Abstract
Background
Cystic or alveolar echinococcosis caused by the larval stages of Echinococcus spp. is a very severe zoonotic helminth infection. Echinococcus shiquicus is a newly discovered species that has only been reported in the Qinghai and Sichuan provinces of the Qinghai-Tibet plateau, China where, to date, it has only been confirmed in Tibetan foxes and wild small mammal populations of the Tibetan plateau. Information on its genetic and evolutionary diversity is scanty. The aim of this study was to investigate the prevalence of E. shiquicus in plateau pikas (Ochotona curzoniae), a known intermediate host, and to determine the genetic variation and phylogenetic relationship of the E. shiquicus population in the Tibet region of China based on mitochondrial DNA.
Methods
Echinococcus shiquicus samples were collected from Damxung and Nyêmo counties (located in Tibet Autonomous Region, China). The mitochondrial cox1 and nad1 gene sequences were analyzed, and the genetic diversity and epidemiology of E. shiquicus in the region were discussed based on the results.
Results
The prevalence of E. shiquicus in pikas in Damxung and Nyêmo counties was 3.95% (6/152) and 6.98% (9/129), respectively. In combination with previous public sequence data, the haplotype analysis revealed 12 haplotypes (H) characterized by two distinct clusters (I and II), and a sequence distance of 99.1–99.9% from the reference haplotype (H1). The diversity and neutrality indices for the entire E. shiquicus populations were: haplotype diversity (Hd) ± standard deviation (SD) 0.862 ± 0.035; nucleotide diversity (Hd ± SD) 0.0056 ± 0.0003; Tajima's D 0.876 (P > 0.05); and Fu’s F 6.000 (P > 0.05).
Conclusions
This was the first analysis of the newly discovered E. shiquicus in plateau pikas in the Tibet Autonomous Region of China. The neutrality indices suggest a deficiency of alleles, indicative of a recent population bottleneck.
Keywords: Echinococcus shiquicus, Epidemiology, Genetic diversity, Tibet region
Background
Echinococcosis or hydatidosis is a serious cosmopolitan parasitic zoonosis [1–3]. Echinococcus shiquicus, a newly described species of the genus Echinococcus, is currently believed to be limited and endemic to the Qinghai-Tibet plateau region of China [4–6] where the Tibetan fox (Vulpes ferrilata) and plateau pika (Ochotona curzoniae) commonly serve as the definitive and intermediate hosts, respectively. E. shiquicus was also recently identified in some rodent species, such as voles, in Sichuan province, which further suggests other small mammal species could serve as potential intermediate hosts to this Echinococcus species [7]. Also, following the detection of E. shiquicus DNA molecules in dog faeces in Sichuan province [7], there are concerns that dogs could have the potential to serve as a definitive host for this helminth. Taking into account the close proximity of dogs to humans and the limited knowledge currently available the zoonotic potential of E. shiquicus, this helminth represents a significant public health concern. However, further study is needed to determine whether this concern is valid.
Since the discovery of this species, several questions have been raised: Is the infection caused by this species zoonotic? What are its epidemiology and geographical distribution? What is the overall difference between this species and other species of Echinococcus? What is its possible economic impact? What is its degree of endemicity? Why does it appear to be geographically restricted to the Qinghai-Tibet Plateau? [5, 8]. The answers to these questions will rely on the accumulation of data over time across the Qinghai-Tibet Plateau.
Tibetan pastoral communities remain the most Echinococcus multi-species endemic regions in China [9]. A major challenge in the past to the study of Echinococcus species was the common misidentification of E. shiquicus as E. multilocularis due to a similar transmission pattern and host range; detailed knowledge of its morphological structure eventually resolved this problem [10]. However, there remains a dearth of information on the epidemiological status of E. shiquicus, even though the problem of misidentification has been resolved with the development and implementation of specific and reliable methods for differentiating species of Echinococcus [11, 12]. Also, the rapid development in sequencing technology, specific genetic markers, especially mitochondrial (mt) DNAs and the development of useful tools/software for resolving the evolutionary and phylogenetic relationships between and within species [13–16] will further benefit our understanding of the phylogenetic relationship existing among E. shiquicus.
Here, we report for the first time E. shiquicus in plateau pikas (Ochotona curzoniae) in the Tibet Autonomous Region (Nyêmo and Damxung counties), China. We have estimated its genetic variation and evolutionary relationship using concatenated cox1–nad1 nucleotide sequence (2505 bp).
Methods
Collection of samples
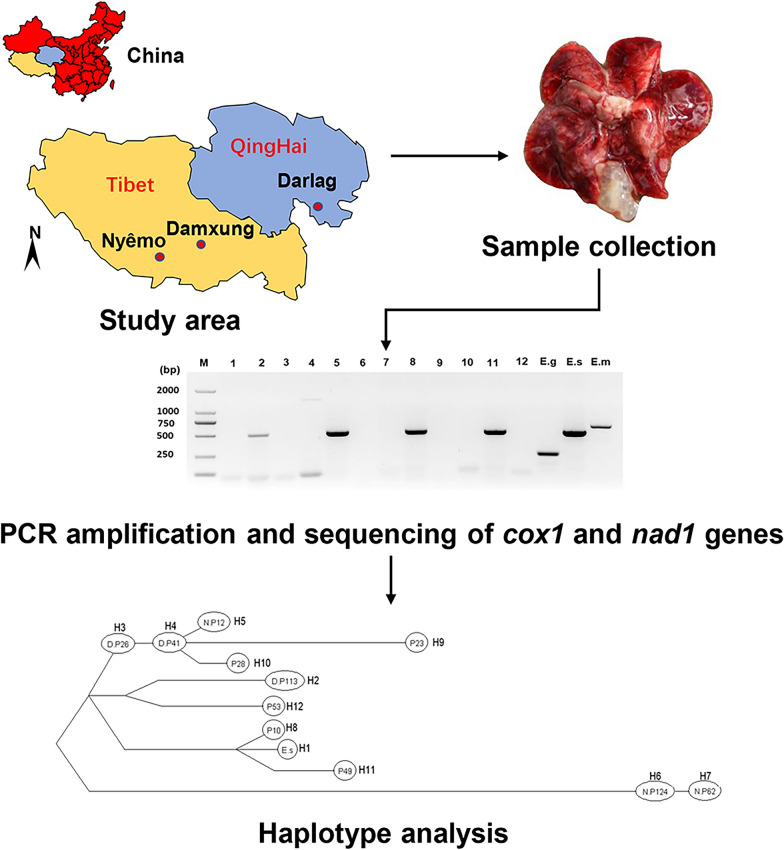
A total of 281 plateau pikas were trapped in Damxung county (n = 152) (91° 1ʹ 42ʺ E, 30° 29ʹ 3ʺ N; altitude elevation 4360 m a.s.l.) and Nyêmo county (n = 129) (90° 17ʹ 2ʺ E, 29° 37ʹ 54ʺ N; altitude elevation 4640 m a.s.l.) in China (Fig. 1) using break-back traps set at the entrance of their dens. All internal organs, including the heart, liver, spleen, lung and kidney, were examined by necropsy for cysts. All cystic-like samples (see Table 2 for cyst location) were examined morphologically on the spot before transportation to the State Key Laboratory at Lanzhou Veterinary Research Institute, CAAS, for molecular identification and further analysis. All samples were stored in liquid nitrogen until DNA extraction.
Fig. 1.
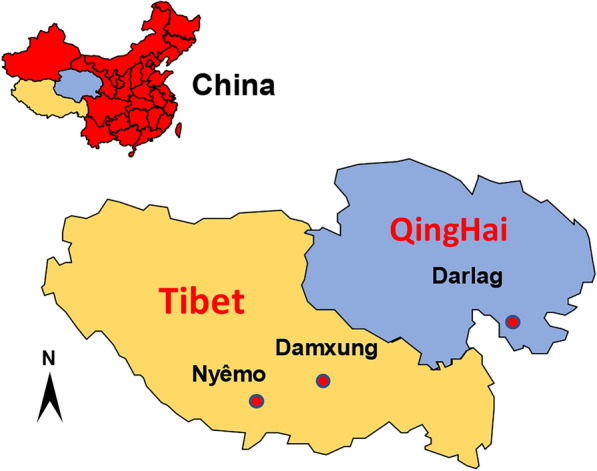
Map showing Damxung and Nyêmo counties in the Tibet Autonomous Region (yellow) and Darlag County in Qinghai (blue).
Table 2.
Characteristics of E. shiquicus isolates
| Sample number | Sample ID | Infected organ or tissue | Location of collection | Haplotype | Sequence distance from H1 (%) | References |
|---|---|---|---|---|---|---|
| 1 | ref. isolate | Lung | Sichuan | H1 | – | Yuan et al. [9] |
| 2 | D.P113 | Lung | Damxung | H2 | 99.6 | This study |
| 3 | D.P149 | Lung | Damxung | H2 | 99.6 | This study |
| 4 | D.P151 | Chest cavity | Damxung | H2 | 99.6 | This study |
| 5 | D.P2 | Lung | Damxung | H2 | 99.6 | This study |
| 6 | D.P26 | Abdomen | Damxung | H3 | 99.8 | This study |
| 7 | D.P41 | Chest | Damxung | H4 | 99.7 | This study |
| 8 | N.P12 | Lung | Nyêmo | H5 | 99.7 | This study |
| 9 | N.P124 | Lung | Nyêmo | H6 | 99.1 | This study |
| 10 | N.P15 | Lung | Nyêmo | H4 | 99.7 | This study |
| 11 | N.P43 | Lung | Nyêmo | H2 | 99.6 | This study |
| 12 | N.P62 | Lung | Nyêmo | H7 | 99.1 | This study |
| 13 | N.P64 | Lung | Nyêmo | H6 | 99.1 | This study |
| 14 | N.P72 | Lung | Nyêmo | H6 | 99.1 | This study |
| 15 | N.P83 | Lung | Nyêmo | H6 | 99.1 | This study |
| 16 | N.P89 | Lung | Nyêmo | H6 | 99.1 | This study |
| 17 | P10 | Lung | Darlag | H8 | 99.9 | Cai et al. [18] |
| 18 | P23 | Lung | Darlag | H9 | 99.5 | Cai et al. [18] |
| 19 | P27 | Lung | Darlag | H6 | 99.1 | Cai et al. [18] |
| 20 | P28 | Lung | Darlag | H10 | 99.6 | Cai et al. [18] |
| 21 | P29 | Lung | Darlag | H8 | 99.9 | Cai et al. [18] |
| 22 | P31 | Lung | Darlag | H9 | 99.5 | Cai et al. [18] |
| 23 | P32 | Lung | Darlag | H6 | 99.1 | Cai et al. [18] |
| 24 | P34 | Lung | Darlag | H9 | 99.5 | Cai et al. [18] |
| 25 | P45 | kidney | Darlag | H6 | 99.1 | Cai et al. [18] |
| 26 | P49 | Lung | Darlag | H11 | 99.8 | Cai et al. [18] |
| 27 | P50 | Lung | Darlag | H9 | 99.5 | Cai et al. [18] |
| 28 | P52L | Lung | Darlag | H6 | 99.1 | Cai et al. [18] |
| 29 | P52S | Spleen | Darlag | H6 | 99.1 | Cai et al. [18] |
| 30 | P53 | Lung | Darlag | H12 | 99.6 | Cai et al. [18] |
| 31 | P54 | Lung | Darlag | H12 | 99.6 | Cai et al. [18] |
| 32 | P55 | Lung | Darlag | H9 | 99.5 | Cai et al. [18] |
| 33 | P6 | Lung | Darlag | H9 | 99.5 | Cai et al. [18] |
| 34 | P7 | Lung | Darlag | H9 | 99.5 | Cai et al. [18] |
| 35 | P70 | Lung | Darlag | H8 | 99.9 | Cai et al. [18] |
| 36 | P77 | Lung | Darlag | H11 | 99.8 | Cai et al. [18] |
DNA extraction and identification of Echinococcus species
Before DNA extraction, samples were removed from liquid nitrogen and placed in an icebox (containing ice) to thaw. A sample of tissues (about 10 mg) of each cyst was cut off and placed in a 1.5-ml micro-centrifuge tube following which it was ground manually with a pestle for 2 min on ice before being digested with proteinase K. Genomic DNA was extracted using the DNeasy Blood and Tissue Kit according to the manufacturer's instructions (Qiagen, Hilden, Germany). The DNA concentration was measured on a microplate reader (Infinite® 200 PRO NanoQuant; Tecan Group Ltd., Männedorf, Switzerland) and then stored at − 20 °C until use.
Amplification and sequencing of Echinococcus shiquicus cox1 and nad1 genes
A multiplex PCR assay previously described [12] was initially used to confirm the species (E. granulosus s.s., E. multilocularis and E. shiquicus) responsible for the infection. Afterward, the complete nad1 and cox1 genes of E. shiquicus from all positive samples were further amplified using specific primers (Table 1) that were designed based on the complete mtDNA sequence of E. shiquicus (GenBank accession no. AB208064 or NC_009460). PCR amplification was carried out in a 50-μl reaction volume (25 μl Premix Taq™ [Takara Bio, Kusatsu, Japan], 1 μl forward and reverse primers [1 μM of each primer] and 1 μl of extracted DNA [100 μg/ml]). The PCR reaction was performed under the following conditions: initial denaturation at 95 °C for 3 min; 35 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s, and extension at 72 °C, for 90 s; followed by a final extension at 72 °C for 10 min. Gel electrophoresis was used to detect PCR products in a 1.5% (w/v) agarose gel stained with GelRed® and visualized under UV light (Molecular Imager® ChemiDoc™ XRS+ with Image Lab™ software; Bio-Rad, Hercules, CA, USA). PCR products were then sequenced (TSINGKE Biological Technology Company, Xi’an, China) using the same primer sets.
Table 1.
Forward and reverse PCR primers used for complete analysis of Echinococcus shiquicus cox1 and nad1 genes based on complete mitochondrial DNA sequence of E. shiquicus (NC_009460 or AB208064)
| Primer name | Primer sequences 5ʹ–3ʹ | Size (bp) | Position in the mtDNAa |
|---|---|---|---|
| E.s-cox1-F | AGTTACTGCTAATAATTTTGTGTCAT | 1837 | 9222 |
| E.s-cox1-R | ATGATGTAAAAGGCAAATAAACC | 10,829 | |
| E.s-nad1-F | TAATGTTGATTATAGAAAATTTTCGTTTTACACGC | 1286 | 7520 |
| E.s-nad1-R | CACAATTTATTATATCAAAGTAACCTGC | 8416 |
E.sE. shiquicus, F forward primer, mt mitochondrial, R reverse primer
aLocation of the primers
Phylogenetic relationship and genetic variation analyses
The EditSeq software module of DNAstar software (DNASTAR, Madison, WI, USA) was used to manually edit, align and find open reading frame (ORF) sequences of the complete nad1 and cox1 genes. Based on the concatenated cox1–nad1 genes, the phylogenetic tree was generated using the software tool Mrbayes v3.1.2 with settings run for 2,500,000 generations, ensuring that the average standard deviation of split frequency was < 0.01 (0.009577) [17]. The concatenated sequence of the E. multilocularis cox1 and nad1 genes (GenBank accession no. AB385610 and AB668376, respectively) was considered as an outgroup [6, 10, 18]. In addition, the cox1-nad1 sequence of E. shiquicus (AB208064 or NC_009460) was chosen as a reference sequence while sequences from Fan et al. [18] were included in our analysis to provide better insight into the genetic diversity of E. shiquicus in the plateau region as this study is the sole study to date to have investigated the genetic diversity of E. shiquicus using longer mitochondrial DNA sequences (2505 bp). The genetic diversities and distances of the concatenated sequences of E. shiquicus were inferred by the haplotype network of TCS 1.21 and MegAlign software [19, 20], respectively, while the number of haplotypes, population diversity and neutrality indices were calculated using DnaSP 5.10 software [21].
Results
Echinococcus shiquicus prevalence and identification
The prevalence of E. shiquicus in plateau pikes in the present study areas, Damxung and Nyêmo counties, was 3.95% (6/152) and 6.98% (9/129), respectively. The multiplex assay revealed that all positive amplified samples were of E. shiquicus expected size (471 bp); no E. multilocularis- and E. granulosus-specific bands were produced or observed. These results confirm that infection was caused by E. shiquicus only (Additional file 1: Fig. S1). Most isolates were collected from the lungs (Fig. 2), with a few collected from the spleen, kidney and other tissues (Table 2).
Fig. 2.
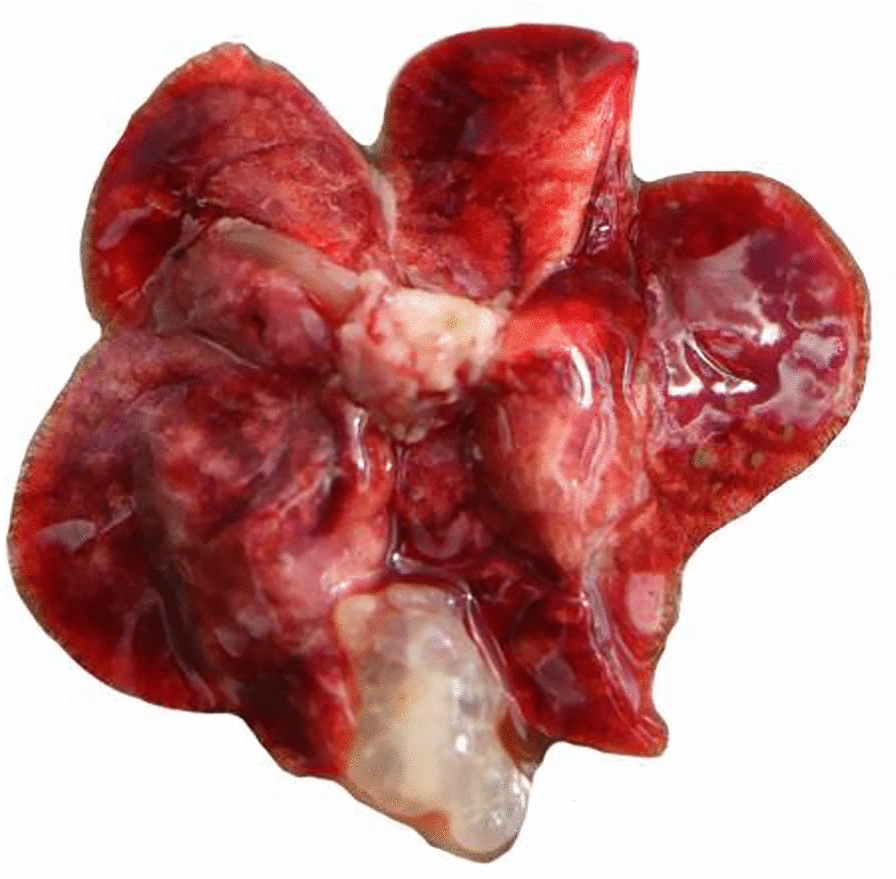
An Echinococcus shiquicus isolate from the lung of plateau pikes (Ochotona curzoniae), in the Tibet Autonomous Region, China
To investigate the genetic diversity, we analysed a total of 35 E. shiquicus nucleotide sequences, including sequences representing the 15 isolates from the current study [Damxung (6) and Nyêmo (9) counties of the Tibet Autonomous region] and 20 complete cox1-nad1 sequences from our previous investigation in Darlag county, Qinghai province [6].
Haplotypes and phylogenetic tree
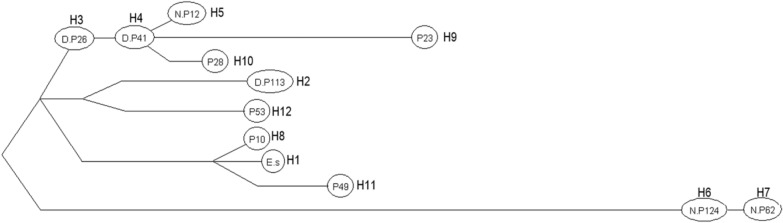
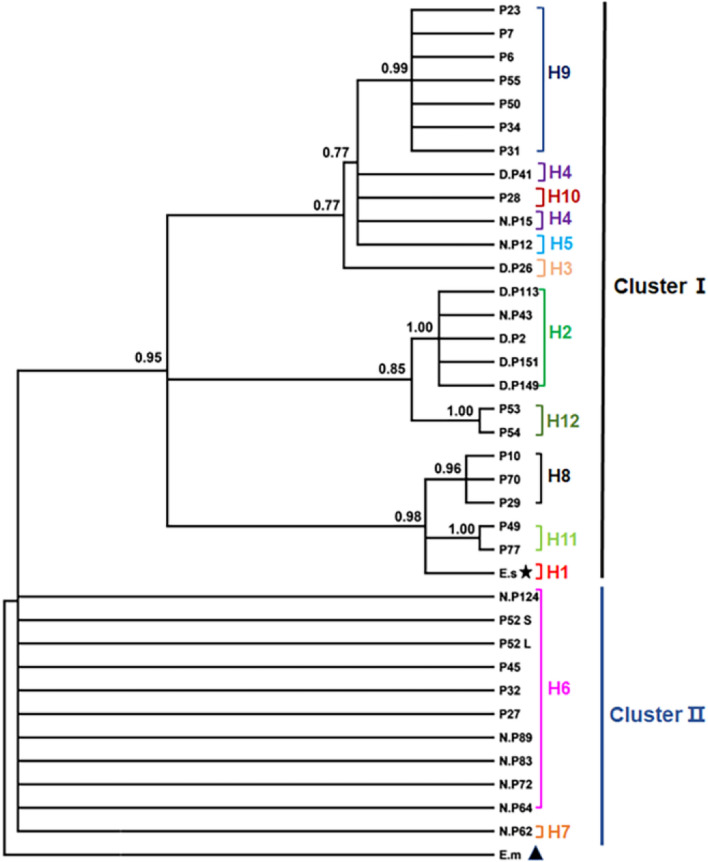
The entire sequences of cox1 and nad1 were amplified and sequenced using E. shiquicus-specific primers (Table 1) with a PCR product size of 1837 and 1286 bp, respectively. Using the concatenated ORF sequence of the cox1 and nad1 genes (2505 bp), the E. shiquicus isolates formed 11 different haplotypes (H2–H12) (Table 2) in addition to the E. shiquicus reference sequence (H1) (GenBank accession no. AB208064 or NC_009460). Of these 11 haplotypes, haplotypes H2–H7 represent haplotypes from this study while haplotypes H6 and H8–H12 represent haplotypes from samples collected in Darlag County. The relationships between these haplotypes and their genetic diversities are shown in Table 2 and Fig. 3, respectively. When compared with the reference sequence (H1), we found H6 to be the farthest genetically distant haplotype from H1 (99.1%) and H8 the closest (99.9%) (Table 2). The Bayesian phylogenetic tree produced two distinct clusters (I and II) of the E. shiquicus population (Fig. 4), with a 0.6% difference in the average genetic distance between cluster I (99.7%) and II (99.1%).
Fig. 3.
The haplotype network and distribution of the different haplotypes. Letters in circles refer to the sample ID (see Table 2). H Haplotype
Fig. 4.
Phylogenetic inference and cluster distribution of E. shiquicus isolates based on the concatenated cox1–nad1 genes. E.m E. multilocularis, E.s E. shiquicus; see Table 2 for other sample ID. (★ = E. shiquicus reference sequence, ▲ = E. multilocularis reference sequence)
Diversity and neutrality indices
Diversity and neutrality indices for E. shiquicus isolates in each location (Darlag, Nyêmo and Damxung) were calculated using the concatenated cox1-nad1 sequences (Table 3). The total number of mutations was 47, and the overall value of haplotype diversity and nucleotide diversity were 0.862 ± 0.035 and 0.0056 ± 0.0003, respectively (Table 3). Haplotype and nucleotide diversities of isolates from Darlag were higher than those from Nyêmo and Damxung. Neutrality indices calculated by Tajima's D (0.876; P > 0.05) and Fu’s F (6.000; P > 0.05) were positive and non-significant, suggesting allele deficiency and a recent population bottleneck event of the E. shiquicus population in the Tibetan region.
Table 3.
Diversity and neutrality indices for E. shiquicus populations in the Tibet Autonomous region
| Location | Diversity | Neutralitya | |||||
|---|---|---|---|---|---|---|---|
| Number of isolates | Total number of mutations | Number of haplotypes | Hd ± SD | Nd ± SD | Tajima's D (P value) | Fu’s F (P value) | |
| Damxung | 6 | 7 | 3 | 0.600 ± 0.215 | 0.0014 ± 0.0005 | 0.888 (0.814) | 2.161 (0.882) |
| Nyêmo | 9 | 27 | 5 | 0.722 ± 0.159 | 0.0045 ± 0.0012 | 0.703 (0.787) | 3.424 (0.932) |
| Darlag | 20 | 41 | 6 | 0.811 ± 0.055 | 0.0057 ± 0.0006 | 0.894 (0.850) | 8.814 (0.998) |
| Overall | 35 | 47 | 11 | 0.862 ± 0.035 | 0.0056 ± 0.0003 | 0.876 (0.867) | 6.000 (0.965) |
Hd Haplotype diversity, Nd nucleotide diversity, SD standard deviation
aNeutrality indices calculated by Tajima's D and Fu’s F were positive and non-significant (P > 0.05)
Discussion
The geographical distribution, epidemiology and zoonotic potential of E. shiquicus remains poorly understood [5]. Although initially reported to be limited geographically to the Qinghai-Tibet Plateau, its presence has since been gradually found in other areas of the plateau [6], thereby increasing the need to intensify efforts to learn more about this species, especially its prevalence, genetic population structure, and host range, in order to understand its transmission pattern and ultimately achieve control of its spread. The use of improved specific detection and discrimination assays [10, 11] in the differential diagnosis of Echinococcus infection in the Qinghai-Tibet plateau cannot be overstated in terms of providing a near-accurate picture of the prevalence of the species in the region. For example, the challenge posed by misidentification was recently discussed by Boufana et al. who investigated the possible presence of E. shiquicus in hosts other than Tibetan foxes and pikas and found for the first time since the discovery of this helminth that dogs could potentially serve as a definitive host for this species [7]. These authors also observed a co-infection with E. shiquicus in a previously identified E. granulosus sample and identified cross-reaction of primers as a major reason for missing the E. shiquicus in the first instance [7].
The results of this study showed a prevalence of E. shiquicus infection in pikas in Damxung and Nyêmo counties of 3.95 and 6.98%, respectively. With the question of the zoonotic potential of E. shiquicus still unanswered [5, 22], the prevalence of E. shiquicus infection observed in the study area and the prevalence of 23.75–37.5% previously reported in Darlag and Shiqu counties of Qinghai and Sichuan provinces, respectively [2, 6, 23], raises concern regarding the significance of E. shiquicus infection for both humans and livestock in the region.
Earlier studies have raised the controversial point of whether or not E. shiquicus has evolved without bottleneck effect [13, 24]. However, the neutrality test in the current study suggests a deficiency of alleles which otherwise indicates that the Tibetan E. shiquicus population could have possibly experienced a bottleneck effect in the course of evolution.
Among the 11 haplotypes identified in sequences reported previously from Qinghai province [6], H2 and H4 were found in Tibet counties and H6 was common to counties in Qinghai and Tibet; however, no haplotype was found to be common to all the three counties. Our results are in agreement with previous observations demonstrating the absence of a common haplotype across different locations in the plateau [13, 24].
In this study, we report the genetic diversity and the phylogenetic status of E. shiquicus population from Tibet in comparison to previous isolates from Qinghai province. Our results demonstrate allele deficiency among the E. shiquicus population indicating a population bottleneck event in the Tibet Autonomous Region. However, we suggest that future studies on population genetics consider using a larger panel of isolates and examining other genetic markers in order to further elucidate the evolutionary status of E. shiquicus in the Qinghai-Tibet Plateau.
Conclusions
This study demonstrates the existence of E. shiquicus in plateau pikas in the Tibet Autonomous Region of China, and the neutrality indices suggest a deficiency of alleles as expected from a recent population bottleneck.
Supplementary information
Additional file 1: Fig. S1. Multiplex PCR results image of Echinococcus shiquicus samples based on three specific primers of E. multilocularis, E. shiquicus and E. granulosus.
Acknowledgements
The authors would like to thank Jian-Zhi Liu (Tibet, China),Chen-Yan Xia (Tibet, China), Gang Yao (Lanzhou, China) and Wen-Jun Tian (Lanzhou, China) for their assistance in sample collection.
Authors' contributions
GQZ and HBY contributed to data collection and in drafting the manuscript. QGZ, LL, YTW, WHL, NZZ and WZJ contributed to sample collection and processing. WZJ, HBY and BQF participated in the design of the study. WZJ and JAO provided comments and modified the manuscript. All authors read and approved the final version of the manuscript.
Funding
This study was supported by National Key Research and Development Plan (2018YFC1602504; 2017YFD0501301) and Central Public-Interest Scientific Institution Basal Research Fund (1610312020016).
Availability of data and materials
All data supporting the conclusions are included in the article. Representative nucleotide sequences of cox1 and nad1 genes from the present study are available in the GenBank database under the accession numbers (MW072804-MW072815).
Ethics approval and consent to participate
Prior to the study, approval was obtained from the research ethics committee of Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences (Reference No. LVRIAEC2010-005). The collection and autopsy of the plateau pikas were handled in strict accordance with good animal practice according to the animal ethics procedures and guidelines for animal husbandry and wildlife protection.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Guo-Qiang Zhu and Hong-Bin Yan contributed equally to this work
Contributor Information
Guo-Qiang Zhu, Email: zhuguoqiang915@163.com.
Hong-Bin Yan, Email: yanhongbin@caas.cn.
Li Li, Email: lili03@caas.cn.
John Asekhaen Ohiolei, Email: asekhaenj@gmail.com.
Yan-Tao Wu, Email: 643058923@qq.com.
Wen-Hui Li, Email: liwenhui@caas.cn.
Nian-Zhang Zhang, Email: zhangnianzhang@caas.cn.
Bao-Quan Fu, Email: fubaoquan@caas.cn.
Wan-Zhong Jia, Email: jiawanzhong@caas.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13071-020-04456-w.
References
- 1.Davidson RK, Romig T, Jenkins E, Tryland M, Robertson LJ. The impact of globalisation on the distribution of Echinococcus multilocularis. Trends Parasitol. 2012;28(6):239–247. doi: 10.1016/j.pt.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 2.Romig T, Ebi D, Wassermann M. Taxonomy and molecular epidemiology of Echinococcus granulosus sensu lato. Vet Parasitol. 2015;213(3–4):76–84. doi: 10.1016/j.vetpar.2015.07.035. [DOI] [PubMed] [Google Scholar]
- 3.Cadavid Restrepo AM, Yang YR, McManus DP, Gray DJ, Giraudoux P, Barnes TS, et al. The landscape epidemiology of echinococcoses. Infect Dis Poverty. 2016;5:13. doi: 10.1186/s40249-016-0109-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiao N, Qiu J, Nakao M, Li T, Yang W, Chen X, et al. Echinococcus shiquicus n. sp., a taeniid cestode from Tibetan fox and plateau pika in China. Int J Parasitol. 2005;35(6):693–701. doi: 10.1016/j.ijpara.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Xiao N, Qiu J, Nakao M, Li T, Yang W, Chen X, et al. Echinococcus shiquicus, a new species from the Qinghai-Tibet plateau region of China: discovery and epidemiological implications. Parasitol Int. 2006;55(Suppl):S233–S236. doi: 10.1016/j.parint.2005.11.035. [DOI] [PubMed] [Google Scholar]
- 6.Fan YL, Lou ZZ, Li L, Yan HB, Liu QY, Zhan F, et al. Genetic diversity in Echinococcus shiquicus from the plateau pika (Ochotona curzoniae) in Darlag County, Qinghai China. Infect Genet Evol. 2016;45:408–414. doi: 10.1016/j.meegid.2016.06.016. [DOI] [PubMed] [Google Scholar]
- 7.Wang X, Liu J, Zuo Q, Mu Z, Weng X, Sun X, et al. Echinococcus multilocularis and Echinococcus shiquicus in a small mammal community on the eastern Tibetan Plateau: host species composition, molecular prevalence, and epidemiological implications. Parasit Vectors. 2018;11(1):302. doi: 10.1186/s13071-018-2873-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boufana B, Qiu J, Chen X, Budke CM, Campos-Ponce M, Craig PS. First report of Echinococcus shiquicus in dogs from eastern Qinghai-Tibet plateau region China. Acta Trop. 2013;127(1):21–24. doi: 10.1016/j.actatropica.2013.02.019. [DOI] [PubMed] [Google Scholar]
- 9.Yuan R, Wu H, Zeng H, Liu P, Xu Q, Gao L, et al. Prevalence of and risk factors for cystic echinococcosis among herding families in five provinces in western China: a cross-sectional study. Oncotarget. 2017;8(53):91568–91576. doi: 10.18632/oncotarget.21229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakao M, McManus DP, Schantz PM, Craig PS, Ito A. A molecular phylogeny of the genus Echinococcus inferred from complete mitochondrial genomes. Parasitology. 2007;134(Pt 5):713–722. doi: 10.1017/S0031182006001934. [DOI] [PubMed] [Google Scholar]
- 11.Boufana B, Umhang G, Qiu J, Chen X, Lahmar S, Boue F, et al. Development of three PCR assays for the differentiation between Echinococcus shiquicus, E. granulosus (G1 genotype), and E. multilocularis DNA in the co-endemic region of Qinghai-Tibet plateau China. Am J Trop Med Hyg. 2013;88(4):795–802. doi: 10.4269/ajtmh.12-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu CN, Lou ZZ, Li L, Yan HB, Blair D, Lei MT, et al. Discrimination between E. granulosus sensu stricto, E. multilocularis and E. shiquicus using a multiplex PCR assay. PLoS Negl Trop Dis. 2015;9(9):e0004084. doi: 10.1371/journal.pntd.0004084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakao M, Li T, Han X, Ma X, Xiao N, Qiu J, et al. Genetic polymorphisms of Echinococcus tapeworms in China as determined by mitochondrial and nuclear DNA sequences. Int J Parasitol. 2010;40(3):379–385. doi: 10.1016/j.ijpara.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakao M, Lavikainen A, Yanagida T, Ito A. Phylogenetic systematics of the genus Echinococcus (Cestoda: Taeniidae) Int J Parasitol. 2013;43(12–13):1017–1029. doi: 10.1016/j.ijpara.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 15.Khademvatan S, Majidiani H, Foroutan M, Hazrati Tappeh K, Aryamand S, Khalkhali HR. Echinococcus granulosusgenotypes in Iran: a systematic review. J Helminthol. 2019;93(2):131–138. doi: 10.1017/S0022149X18000275. [DOI] [PubMed] [Google Scholar]
- 16.Cucher MA, Macchiaroli N, Baldi G, Camicia F, Prada L, Maldonado L, et al. Cystic echinococcosis in South America: systematic review of species and genotypes of Echinococcus granulosus sensu lato in humans and natural domestic hosts. Trop Med Int Health. 2016;21(2):166–175. doi: 10.1111/tmi.12647. [DOI] [PubMed] [Google Scholar]
- 17.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17(8):754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 18.Cai QG, Han XM, Yang YH, Zhang XY, Ma LQ, Karanis P, et al. Lasiopodomys fuscus as an important intermediate host for Echinococcus multilocularis: isolation and phylogenetic identification of the parasite. Infect Dis Poverty. 2018;7(1):27. doi: 10.1186/s40249-018-0409-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000;9(10):1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- 20.Clewley JP, Arnold C. MEGALIGN. The multiple alignment module of LASERGENE. Methods Mol Biol. 1997;70:119–129. [PubMed] [Google Scholar]
- 21.Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19(18):2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- 22.Xiao N, Qiu JM, Nakao M, Li TY, Chen XW, Schantz PM, et al. Biological features of a new Echinococcus species (Echinococcus shiquicus) in the east of Qinghai-Tibet Plateau. Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi. 2008;26(4):307–312. [PubMed] [Google Scholar]
- 23.Addy F, Wassermann M, Kagendo D, Ebi D, Zeyhle E, Elmahdi IE, et al. Genetic differentiation of the G6/7 cluster of Echinococcus canadensis based on mitochondrial marker genes. Int J Parasitol. 2017;47(14):923–931. doi: 10.1016/j.ijpara.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 24.Ma J, Wang H, Lin G, Craig PS, Ito A, Cai Z, et al. Molecular identification of Echinococcus species from eastern and southern Qinghai, China, based on the mitochondrial cox1 gene. Parasitol Res. 2012;111(1):179–184. doi: 10.1007/s00436-012-2815-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig. S1. Multiplex PCR results image of Echinococcus shiquicus samples based on three specific primers of E. multilocularis, E. shiquicus and E. granulosus.
Data Availability Statement
All data supporting the conclusions are included in the article. Representative nucleotide sequences of cox1 and nad1 genes from the present study are available in the GenBank database under the accession numbers (MW072804-MW072815).