockout efficiency and may reduce the adverse effects associated with high concentrations of Ca2+ in vivo. Therefore, we developed a convenient, safe, and effective siRNA delivery system which can effectively encapsulate and protect siRNA from degradation. The pH-responsive behavior of BSA-Ca2+-siRNA promotes effective siRNA release into the cytoplasm, leading to durable and efficient gene silencing. Therefore, BSA-Ca2+ nanoparticles might be used as a platform to deliver siRNA into the cytoplasm for multiple gene targets and as a nanomaterial with high biocompatibility for intravenously injected drugs.
Funding Statement
The authors are grateful for grants from the School of Stomatology, Air Force Medical University. This work was also supported by a grant from the Nature Science Foundation of China (NSFC No. 81771069) and the Fundamental Research Funds for the Central Universities (xzy012019097).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Yalcin E, Kara G, Celik E, et al. Preparation and characterization of novel albumin-sericin nanoparticles as siRNA delivery vehicle for laryngeal cancer treatment. Prep Biochem Biotechnol. 2019;49:659–670. doi: 10.1080/10826068.2019.1599395 [DOI] [PubMed] [Google Scholar]
- 2.Ruvinov E, Kryukov O, Forti E, Korin E, Goldstein M, Cohen S. Calcium-siRNA nanocomplexes: what reversibility is all about. J Control Release. 2015;203:150–160. doi: 10.1016/j.jconrel.2015.02.029 [DOI] [PubMed] [Google Scholar]
- 3.Putnam D. Design and development of effective siRNA delivery vehicles. Proc Natl Acad Sci U S A. 2014;111:3903–3904. doi: 10.1073/pnas.1401746111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xue HY, Liu S, Wong HL. Nanotoxicity: a key obstacle to clinical translation of siRNA-based nanomedicine. Nanomedicine (Lond). 2014;9:295–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldshtein M, Forti E, Ruvinov E, Cohen S. Mechanisms of cellular uptake and endosomal escape of calcium-siRNA nanocomplexes. Int J Pharm. 2016;515:46–56. doi: 10.1016/j.ijpharm.2016.10.009 [DOI] [PubMed] [Google Scholar]
- 6.Xu B, Xu Y, Su G, Zhu H, Zong L. A multifunctional nanoparticle constructed with a detachable albumin outer shell and a redox-sensitive inner core for efficient siRNA delivery to hepatocellular carcinoma cells. J Drug Target. 2018;26:941–954. doi: 10.1080/1061186X.2018.1455840 [DOI] [PubMed] [Google Scholar]
- 7.Song W, Yang C, Svend Le DQ, Zhang Y, Kjems J. Calcium-microRNA complex-functionalized nanotubular implant surface for highly efficient transfection and enhanced osteogenesis of mesenchymal stem cells. ACS Appl Mater Interfaces. 2018;10:7756–7764. doi: 10.1021/acsami.7b18289 [DOI] [PubMed] [Google Scholar]
- 8.Mammadova-Bach E, Nagy M, Heemskerk JWM, Nieswandt B, Braun A. Store-operated calcium entry in thrombosis and thrombo-inflammation. Cell Calcium. 2019;77:39–48. doi: 10.1016/j.ceca.2018.11.005 [DOI] [PubMed] [Google Scholar]
- 9.Dalal PJ, Muller WA, Sullivan DP. Endothelial cell calcium signaling during barrier function and inflammation. Am J Pathol. 2020;190:535–542. doi: 10.1016/j.ajpath.2019.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elzoghby AO, Samy WM, Elgindy NA. Albumin-based nanoparticles as potential controlled release drug delivery systems. J Control Release. 2012;157:168–182. [DOI] [PubMed] [Google Scholar]
- 11.Bahrami M, Morris MB, Day ML. Amino acid supplementation of a simple inorganic salt solution supports efficient in vitro maturation (IVM) of bovine oocytes. Sci Rep. 2019;9:11739. doi: 10.1038/s41598-019-48038-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tarhini M, Greige-Gerges H, Elaissari A. Protein-based nanoparticles: from preparation to encapsulation of active molecules. Int J Pharm. 2017;522:172–197. doi: 10.1016/j.ijpharm.2017.01.067 [DOI] [PubMed] [Google Scholar]
- 13.Verma ML, Dhanya BS, Rani V, et al. Carbohydrate and protein based biopolymeric nanoparticles: current status and biotechnological applications. Int J Biol Macromol. 2020;154:390–412. doi: 10.1016/j.ijbiomac.2020.03.105 [DOI] [PubMed] [Google Scholar]
- 14.Nair AB, Kaushik A, Attimarad M, Al-Dhubiab BE. Enhanced oral bioavailability of calcium using bovine serum albumin microspheres. Drug Deliv. 2012;19:277–285. doi: 10.3109/10717544.2012.704094 [DOI] [PubMed] [Google Scholar]
- 15.Glimcher LH, Jones DC, Wein MN. Control of postnatal bone mass by the zinc finger adapter protein Schnurri-3. Ann N Y Acad Sci. 2007;1116:174–181. doi: 10.1196/annals.1402.044 [DOI] [PubMed] [Google Scholar]
- 16.Zhi X, Chen C. WWP1: a versatile ubiquitin E3 ligase in signaling and diseases. Cell Mol Life Sci. 2012;69:1425–1434. doi: 10.1007/s00018-011-0871-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao L, Huang J, Zhang H, et al. Tumor necrosis factor inhibits mesenchymal stem cell differentiation into osteoblasts via the ubiquitin E3 ligase Wwp1. Stem Cells. 2011;29:1601–1610. doi: 10.1002/stem.703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tucker WO, Kinghorn AB, Fraser LA, Cheung YW, Tanner JA. Selection and characterization of a DNA aptamer specifically targeting human HECT ubiquitin ligase WWP1. Int J Mol Sci. 2018;19:763. doi: 10.3390/ijms19030763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shu L, Zhang H, Boyce BF, Xing XL. Ubiquitin E3 ligase Wwp1 negatively regulates osteoblast function by inhibiting osteoblast differentiation and migration. J Bone Miner Res. 2013;28:1925–1935. doi: 10.1002/jbmr.1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi E, Lim D-K, Kim S. Calcium-doped mesoporous silica nanoparticles as a lysosomolytic nanocarrier for amine-free loading and cytosolic delivery of siRNA. J Ind Eng Chem. 2020;81:71–80. doi: 10.1016/j.jiec.2019.08.054 [DOI] [Google Scholar]
- 21.Yang C, Gao S, Dagnæs-Hansen F, Jakobsen M, Kjems J. Impact of PEG chain length on the physical properties and bioactivity of PEGylated chitosan/siRNA nanoparticles in vitro and in vivo. ACS Appl Mater Interfaces. 2017;9:12203–12216. doi: 10.1021/acsami.6b16556 [DOI] [PubMed] [Google Scholar]
- 22.Ma B, Niu C, Zhou Y, et al. The disulfide bond of the peptide thanatin is dispensible for its antimicrobial activity in vivo and in vitro. Antimicrob Agents Chemother. 2016;60:4283–4289. doi: 10.1128/AAC.00041-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin PH, Zhang W, Lu WC. Theoretical study of hydrated Ca2+-amino acids (glycine, threonine and phenylalanine) clusters. Comput Theor Chem. 2013;1021:164–170. doi: 10.1016/j.comptc.2013.07.005 [DOI] [Google Scholar]
- 24.Anders CB, Chess JJ, Wingett DG, Punnoose A. Serum proteins enhance dispersion stability and influence the cytotoxicity and dosimetry of ZnO nanoparticles in suspension and adherent cancer cell models. Nanoscale Res Lett. 2015;10:448. doi: 10.1186/s11671-015-1158-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu Y, Jin X, Ping Q, et al. A novel lipoprotein-mimic nanocarrier composed of the modified protein and lipid for tumor cell targeting delivery. J Control Release. 2010;146:299–308. doi: 10.1016/j.jconrel.2010.05.022 [DOI] [PubMed] [Google Scholar]
- 26.Peng Q, Zhang S, Yang Q, et al. Preformed albumin corona, a protective coating for nanoparticles based drug delivery system. Biomaterials. 2013;34(33):8521–8530. doi: 10.1016/j.biomaterials.2013.07.102 [DOI] [PubMed] [Google Scholar]
- 27.Canton J, Schlam D, Breuer C, et al. Calcium-sensing receptors signal constitutive macropinocytosis and facilitate the uptake of NOD2 ligands in macrophages. Nat Commun. 2016;7(1):11284. doi: 10.1038/ncomms11284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dominska M, Dykxhoorn DM. Breaking down the barriers: siRNA delivery and endosome escape. J Cell Sci. 2010;123:1183–1189. doi: 10.1242/jcs.066399 [DOI] [PubMed] [Google Scholar]
- 29.Chou LY, Ming K, Chan WC. Strategies for the intracellular delivery of nanoparticles. Chem Soc Rev. 2011;40:233–245. doi: 10.1039/C0CS00003E [DOI] [PubMed] [Google Scholar]
- 30.Han J, Wang Q, Zhang Z, Gong T, Sun X. Cationic bovine serum albumin based self-assembled nanoparticles as siRNA delivery vector for treating lung metastatic cancer. Small. 2014;10(3):524–535. doi: 10.1002/smll.201301992 [DOI] [PubMed] [Google Scholar]
- 31.Xu K, Zhao Z, Zhang J, et al. Albumin-stabilized manganese-based nanocomposites with sensitive tumor microenvironment responsivity and their application for efficient SiRNA delivery in brain tumors. J Mater Chem B. 2020;8(7):1507–1515. doi: 10.1039/C9TB02341K [DOI] [PubMed] [Google Scholar]
- 32.Zhang J, Li ZG, Si YM, Chen B, Meng J. The difference on the osteogenic differentiation between periodontal ligament stem cells and bone marrow mesenchymal stem cells under inflammatory microenvironments. Differentiation. 2014;88:97–105. doi: 10.1016/j.diff.2014.10.001 [DOI] [PubMed] [Google Scholar]