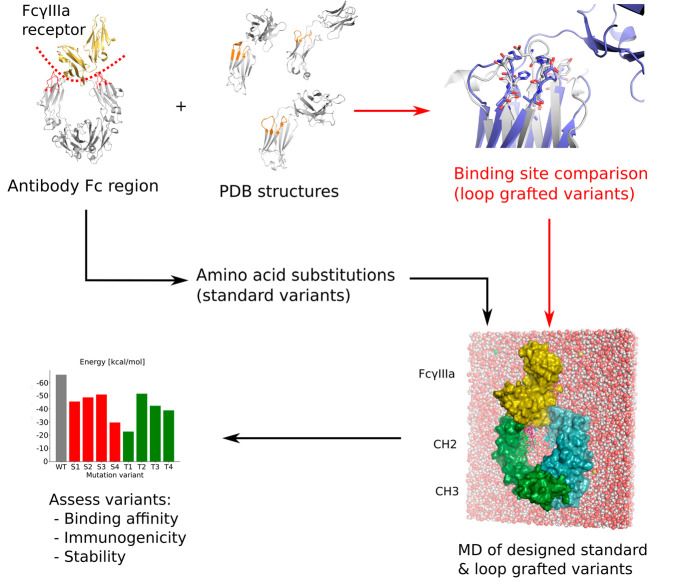
Abstract
Reduction of the affinity of the fragment crystallizable (Fc) region with immune receptors by substitution of one or a few amino acids, known as Fc-silencing, is an established approach to reduce the immune effector functions of monoclonal antibody therapeutics. This approach to Fc-silencing, however, is problematic as it can lead to instability and immunogenicity of the developed antibodies. We evaluated loop grafting as a novel approach to Fc-silencing in which the Fc loops responsible for immune receptor binding were replaced by loops of up to 20 amino acids from similar local environments in other human and mouse antibodies. Molecular dynamics simulations of the designed variants of an Fc region in a complex with the immune receptor FcγIIIa confirmed that loop grafting potentially leads to a significant reduction in the binding of the antibody variants to the receptor, while retaining their stability. In comparison, standard variants with less than eight substituted amino acids showed possible instability and a lower degree of Fc-silencing due to the occurrence of compensatory interactions. The presented approach to Fc-silencing is general and could be used to modulate undesirable side effects of other antibody therapeutics without affecting their stability or increasing their immunogenicity.
Introduction
Biological drugs are increasingly used in modern medicine as an effective alternative to conventional small-molecule drugs. The concept of biological drugs refers to all medicines derived from biological sources such as plants or blood. However, more narrowly, this term applies to recombinant therapeutic proteins. Perhaps the most well-known therapeutics from this group are monoclonal immunoglobulin G1 (IgG1) antibodies. In recent decades therapeutic antibodies have shown great success in the treatment of cancer. The main advantage of monoclonal antibodies compared to classical chemotherapeutic agents is their ability to specifically bind to target proteins that are expressed on the surface of the cells of the immune system, activating them to fight cancer.1
Immunoglobulin G antibody is a heterotetramer protein composed of two heavy chains and two light chains and has a dual role in the immune system.2 A part of an antibody is the so-called fragment crystallizable (Fc) region, whose role is to interact with various native receptors. The Fc region is further divided into CH2 and CH3 domains, and the CH2 domain interacts directly with various immune Fcγ receptors (FcγRs). Binding to FcγRI, FcγRIIa, FcγRIIc, FcγRIIIa, FcγRIIIb, and to the complement component 1q (C1q) induces pro-inflammatory responses, such as antibody-dependent cellular cytotoxicity and complement-dependent cytotoxicity, while the binding to the FcγRIIb induces an anti-inflammatory response.2,3
In the therapeutic use of recombinant monoclonal antibodies, the involvement of the Fc mediated cytotoxicity may be considered desirable or undesirable.4−9 Activation of this cytotoxic response is undesirable in the case of antibodies with known Fc-mediated side effects, e.g., platelet aggregation,10 or those that are designed to block receptors without activating the immune system, the so-called “benign blocker” antibodies, or in the case of antibody-drug conjugates, where unforeseen interactions with nontumor cells can be harmful.11 Unwanted cytotoxic responses can be eliminated at least partially through so-called Fc-silencing. This involves substituting amino acid residues in the binding site for FcγRs, specifically in the loops BC, C′E, and FG in the CH2 domain, to reduce the binding to FcγRs. The effects of such substitutions involving a single amino acid or several amino acids within the Fc region of IgGs have been studied.5,12−14 Various Fc variants can increase or decrease the antibody’s binding affinity for different FcγRs and consequently increase or decrease its effector function. Mutations of amino acid residues in the hinge region connecting the CH2 and CH1 domains can achieve a similar effect.15 However, such artificial amino acid sequences may be immunogenic or exhibit diminished stability. Thus, mutations based on the natural difference in affinity to FcγRs between IgG1 (high affinity) versus IgG2 and IgG4 (low affinity) antibody subclasses were explored.16 Another way to achieve the silencing of IgG1-based therapeutic antibodies is to use isolated Fab regions that do not cause cytotoxic responses through the activation of FcγRs. This approach however can lead to impaired serum half-life.17 Removal of glycan from IgG also results in lower affinity of binding to FcγRs but can subsequently impair other properties of such antibodies.3,18,19
Loop grafting involves substitution of loops, so that a loop is transplanted from a donor protein to the acceptor protein.20 Loop grafting adds properties of the donor protein to the acceptor protein and has been used to prepare humanized antibodies with reduced immunogenicity21 and to modulate binding in the CH3 domain.22,23 However, loop grafting has not yet been used in the CH2 domain to achieve Fc-silencing.
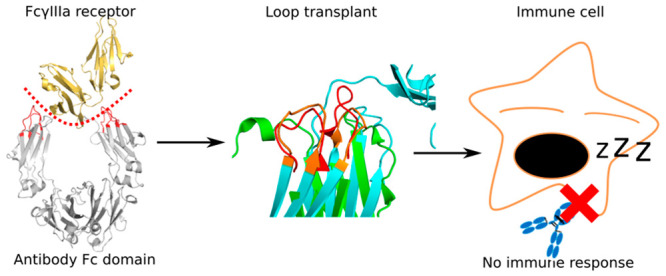
We present a novel approach to Fc-silencing based on loop grafting (Figure 1). Using the ProBiS algorithm24−28 that enables accurate detection of local structural similarities in proteins independent of their fold-similarity, the loops were transplanted to the Fc region of a therapeutic antibody from similar local environments in different antibodies. Since the grafted loops have been optimized by evolution for stability but not for FcγRs binding, the designed Fc variants should be stable and Fc-silent. We applied the new approach to the Fc region of an IgG1 type therapeutic antibody, and using molecular dynamics simulations, we showed that the developed Fc variants potentially exhibit a considerably higher level of Fc-silencing on average compared to variants prepared using the standard approach of amino acid substitutions. Two of the loop grafted variants also retained similar stability to that of the wild-type Fc region making them promising candidates for achieving completely Fc-silent and stable immunotherapeutics.
Figure 1.
Workflow for the design and evaluation of Fc silent antibodies used in this study. The new approach is marked in red.
Materials and Methods
The glycosylated human IgG1 antibody Fc region noncovalently bound to the human FcγIIIa receptor (FcγRIIIa) structure (PDB ID: 3SGJ)29 was used as the reference X-ray crystal structure.
Loop Grafting for Fc-Silencing
As the database of template protein structures we used the Protein Data Bank (PDB) (downloaded Jan 2019),30 in which we replaced the protein structures that consisted of Cα atoms with full-atom structural models from the ModBase database.31 The structures were filtered for antibodies by performing a text search in each of the downloaded PDB files using antibody and immunoglobulin keywords, and only files that contained either of these keywords were kept. The binding site for FcγRIIIa on the Fc region (3SGJ, chain A) was defined as the amino acid residues of the Fc region that were within 7 Å of the FcγRIIIa (chain C). Using the ProBiS algorithm,24−27 this binding site was then structurally compared in terms of its geometry and physicochemical properties with the template protein structures, in which structurally similar regions were identified. Rotation-translation matrices that superimposed the query binding site to the similar local structures found in other proteins were generated. As candidates for grafting, we considered loops that were structurally similar to the query BC and FG loops in their start and end sections but whose middle sections differed from these query loops. Based on the generated superimpositions, we then transposed the found similar loops to the query Fc region structure and attached them in place of the original loops that had been cut out of the Fc region. In each loop grafted Fc variant, we replaced the BC loop (residues 265–274) and the FG loop (residues 323–332), since these formed most of the interactions with the FcγRIIIa.
Standard Approach to Fc-Silencing
For comparison to loop grafting, we produced four Fc variants, each consisting of one or several amino acid substitutions that are known to result in Fc-silencing. The substitutions used were L234F,32 G237A,33 and P238S12 in the hinge region that are known to reduce binding affinity to all FcγRs and S239A, which reduces affinity to FcγRIIIa only.12 The substitution D265A12 in the BC loop reduces binding to all FcγRs, and S267A12 shows increased binding to FcγRIIb, which represents an additional Fc-silencing mechanism complementary to the reduction of FcγRIIIa binding. H268A improves binding to FcγRIIs and reduces binding to FcγRIIIa,12 and D270A12 reduces binding to FcγRIIs and FcγRIIIa. Substitution of A327G12 in the FG loop reduces binding to FcγRIIIa but has no effect on FcγRII, and P329S12 reduces binding to all FcγRs, while the combination of A330S and P331S33 substitutions eliminates affinity for all FcγRs and the C1q complement protein.
Molecular Dynamics Simulations of the Fc-Fcγ Receptor Complexes
To assess the performance of both the novel and the standard approach to Fc-silencing, we employed molecular dynamics (MD) simulations. These allowed determination of the quality of the designed Fc variants measured in terms of their remaining receptor binding affinity and their stability. Molecular dynamics simulations were conducted using the CHARMM molecular simulation program.34 The standard CHARMM36 force field parameters35 were used for the protein part of the molecule and for carbohydrates.36 Disulfide bonds were modeled as they appear in the crystal structure 3SGJ, that is, two disulfide bonds within each chain of the Fc region, two within the FcγRIIIa, and one in the hinge region connecting the chains A and B of the Fc region. The complexes were solvated using the rigid TIP3P water model, where the dimensions of the water box were 155 × 120 × 105 Å. The systems were neutralized with sodium chloride corresponding to a 0.1 M concentration. Before each production run, the systems were equilibrated for 1 ns using the constant number of particles, volume, and temperature (NVT) ensemble. Production runs were performed using a constant number of particles, pressure, and temperature (NPT) ensemble, in which each simulation continued for at least 200 ns; analysis was performed from the 100th ns to the end of each simulation. In this time interval the complexes have reached equilibrium judging from their RMSD plots. From the trajectories obtained by MD simulations we evaluated and ranked the Fc variants according to their stability and the strength of their binding to FcγRIIIa.
Stability Analysis of the Designed Fc Variants
To estimate the structural stability of the CH2 domain, we calculated the root-mean-square fluctuation (RMSF) of each of its Cα atom positions with respect to its average position during the MD simulation. Root mean square deviation (RMSD) of the whole Fc region provided the estimate of the conformational changes of the designed Fc variants induced by the differences in binding to FcγRIIIa. It was calculated between the initial structure of the Fc region (chains A and B) and the snapshots of the Fc region at regular time intervals from the MD simulation trajectories.
Interactions and Binding Affinity of the Designed Fc Variants
Molecular dynamics simulations enabled us to observe the behavior of the designed variants in a complex with the FcγRIIIa and to determine the interactions and their importance for complex association. The interactions of the Fc region variants with the FcγRIIIa were estimated by calculating the average number of hydrogen bonds between the Fc region and the receptor over the time of each simulation using the COOR HBOND command in CHARMM. An interaction was considered to be a hydrogen bond if the distance between a hydrogen atom and an acceptor atom was less than 2.4 Å. Using the COOR CONT command we calculated the van der Waals (vdW) interactions as the average number of Fc region’s atoms <3 Å from the FcγRIIIa atoms. The obtained average values were used to compare the designed Fc variants with the control simulation of the wild-type Fc region in a complex with the FcγRIIIa. The stability of individual hydrogen bonds was estimated using occupancy, representing the fraction of the total simulation time in which an interaction is present. Cumulative occupancies were reported, which are the sum of individual hydrogen bond occupancies between a residue and another residue, where a value >1.0 denotes that more hydrogen bonds occurred between the two residues. We also calculated the relative binding free energies for the Fc-FcγRIIIa complexes using the Molecular Mechanical/Generalized Born Surface Area (MM/GBSA) approach.37,38 Here, the binding free energy (ΔGbind) is calculated as the sum of the changes in the molecular mechanical energy of the gas phase, ΔEMM, the solvation free energy, ΔGsol, and the conformational entropy of the system during binding, −T·ΔS:
| 1 |
| 2 |
| 3 |
In eq 2, ΔEMM is the sum of ΔEinternal (bond, angle, and dihedral energy), ΔEelectrostatic (electrostatic energy), and ΔEvdW (van der Waals energy); in eq 3, ΔGsol is the sum of electrostatic solvation energy, ΔGGB (polar contribution), and nonelectrostatic solvation component, ΔGSA (nonpolar contribution). The polar contribution to the desolvation free energy was calculated using the analytical Generalized Born using Molecular Volume (GBMV) model,39,40 while the nonpolar energy was estimated using the solvent accessible surface area calculation implemented in the GB module. We assumed that the entropy changes upon binding are similar in all complexes, since they all have similar binding sites. Accordingly, we neglected the entropy term (−T·ΔS) when calculating the relative binding free energies.
Results
To evaluate loop grafting as a novel approach for Fc-silencing, we constructed Fc variants T1–T4 using loop grafting by local structural alignment. For comparison, we also constructed Fc variants S1–S4 using a standard approach by combining amino acid substitutions known to reduce receptor binding. The stability and FcγIIIa receptor (FcγRIIIa) interactions of the designed variants were determined using molecular dynamics simulations and compared with each other and to the wild-type (WT) Fc region in complex with the FcγRIIIa.
Loop Grafting between Similar Local Environments
We found loops in antibody domains other than Fc regions with similar physicochemical properties at their N- and C-terminal ends to the BC and FG loops in the Fc region (Figure 2A–D; Table S1, Supporting Information). In the T1 variant, we grafted the loops from the constant light chain (CL) domain of the Fab region of an IgG1 monoclonal mouse antibody (PDB ID: 5WTT),41 and in the T2 variant we obtained the loops from the mouse IgG2b-derived constant heavy chain domain (CH1) of the Fab region of a chimeric monoclonal antibody structure (5OCK).42 The T3 variant was constructed by grafting the loops from the human Fab fragment, specifically from the immunoglobulin light chain lambda constant domain (CL-lambda) of a monoclonal IgM rheumatoid factor (2J6E),43 and in variant T4, we grafted the loops from the light chain of the Fab region of a human secretory IgA2 antibody (3CM9).44
Figure 2.
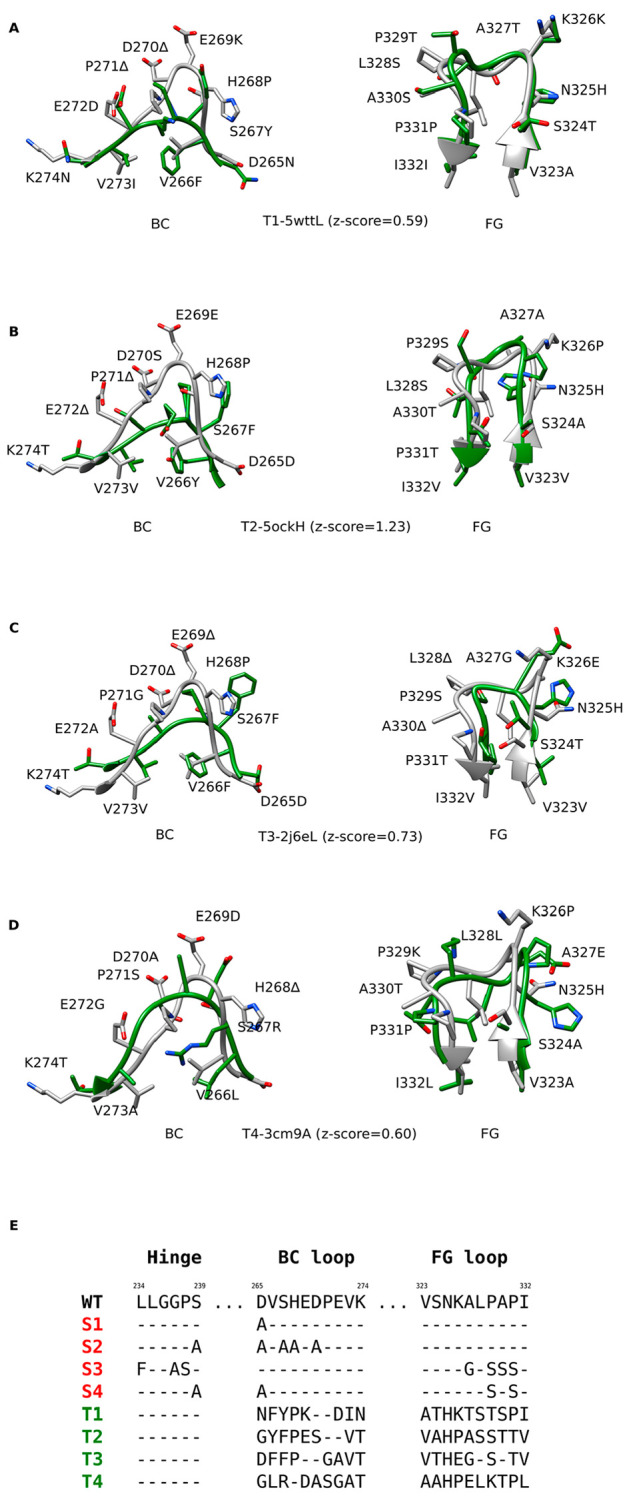
Designed Fc variants. A–D) Structural superpositions of the wild-type Fc region BC and FG loops (white) with the loops from different antibodies (green), based on which the loop grafted variants T1–T4 were constructed. Z-scores denote the degrees of local structural similarity between the binding site for FcγRIIIa on the Fc region and other antibody structures (PDB and Chain IDs) determined by ProBiS. E) Alignments of the wild-type (WT) amino acid sequence with the sequences of standard Fc variants (S1–S4, red) and the sequences of the loop grafted Fc variants (T1–T4, green). The sequence alignments for the latter were determined based on the structural alignments between the wild-type and the grafted loops.
Benchmarking the MM/GBSA Method for Predicting Binding Affinity of Fc Variants for FcγRIIIa
To evaluate the ability of the MM/GBSA method to predict experimental binding affinities, we have selected known Fc variants from Table 1 in ref (12), each of which has an experimentally measured ratio of binding (rexp) to FcγRIIIa relative to that of the WT Fc region (Table S11). The calculated binding free energies are generally consistent (r2 = 0.45) with the experimental ratios (Figure 3). This correlation is comparable to previous benchmarks of this method.45 The variant D265A, which is known to significantly reduce the binding to FcγRIIIa (rexp = 0.09), has the second highest calculated binding free energy of all standard variants at −45.6 kcal/mol. On the other hand, the variant K334A that most improves the binding to FcγRIIIa (rexp = 1.39) has the binding free energy of −66.2 kcal/mol, which is slightly lower than the wild-type Fc region. A fitted model
| 4 |
allows us to predict the binding ratios (rpred) of the constructed standard and loop grated variants described below.
Figure 3.
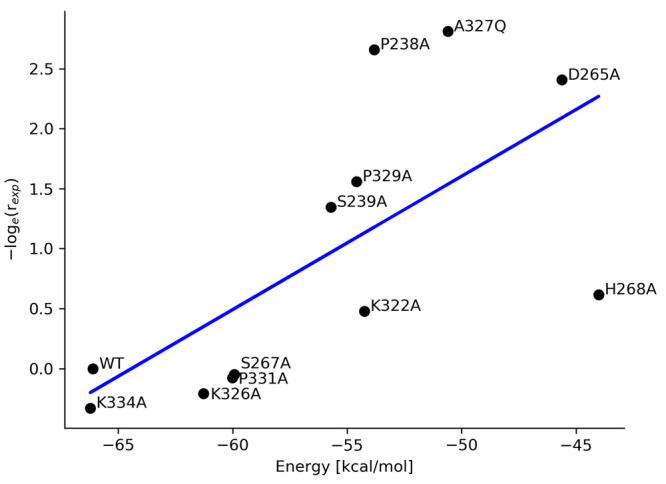
Comparison of the calculated binding free energies (ΔGbind) with experimental binding ratios (rexp) of standard Fc variants to FcγRIIIa with respect to the wild-type Fc region from Table 1, ref (12). Each dot represents an energy value calculated from a 150 ns MD simulation of an Fc variant in complex with FcγRIIIa; the blue line is the fitted model.
To test whether interaction energy would be sufficient to predict the binding ratios, we calculated the interaction energy between Fc variants and FcγRIIIa after initial minimization (Figure S1). The r2 of 0.07 indicates that this energy does not correlate with experimental binding ratios; however, other specialized molecular modeling tools that use empirical force fields may give better results.46
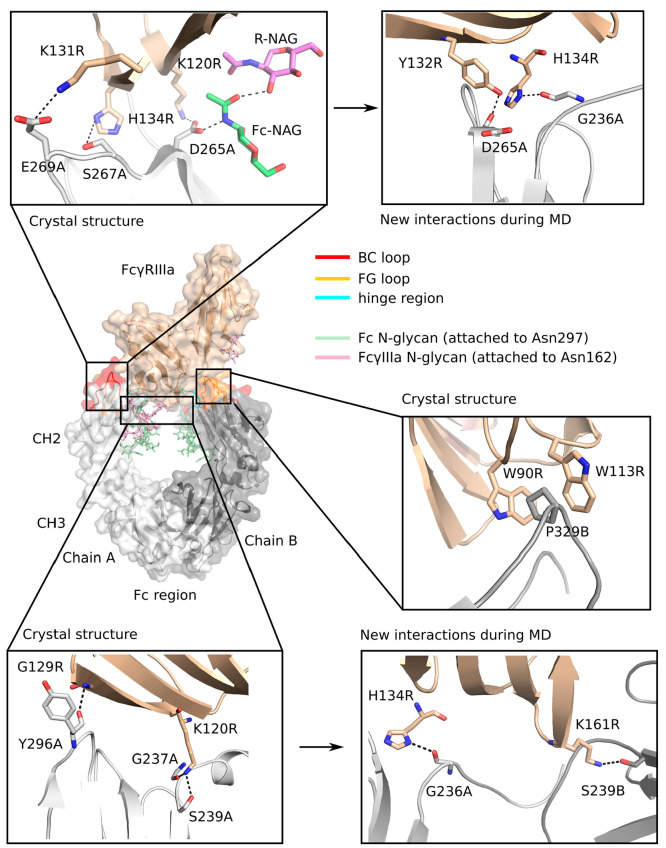
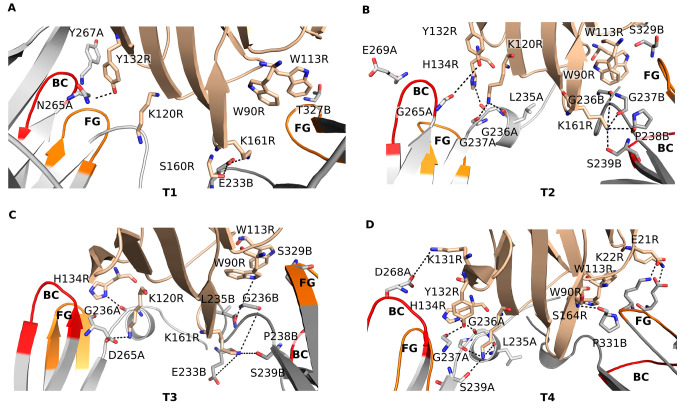
Interactions in the Wild-Type Fc-FcγIIIa Receptor Complex
We identified the interactions between the wild-type CH2 domain of the Fc region and the FcγRIIIa, as can be seen from the crystal structure 3SGJ and during MD simulation (Figure 4). In the crystal structure, hydrogen bonds are formed between the main chain oxygen of Gly237 of chain A (Gly237A) and the side chains of Ser239A and Asp265A in the CH2 domain and Lys120 in the FcγRIIIa (Lys120R). The main chain oxygen of Asp265A hydrogen bonds with the side chain nitrogen of His134R, and the main chain oxygen of Tyr296A interacts with the main chain amino group of Gly129R. A salt bridge is formed between Glu269A and Lys131R, and a stacking interaction occurs between Pro329 in chain B of the CH2 domain (Pro329B) and Trp90R and Trp113R so that Pro329B is stacked between the two tryptophans. Stacking interactions between prolines and tryptophans are known to stabilize protein–protein complexes.33,47 In addition, the N-glycan covalently attached to Asn297A and the N-glycan attached to Asn162R form a hydrogen bond by their N-acetyl-d-glucosamine sugar moieties. Hydrogen bonds are also formed between Asp265 in chains A and B and the amino group of the N-acetyl-d-glucosamines attached to Asn297.
Figure 4.
Interactions in the wild-type complex between the Fc region and the FcγIIIa receptor seen in the crystal structure (PDB ID: 3SGJ) and formed during molecular dynamics simulation.
During the MD simulation of the WT complex, new interactions were observed that are not seen in the crystal structure (Figure 4; Table S2, Supporting Information). Hydrogen bonds were formed between the main chain oxygen of Asp265A and the hydroxyl group of Tyr132R (occupancy: 0.92), between the main chain oxygen of Gly236A and the side chain nitrogen of His134R (0.84), and between the side chain of Ser239B and the side chain of Lys161R (0.69). Some interactions that were visible in the crystal structure were absent or occurred with low occupancy during the MD simulation. Included among these were the interactions of the main chain oxygen of Gly237A and the side chain of Ser239A with the side chain of Lys120R (occupancies 0.07 and 0.39, respectively) and the interaction between the main chain oxygen of Asp265A and the side chain nitrogen of His134R which was not observed during the entire MD simulation time.
Interactions in the Standard Fc Variants
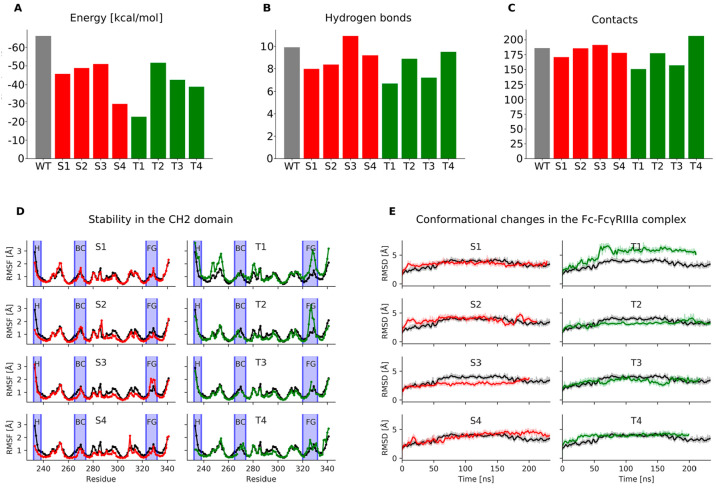
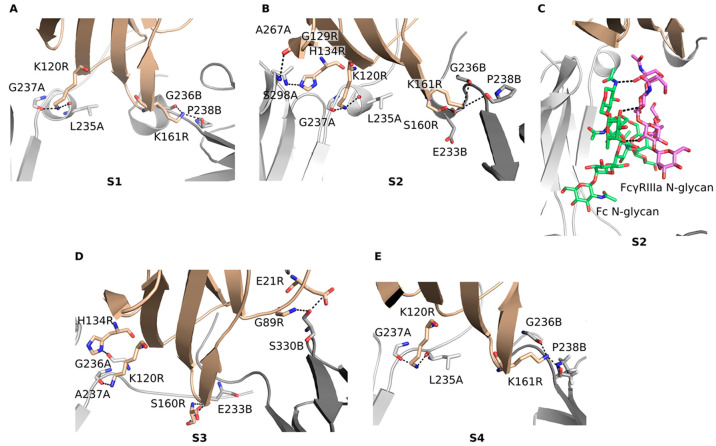
To test the effects of substitutions in the BC loop, hinge and BC loop, hinge and FG loop, and hinge and BC and FG loops, we designed standard variants S1–S4, respectively, which are combinations of single amino acid substitutions known to result in Fc-silencing. The S1 variant (D265A) had the binding free energy of −45.6 ± 16.1 kcal/mol, the second highest among the standard variants and significantly higher than −66.1 ± 18.7 kcal/mol of the WT (Figure 2E; Figure 5A). The predicted binding ratio rpred for this variant was 0.12. It also formed fewer hydrogen bonds than other standard variants and the WT (Figure 5B), indicating strong Fc-silencing in agreement with experiment.48 This reduction in FcγRIIIa binding was due to the loss of two salt bridge interactions between Asp265 in the CH2 domain and Lys120R and Lys161R in the receptor (Figure 6A; Table S3, Supporting Information). Another hydrogen bond was lost between the side chain of Ser239B and Lys161R due to the structural rearrangement of the region. In addition, the hydrogen bond between the main chain oxygen of Tyr296A and the main chain amino group of Gly129R had a much lower occupancy compared to the WT (WT: 0.60, S1: 0.15). However, these lost interactions were partially offset by the formation of two high occupancy hydrogen bonds between the main chain oxygens of Leu235A and Gly237A and the side chain of Lys120R (WT: 0.05, S1: 0.65; WT: 0.07, S1: 0.50) and two new hydrogen bonds between the main chain oxygens of Gly236B and Pro238B and the side chain of Lys161R (WT: 0.10, S1: 0.79; WT: 0.10, S1: 0.80). Due to these hydrogen bonds, the hinge region between Pro232 and Gly236 changed from a disordered region seen in the WT crystal structure to an α helix, which was observed in all standard variants. The variant S1 proved to be the most stable among the standard variants (Figure 5D,E) as its stability measured in terms of the RMSF and RMSD was comparable to that of the WT with the RMSF fluctuations in the BC and FG loops only slightly above those observed in the WT.
Figure 5.
Fc-silencing potential and structural stability of the designed standard (red) and loop grafted (green) Fc variants in comparison to the wild-type Fc-FcγIIIa receptor complex (gray columns in A–C, black lines in D and E). A) Relative binding free energies between the Fc region and the FcγIIIa receptor, where the height of columns indicates binding affinity for the receptor (lower columns indicate reduced affinity); B) Average number of hydrogen bonds and C) van der Waals contacts formed between the Fc region and the FcγIIIa receptor. The N-glycan bound to Asn297 in the Fc region and the N-glycan bound to Asn162 in the FcγIIIa receptor were included in this calculation; D) Root mean square fluctuations measuring the stability of individual amino acid residues in the CH2 domain. Blue vertical ribbons indicate the hinge region and the BC and FG loops; E) Root mean square deviations measuring the conformational changes in the Fc region during molecular dynamics simulations.
Figure 6.
Compensatory interactions formed by the standard Fc variants with the FcγIIIa receptor (A–E).
The S2 variant (S239A/D265A/S267A/H268A/D270A) had a binding free energy of −48.8 ± 17.5 kcal/mol (rpred = 0.18), and compared to the WT it formed less hydrogen bonds and slightly less vdW contacts with the receptor (Figure 5A–C). It also formed more hydrogen bonds with the receptor than the S1 variant, which was unexpected given that it had five substituted amino acid residues compared to one in the S1 variant (Figure 6B; Table S4, Supporting Information). However, the main chain oxygen of Leu235A in this variant interacted much more strongly with the side chain of Lys120R (WT: 0.05, S2: 0.84). This compensation was due to the rearrangement of the binding site caused by the elimination of the interaction of Asp265A with Lys120R, which brought the Leu235A and Lys120R closer together. The N-glycan attached to Asn297 hydrogen bonded with the fucose moiety with the manose of the receptor’s N-glycan (S2: 1.48) (Figure 6C). This interaction was not observed in the WT, where the hydrogen bond of the Fc N-glycan’s first sugar moiety N-acetyl-d-glucosamine with Asp265 (Figure 4) may have restricted its ability to move closer to and form stable contacts with the N-glycan of the receptor. Other new hydrogen bonds that compensated for the lost interactions were the interaction of Gly237A with Lys120R (WT: 0.07, S2: 0.66), Ala267A with His134R (WT: 0.0, S2: 0.72), Ser298A with Gly129R (WT: 0.02, S2: 0.27), Glu233B with Ser160R (WT: 0.09, S2: 0.27), Gly236B with Lys161R (WT: 0.1, S2: 0.82), and Pro238B with Lys161R (WT: 0.1, S2: 0.91). While the S2 variant was stable overall, an increased flexibility was observed at His285 (Figure 5D) located in the binding site for the second neonatal Fc receptor (FcRn) in the FcRn dimer.49,50 Reduced FcRn interactions are known to lead to increased serum clearance,50 and increased flexibility of this histidine may result in lower serum half-life and reduced therapeutic applicability of this variant.
The S3 variant (L234F/G237A/P238S/A327G/P329S/A330S/P331S) formed more hydrogen bond interactions and contacts with the receptor than the WT, and its binding energy of −51.0 ± 20.1 kcal/mol (rpred = 0.22) was the lowest of the developed variants (Figure 5A). It is therefore unlikely that the S3 variant is completely Fc silent. A reason for this may be that Asp265, the residue that forms the most hydrogen bonds with the receptor, was not substituted in this variant. New hydrogen bonds formed, while the existing ones were retained (Figure 6D; Table S5, Supporting Information). Due to the substitutions of P329S and P331S in the FG loop, the flexibility of the FG loop increased (Figure 5D) allowing this loop to move closer to the receptor and form new contacts that were previously not possible. In addition, P329S substitution eliminated the stacking interactions of this proline with Trp90R and Trp113R, which may have reduced the binding of the S3 variant with the receptor. Despite the substitutions L234F, G237A, and P238S, the hinge region showed no difference in stability compared to the WT.
In the S4 variant (S239A/D265A/P329S/A330S), which was a combination of substitutions in the hinge region and in the BC and FG loops, a significant increase in the binding free energy of −29.6 ± 17.5 kcal/mol (rpred = 0.02) compared to the WT was observed (Figure 5A–C), indicating that this variant reduced binding to receptor the most among the standard variants. In addition to hydrogen bonds that were removed due to the substituted residues, the interactions of Ser239 in both chains with Lys120R and Lys161R were completely removed (Figure 6E; Table S6, Supporting Information), and the interaction of the main chain oxygen of Tyr296A with Gly129R was significantly reduced (WT: 0.60, S4: 0.1). Compensatory interactions were somewhat less pronounced in this variant. Nevertheless, the main chain oxygens of Leu235A and Gly237A interacted more strongly with the side chain of Lys120R (WT: 0.05, S4: 0.83; WT: 0.07, S4: 0.67) as well as the interactions of the main chain oxygens of Gly236B and Pro238B with the side chain of Lys161R increased considerably (WT: 0.1, S4: 0.70; WT: 0.1, S2: 0.84). The overall stability of the CH2 domain was comparable to that of the WT, with the exception of His310 located in the FcRn binding site, whose flexibility increased (Figure 5D). Since mutations of His310 are known to reduce binding to FcRn and increase clearance,51 this increased flexibility of this residue could be an issue if the S4 variant was to be used in vivo.
Interactions in the Loop Grafted Fc Variants
The T1 variant showed the highest level of Fc-silencing of all the designed variants with its binding free energy of −22.7 ± 25.5 kcal/mol (rpred = 0.01) significantly higher than that of the WT and of other variants (Figure 5A). It also lacked most hydrogen bond interactions and vdW contacts with the FcγRIIIa that were present in the WT (Figure 5B,C). In the grafted BC loop, the replacement of Asp265A by Asn265A eliminated the salt bridge with Lys120R (Figure 2A,E; Figure 7A; Table S7, Supporting Information); however, a weaker hydrogen bond formed with Tyr132R (WT: 0.92, T1: 0.28). The replacement of Glu269A by Tyr267A led to the loss of a salt bridge interaction with Lys131R, and the replacement of Pro329B by Thr327B in the FG loop eliminated the stacking interactions of this proline with Trp90R and Trp113R. In compensation, two new hydrogen bonds were formed between Glu233B in the hinge region and Ser160R and Lys161R (WT: 0.02, T1: 0.72; WT: 0.02, T1: 0.31). The T1 variant experienced considerable conformational changes during MD simulation as seen from its RMSD plot (Figure 5E). The high fluctuations were due to the change in the relative positions between the CH2 and CH3 domains and between the CH2 and FcγRIIIa and less due to the changes within the CH2 domain, which remained relatively stable as seen from the RMSF plot (Figure 5F). The grafted FG loop however fluctuated more than the respective loop in the WT. The T1 variant thus showed significant Fc-silencing potential but could potentially have lower stability.
Figure 7.
Interactions with the FcγIIIa receptor of the Fc variants predicted to be Fc-silenced obtained with loop grafting (A–D).
The T2 variant had an increased average number of hydrogen bonds and vdW contacts as well as a lower binding free energy of −51.6 ± 17.4 kcal/mol (rpred = 0.24) compared to the T1 variant (Figure 5A). The interactions formed by Asp265 in the WT were mostly eliminated due to the replacement of this aspartate with Gly265 (Figure 7B; Table S8, Supporting Information). An exception was the hydrogen bond between the main chain oxygen of Gly265A and the side chain nitrogen of Tyr132R, which was nevertheless weaker than the respective WT interaction (WT: 0.92, T2: 0.8). The salt bridge between Glu269A and Lys131R did not form (WT: 0.48, T2: not observed) due to the two deletions in the BC loop sequence that shifted this glutamate away from the lysine (Figure 2B). The stacking interactions between the Pro329B in the original FG loop and the Trp90R and Trp113R were also removed due to the replacement of this residue with Ser329B. The interactions of Gly237B and Ser239B with Lys161R (WT: 0.67, T2: 0.24; WT: 0.69, T2: 0.2) were weakened but not completely eliminated (Figure 6C,D). In addition to this relatively weak reduction of WT interactions, compensatory hydrogen bonding interactions were formed between the main chain oxygens of Leu235A and Gly237A and the side chain of Lys120R (WT: 0.05, T2: 0.49; WT: 0.07, T2: 0.44) and between the main chain oxygen of Gly236A and the side chain of His134R (WT: 0.85, T2: 0.96) as well as between the main chain oxygens of Gly236B and Pro238B and the side chain of Lys161R (WT: 0.1, T2: 0.34; WT: 0.1, T2: 0.25). The T2 variant showed similar overall stability to that of the WT, with the exception of the grafted FG loop (Figure 5D), which was more flexible.
The T3 variant formed on average fewer hydrogen bonds and contacts with the FcγRIIIa than the WT. Its binding free energy of −42.4 ± 19.6 kcal/mol (rpred = 0.09) was significantly higher than that of the WT making this variant a promising Fc-silenced candidate. Its overall stability and the stability of its grafted FG loop were similar as the corresponding stability of the WT (Figure 5D,E). With the exception of Asp265A in the grafted BC loop, which retained a hydrogen bond with Lys120R, the grafted BC loop did not form any other significant interactions with the receptor (Figure 7C; Table S9, Supporting Information). The stacking of Pro329B with Trp90R and Trp113R was also eliminated due to the replacement of this proline by a serine in the grafted FG loop (Figure 2C,E). Some interactions were considerably weaker compared to the WT. These were the interaction of the main chain oxygen of Gly236B with the side chain of Lys161R (WT: 0.67, T3: 0.24) and the interaction between Ser239B and Lys161R (WT: 0.69, T3: 0.14). Compensatory interactions were formed; however, their scope was limited compared to the standard variants. Nevertheless, the hydrogen bond between Gly236A and His134R became stronger (WT: 0.84, T3: 0.97) as well as the salt bridge between Glu233B and Lys161R (WT: 0.02, T3: 1.03) and the hydrogen bond between Leu235B and Trp90R (WT: not observed, T3: 0.59).
The T4 variant’s binding free energy of −38.9 ± 19.2 kcal/mol (rpred = 0.06) was the second highest among the loop grafted variants (Figure 5A), and its stability was comparable to that of the WT over the entire CH2 domain (Figure 5D,E). During the MD simulation it maintained some polar interactions with the FcγRIIIa that were present in the WT (Figure 7D; Table S10, Supporting Information), but these occurred with lower occupancies than in the WT. The interactions between Pro329B and the tryptophans 90 and 113 of the receptor were removed due to the substitution of this proline with Lys329B (Figure 2D,E). Other interactions that were significantly weakened compared to the WT were the interaction between the main chain oxygen of Gly236A and the side chain nitrogen of His134R (WT: 0.85, T4: 0.49), the interaction between the side chain of Ser239A and the side chain of Lys120R (WT: 0.39, T4: 0.09), and the interaction between the main chain oxygen of Gly265A, which replaced Asp265A in the BC loop, and the side chain of Tyr132R (WT: 0.92, T4: 0.58). Increased occupancies were observed for the interaction of the main chain oxygen of Leu235A with the side chain of Lys120R (WT: 0.05, T4: 1.0) and the interaction of the main chain oxygen of Gly237A with the side chain of Lys120R (WT: 0.07, T4: 0.34). In addition, the Asp268A in the grafted BC loop formed a salt bridge with Lys131R (WT: not observed, T4: 0.41) similar to Glu269A in the WT. In the grafted FG loop, Glu327B formed a salt bridge with Lys22R (WT: not observed, T4: 0.41), Lys329B formed a salt bridge with Glu21R (WT: not observed, T4: 0.49), and Pro331B interacted with its main chain oxygen with the main chain amino group of Ser164R (WT: not observed, T4: 0.49).
Discussion
Silencing of the Fc region has an important role in the rapid development of various antibody- and T cell-based therapeutic platforms for the treatment of cancers and other currently incurable diseases.15,16,52 We propose a novel approach to achieve Fc-silencing based on the replacement of the BC and FG loops in the Fc region of therapeutic antibodies with loops from non-Fc antibody domains. The proposed approach enables high degree of Fc-silencing and stability of the constructed Fc variants comparable to the wild-type protein as shown by molecular dynamics simulations.
Among the standard Fc variants, the S4 variant showed the highest reduction in FcγRIIIa binding (Figure 5), followed by the S1 variant. The S1 variant is known to reduce FcγRIIIa interaction to only 9% of the WT activity and is one of the most effective Fc-silencing substitutions currently available.12 The S1 variant’s calculated stability is comparable to that of the WT over the entire CH2 domain (Figure 5D). The S2 and S3 variants were not completely Fc-silent according to their binding free energies (Figure 5A), which was likely due to the compensation of the lost interactions through the formation of new hydrogen bonds with the FcγRIIIa (Figure 6). The S3 variant also had an increased flexibility in its FG loop indicating a potential structural instability of this variant (Figure 5D), while the S2 and S4 variants showed increased flexibility in their neonatal Fc receptor (FcRn) binding sites (Figure 6). Binding of the Fc region to FcRn rescues an antibody from being excreted from the serum,49−51 and thus this instability potentially reduces the serum half-life and therapeutic applicability of the S2 and S4 variants. The S1 variant showed the best compromise between Fc-silencing and stability, suggesting that a single substitution D265A can be advantageous to the combinations of several artificial substitutions, which may introduce instability into the designed Fc regions.
The loop grafted Fc variants formed fewer hydrogen bonds and vdW contacts with the FcγRIIIa compared to the WT as well as their binding free energies were considerably higher on average than the energies of the WT and most of the standard variants (Figure 5A–C). The variants T3 and T4 in which the grafted loops originated from other human antibodies had a similar stability to the WT over their entire CH2 domains and their FG loops (Figure 5D). The grafted FG loops in the T3 and T4 variants were likely stabilized by the deletions that reduced the flexibility and by the conservation of the two rigid prolines, respectively. The T3 and T4 variants also had increased stability compared to the S2, S3, and S4 variants and were stable in their FcRn binding sites. We thus propose that the T3 and T4 variants are stable and highly Fc-silent.
In addition, since in the T3 and T4 variants the grafted loops were from human antibodies, their immunogenicity should not be increased. However, grafting murine loops may cause undesired T cell immunogenicity, and even grafting fully human loops may do so, due to epitopes at the junctions where the loop is spliced in. In order to test this, we predicted MHC-II binding for the constructed variants using the IEDB tool (Table 1).53 The T4 variant had the lowest predicted binding to MHC-II, suggesting that it is potentially significantly less immunogenic than the other constructed variants and the wild-type Fc region. Immunogenicity could be an issue with the standard variants S3 and S4 and the loop grafted variants T1 and T2 in which the loops were obtained from murine antibodies, which all showed increased binding to MHC-II molecules. The T1 and T2 variants also had an increased flexibility in their FG loops, due to the replacement of one of the FG loop’s prolines with a serine residue. Notably, we constructed four loop grafted variants, but many possible replacement loops remain unexplored (Table S1, Supporting Information).
Table 1. Prediction of Binding to MHC-II Alleles for Constructed Fc Variants Using the IEDB Toola,e.
| Fc variant | hinge region | BC loop | FG loop |
|---|---|---|---|
| percentile rank (IC50)b | percentile rank (IC50)b | percentile rank (IC50)b | |
| WT | 9.3 (161 nM) | 13 (776 nM) | 2.9 (166 nM)c |
| S1 | 9.3 (161 nM) | 13 (1029 nM) | 2.9 (166 nM)c |
| S2 | 9.1 (159 nM) | 13 (1029 nM) | 2.9 (166 nM)c |
| S3 | 5.1 (23 nM) | 13 (776 nM) | 5.9 (346 nM)c |
| S4 | 9.1 (159 nM) | 13 (1029 nM) | 1.9 (103 nM)d |
| T1 | 9.3 (161 nM) | 4 (352 nM) | 12 (102 nM) |
| T2 | 9.3 (161 nM) | 11 (476 nM) | 16 (517 nM) |
| T3 | 9.3 (161 nM) | 7 (193 nM) | 16 (283 nM) |
| T4 | 9.3 (161 nM) | 13 (1042 nM) | 15 (255 nM) |
Seven-allele HLA reference set was selected; for other parameters the default values were used.
Each percentile rank and IC50 value represents the most immunogenic peptide of length 15 in a set of peptides, in which each overlaps with at least one amino acid with the hinge region, BC or FG loops. The lower values indicate tighter binding of an Fc peptide to MHC-II and higher potential immunogenicity; smm_align_ic50 values are reported where available.
IC50 is calculated using the netmhciipan_ic50 method.
IC50 is calculated using the nn_align_ic50 method.
The IEDB tool can be found at http://tools.iedb.org/mhcii.
The fact that we did not see unbinding in the MD simulations is not uncommon, as the residence time of protein–protein complexes can be in the order of microseconds or more.54 A reason for this may also be that the Fc variants were modeled based on the WT, and therefore they assume a similar conformation at the beginning of the simulation. Thus, they can remain bound to the receptor longer than in a physical system, where such a conformation may not be accessible. The calculated binding free energies, H-bonds and vdW contacts, are thus estimates of the relative binding affinity of the Fc-FcγRIIIa complex, rather than absolute values.
Loop grafting thus shows a high potential as a new approach for the design of stable Fc-silent variants that could be used as building blocks of novel antibody therapeutics. Significant reduction of immunogenicity is predicted in the T4 variant in which the replacement loops originated from human antibodies.
Conclusion
Binding of therapeutic antibodies to Fcγ receptors is responsible for serious side effects and unwanted actions. Substitutions in the Fc region are used to achieve Fc-silent antibodies, but these may have decreased stability or increased immunogenicity. To mitigate this problem, we evaluated loop grafting as a new approach for the design of stable Fc-silent antibody variants with reduced binding to the FcγIIIa receptor. The developed loop grafted variants on average formed fewer hydrogen bonds and vdW contacts with the FcγIIIa receptor, while their binding free energies were considerably higher than in the wild-type Fc region and in most of the standard variants prepared using individual amino acid substitutions. The main cause of incomplete Fc-silencing in standard variants was the occurrence of compensatory interactions, which were to a lesser extent also observed in loop grafted variants. The variants T3 and T4 in which the grafted loops originated from human antibodies had a similar stability to the wild-type Fc region over their entire CH2 domains and lower affinity toward the FcγIIIa receptor in comparison to the S1 variant (D265A) variant, whose high Fc-silencing potential is known. We propose that the variants T3 (D265D, V266F, S267F, H268P, E269Δ, D270Δ, P271G, E272A, V273V, K274T, V323V, S324T, N325H, K326E, A327G, L328Δ, P329S, A330Δ, P331T, I332V) and T4 (D265G, V266L, S267R, H268Δ, E269D, D270A, P271S, E272G, V273A, K274T, V323A, S324A, N325H, K326P, A327E, L328L, P329K, A330T, P331P, I332L) are stable and completely Fc-silent. In addition, the loop grafted T4 variant is potentially the least immunogenic among the constructed variants and less immunogenic than the wild-type protein. With further experimental confirmation, the presented approach to Fc-silencing should prove an invaluable tool in the design of novel safer biological drugs.
Acknowledgments
Funding from the Slovenian Research Agency under project number L7-8269: “New approaches for better biopharmaceuticals” is acknowledged.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jcim.9b01198.
Table S1, antibody structures that share local similarities to Fc region; Tables S2–S10, interactions between Fc region and FcγIIIa receptor determined by molecular dynamics simulations; Figure S1, comparison of calculated interaction energies with experimental binding ratios; and Table S11, comparison of binding free energies calculated using MM/GBSA method with experimental binding ratios (PDF)
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Weiner L. M.; Rishi S.; Wang S. Monoclonal Antibodies: Versatile Platforms for Cancer Immunotherapy. Nat. Rev. Immunol. 2010, 10, 317–327. 10.1038/nri2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson Q. M.; Barb A. W. A Perspective on the Structure and Receptor Binding Properties of Immunoglobulin G Fc. Biochemistry 2015, 54, 2931–2942. 10.1021/acs.biochem.5b00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogarth P. M.; Pietersz G. A. Fc Receptor-Targeted Therapies for the Treatment of Inflammation, Cancer and Beyond. Nat. Rev. Drug Discovery 2012, 11, 311–331. 10.1038/nrd2909. [DOI] [PubMed] [Google Scholar]
- Mimoto F.; Katada H.; Kadono S.; Igawa T.; Kuramochi T.; Muraoka M.; Wada Y.; Haraya K.; Miyazaki T.; Hattori K. Engineered Antibody Fc Variant with Selectively Enhanced FcγRIIb Binding over Both FcγRIIaR131 and FcγRIIaH131. Protein Eng., Des. Sel. 2013, 26, 589–598. 10.1093/protein/gzt022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z.; Gunasekaran K.; Wang W.; Razinkov V.; Sekirov L.; Leng E.; Sweet H.; Foltz I.; Howard M.; Rousseau A.-M.; Kozlosky C.; Fanslow W.; Yan W. Asymmetrical Fc Engineering Greatly Enhances Antibody-Dependent Cellular Cytotoxicity (ADCC) Effector Function and Stability of the Modified Antibodies. J. Biol. Chem. 2014, 289, 3571–3590. 10.1074/jbc.M113.513366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart R.; Thom G.; Levens M.; Güler-Gane G.; Holgate R.; Rudd P. M.; Webster C.; Jermutus L.; Lund J. A Variant Human IgG1-Fc Mediates Improved ADCC. Protein Eng., Des. Sel. 2011, 24, 671–678. 10.1093/protein/gzr015. [DOI] [PubMed] [Google Scholar]
- Strohl W. R. Optimization of Fc-Mediated Effector Functions of Monoclonal Antibodies. Curr. Opin. Biotechnol. 2009, 20, 685–691. 10.1016/j.copbio.2009.10.011. [DOI] [PubMed] [Google Scholar]
- Kellner C.; Otte A.; Cappuzzello E.; Klausz K.; Peipp M. Modulating Cytotoxic Effector Functions by Fc Engineering to Improve Cancer Therapy. Transfus. Med. Hemother. 2017, 44, 327–336. 10.1159/000479980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang T. H.; Jung S. T. Boosting Therapeutic Potency of Antibodies by Taming Fc Domain Functions. Exp. Mol. Med. 2019, 51, 1–9. 10.1038/s12276-019-0345-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollreisz A.; Assinger A.; Hacker S.; Hoetzenecker K.; Schmid W.; Lang G.; Wolfsberger M.; Steinlechner B.; Bielek E.; Lalla E.; Klepetko W.; Volf I.; Ankersmit H. J. Intravenous Immunoglobulins Induce CD32-Mediated Platelet Aggregation in Vitro. Br. J. Dermatol. 2008, 159, 578–584. 10.1111/j.1365-2133.2008.08700.x. [DOI] [PubMed] [Google Scholar]
- McDonagh C. F.; Kim K. M.; Turcott E.; Brown L. L.; Westendorf L.; Feist T.; Sussman D.; Stone I.; Anderson M.; Miyamoto J.; Lyon R.; Alley S. C.; Gerber H.-P.; Carter P. J. Engineered Anti-CD70 Antibody-Drug Conjugate with Increased Therapeutic Index. Mol. Cancer Ther. 2008, 7, 2913–2923. 10.1158/1535-7163.MCT-08-0295. [DOI] [PubMed] [Google Scholar]
- Shields R. L.; Namenuk A. K.; Hong K.; Meng Y. G.; Rae J.; Briggs J.; Xie D.; Lai J.; Stadlen A.; Li B.; Fox J. A.; Presta L. G. High Resolution Mapping of the Binding Site on Human IgG1 for FcγRI, FcγRII, FcγRIII, and FcRn and Design of IgG1 Variants with Improved Binding to the FcγR. J. Biol. Chem. 2001, 276, 6591–6604. 10.1074/jbc.M009483200. [DOI] [PubMed] [Google Scholar]
- Borrok M. J.; Mody N.; Lu X.; Kuhn M. L.; Wu H.; Dall’Acqua W. F.; Tsui P. An “Fc-Silenced” IgG1 Format with Extended Half-Life Designed for Improved Stability. J. Pharm. Sci. 2017, 106, 1008–1017. 10.1016/j.xphs.2016.12.023. [DOI] [PubMed] [Google Scholar]
- Saunders K. O. Conceptual Approaches to Modulating Antibody Effector Functions and Circulation Half-Life. Front. Immunol. 2019, 10, 1296. 10.3389/fimmu.2019.01296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arduin E.; Arora S.; Bamert P. R.; Kuiper T.; Popp S.; Geisse S.; Grau R.; Calzascia T.; Zenke G.; Kovarik J. Highly Reduced Binding to High and Low Affinity Mouse Fc Gamma Receptors by L234A/L235A and N297A Fc Mutations Engineered into Mouse IgG2a. Mol. Immunol. 2015, 63, 456–463. 10.1016/j.molimm.2014.09.017. [DOI] [PubMed] [Google Scholar]
- An Z.; Forrest G.; Moore R.; Cukan M.; Haytko P.; Huang L.; Vitelli S.; Zhao J. Z.; Lu P.; Hua J.; Gibson C. R.; Harvey B. R.; Montgomery D.; Zaller D.; Wang F.; Strohl W. IgG2m4, an Engineered Antibody Isotype with Reduced Fc Function. MAbs 2009, 1, 572–579. 10.4161/mabs.1.6.10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan R. J.; Jones A. L. Fab Antibody Fragments. Drug Saf. 2004, 27, 1115–1133. 10.2165/00002018-200427140-00004. [DOI] [PubMed] [Google Scholar]
- Kao D.; Danzer H.; Collin M.; Gross A.; Eichler J.; Stambuk J.; Lauc G.; Lux A.; Nimmerjahn F. A Monosaccharide Residue Is Sufficient to Maintain Mouse and Human IgG Subclass Activity and Directs IgG Effector Functions to Cellular Fc Receptors. Cell Rep. 2015, 13, 2376–2385. 10.1016/j.celrep.2015.11.027. [DOI] [PubMed] [Google Scholar]
- Feige M. J.; Nath S.; Catharino S. R.; Weinfurtner D.; Steinbacher S.; Buchner J. Structure of the Murine Unglycosylated IgG1 Fc Fragment. J. Mol. Biol. 2009, 391, 599–608. 10.1016/j.jmb.2009.06.048. [DOI] [PubMed] [Google Scholar]
- Smith J. W.; Tachias K.; Madison E. L. Protein Loop Grafting to Construct a Variant of Tissue-Type Plasminogen Activator That Binds Platelet Integrin AIIbβ3. J. Biol. Chem. 1995, 270, 30486–30490. 10.1074/jbc.270.51.30486. [DOI] [PubMed] [Google Scholar]
- Kashmiri S. V.; De Pascalis R.; Gonzales N. R.; Schlom J. SDR Grafting—a New Approach to Antibody Humanization. Methods 2005, 36, 25–34. 10.1016/j.ymeth.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Lobner E.; Traxlmayr M. W.; Obinger C.; Hasenhindl C. Engineered IgG1-Fc–One Fragment to Bind Them All. Immunol. Rev. 2016, 270, 113–131. 10.1111/imr.12385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traxlmayr M. W.; Wozniak-Knopp G.; Antes B.; Stadlmayr G.; Rüker F.; Obinger C. Integrin Binding Human Antibody Constant Domains—Probing the C-Terminal Structural Loops for Grafting the RGD Motif. J. Biotechnol. 2011, 155, 193–202. 10.1016/j.jbiotec.2011.06.042. [DOI] [PubMed] [Google Scholar]
- Konc J.; Miller B. T.; Štular T.; Lešnik S.; Woodcock H. L.; Brooks B. R.; Janežič D. ProBiS-CHARMMing: Web Interface for Prediction and Optimization of Ligands in Protein Binding Sites. J. Chem. Inf. Model. 2015, 55, 2308–2314. 10.1021/acs.jcim.5b00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konc J.; Janežič D. ProBiS-2012: Web Server and Web Services for Detection of Structurally Similar Binding Sites in Proteins. Nucleic Acids Res. 2012, 40, W214–W221. 10.1093/nar/gks435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konc J.; Janežič D. ProBiS-Ligands: A Web Server for Prediction of Ligands by Examination of Protein Binding Sites. Nucleic Acids Res. 2014, 42, W215–W220. 10.1093/nar/gku460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konc J.; Janežič D. ProBiS Algorithm for Detection of Structurally Similar Protein Binding Sites by Local Structural Alignment. Bioinformatics 2010, 26, 1160–1168. 10.1093/bioinformatics/btq100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konc J. Binding Site Comparisons for Target-Centered Drug Discovery. Expert Opin. Drug Discovery 2019, 14, 445–454. 10.1080/17460441.2019.1588883. [DOI] [PubMed] [Google Scholar]
- Ferrara C.; Grau S.; Jäger C.; Sondermann P.; Brünker P.; Waldhauer I.; Hennig M.; Ruf A.; Rufer A. C.; Stihle M.; Umaña P.; Benz J. Unique Carbohydrate–Carbohydrate Interactions Are Required for High Affinity Binding between FcγRIII and Antibodies Lacking Core Fucose. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 12669–12674. 10.1073/pnas.1108455108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley S. K.; Berman H. M.; Christie C.; Duarte J. M.; Feng Z.; Westbrook J.; Young J.; Zardecki C. RCSB Protein Data Bank: Sustaining a Living Digital Data Resource That Enables Breakthroughs in Scientific Research and Biomedical Education. Protein Sci. 2018, 27, 316–330. 10.1002/pro.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieper U.; Webb B. M.; Dong G. Q.; Schneidman-Duhovny D.; Fan H.; Kim S. J.; Khuri N.; Spill Y. G.; Weinkam P.; Hammel M.; Tainer J. A.; Nilges M.; Sali A. ModBase, a Database of Annotated Comparative Protein Structure Models and Associated Resources. Nucleic Acids Res. 2014, 42, D336–D346. 10.1093/nar/gkt1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oganesyan V.; Gao C.; Shirinian L.; Wu H.; Dall’Acqua W. F. Structural Characterization of a Human Fc Fragment Engineered for Lack of Effector Functions. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2008, 64, 700–704. 10.1107/S0907444908007877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafa O.; Gilliland G. L.; Brezski R. J.; Strake B.; Wilkinson T.; Lacy E. R.; Scallon B.; Teplyakov A.; Malia T. J.; Strohl W. R. An Engineered Fc Variant of an IgG Eliminates All Immune Effector Functions via Structural Perturbations. Methods 2014, 65, 114–126. 10.1016/j.ymeth.2013.06.035. [DOI] [PubMed] [Google Scholar]
- Brooks B. R.; Brooks C. L. III; Mackerell A. D. Jr; Nilsson L.; Petrella R. J.; Roux B.; Won Y.; Archontis G.; Bartels C.; Boresch S.; Caflisch A.; Caves L.; Cui Q.; Dinner A. R.; Feig M.; Fischer S.; Gao J.; Hodoscek M.; Im W.; Kuczera K.; Lazaridis T.; Ma J.; Ovchinnikov V.; Paci E.; Pastor R. W.; Post C. B.; Pu J. Z.; Schaefer M.; Tidor B.; Venable R. M.; Woodcock H. L.; Wu X.; York D. M.; Karplus M. CHARMM: The Biomolecular Simulation Program. J. Comput. Chem. 2009, 30, 1545–1614. 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.; MacKerell A. D. Jr CHARMM36 All-Atom Additive Protein Force Field: Validation Based on Comparison to NMR Data. J. Comput. Chem. 2013, 34, 2135–2145. 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guvench O.; Greene S. N.; Kamath G.; Brady J. W.; Venable R. M.; Pastor R. W.; Mackerell A. D. Jr Additive Empirical Force Field for Hexopyranose Monosaccharides. J. Comput. Chem. 2008, 29, 2543–2564. 10.1002/jcc.21004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan J.; Cheatham T. E.; Cieplak P.; Kollman P. A.; Case D. A. Continuum Solvent Studies of the Stability of DNA, RNA, and Phosphoramidate-DNA Helices. J. Am. Chem. Soc. 1998, 120, 9401–9409. 10.1021/ja981844+. [DOI] [Google Scholar]
- Kollman P. A.; Massova I.; Reyes C.; Kuhn B.; Huo S.; Chong L.; Lee M.; Lee T.; Duan Y.; Wang W.; Donini O.; Cieplak P.; Srinivasan J.; Case D. A.; Cheatham T. E. Calculating Structures and Free Energies of Complex Molecules: Combining Molecular Mechanics and Continuum Models. Acc. Chem. Res. 2000, 33, 889–897. 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- Lee M. S.; Salsbury F. R.; Brooks C. L. Novel Generalized Born Methods. J. Chem. Phys. 2002, 116, 10606–10614. 10.1063/1.1480013. [DOI] [Google Scholar]
- Lee M. S.; Feig M.; Salsbury F. R.; Brooks C. L. New Analytic Approximation to the Standard Molecular Volume Definition and Its Application to Generalized Born Calculations. J. Comput. Chem. 2003, 24, 1348–1356. 10.1002/jcc.10272. [DOI] [PubMed] [Google Scholar]
- Zhong C.; Huo R.; Hu K.; Shen J.; Li D.; Li N.; Ding J. Molecular Basis for the Recognition of CCN1 by Monoclonal Antibody 093G9. J. Mol. Recognit. 2017, 30, e2645 10.1002/jmr.2645. [DOI] [PubMed] [Google Scholar]
- Ge C.; Xu B.; Liang B.; Lönnblom E.; Lundström S. L.; Zubarev R. A.; Ayoglu B.; Nilsson P.; Skogh T.; Kastbom A.; Malmström V.; Klareskog L.; Toes R. E. M.; Rispens T.; Dobritzsch D.; Holmdahl R. Structural Basis of Cross-Reactivity of Anti–Citrullinated Protein Antibodies. Arthritis Rheumatol. 2019, 71, 210–221. 10.1002/art.40698. [DOI] [PubMed] [Google Scholar]
- Duquerroy S.; Stura E. A.; Bressanelli S.; Fabiane S. M.; Vaney M. C.; Beale D.; Hamon M.; Casali P.; Rey F. A.; Sutton B. J.; Taussig M. J. Crystal Structure of a Human Autoimmune Complex between IgM Rheumatoid Factor RF61 and IgG1 Fc Reveals a Novel Epitope and Evidence for Affinity Maturation. J. Mol. Biol. 2007, 368, 1321–1331. 10.1016/j.jmb.2007.02.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner A.; Almogren A.; Furtado P. B.; Kerr M. A.; Perkins S. J. The Nonplanar Secretory IgA2 and Near Planar Secretory IgA1 Solution Structures Rationalize Their Different Mucosal Immune Responses. J. Biol. Chem. 2009, 284, 5077–5087. 10.1074/jbc.M807529200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genheden S.; Ryde U. The MM/PBSA and MM/GBSA Methods to Estimate Ligand-Binding Affinities. Expert Opin. Drug Discovery 2015, 10, 449–461. 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schymkowitz J.; Borg J.; Stricher F.; Nys R.; Rousseau F.; Serrano L. The FoldX Web Server: An Online Force Field. Nucleic Acids Res. 2005, 33, W382–W388. 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedermannova L.; Riley K. E.; Berka K.; Hobza P.; Vondrasek J. Another Role of Proline: Stabilization Interactions in Proteins and Protein Complexes Concerning Proline and Tryptophane. Phys. Chem. Chem. Phys. 2008, 10, 6350–6359. 10.1039/b805087b. [DOI] [PubMed] [Google Scholar]
- Baudino L.; Shinohara Y.; Nimmerjahn F.; Furukawa J.-I.; Nakata M.; Martínez-Soria E.; Petry F.; Ravetch J. V.; Nishimura S.-I.; Izui S. Crucial Role of Aspartic Acid at Position 265 in the CH2 Domain for Murine IgG2a and IgG2b Fc-Associated Effector Functions. J. Immunol. 2008, 181, 6664–6669. 10.4049/jimmunol.181.9.6664. [DOI] [PubMed] [Google Scholar]
- Burmeister W. P.; Huber A. H.; Bjorkman P. J. Crystal Structure of the Complex of Rat Neonatal Fc Receptor with Fc. Nature 1994, 372, 379–383. 10.1038/372379a0. [DOI] [PubMed] [Google Scholar]
- Ghetie V.; Popov S.; Borvak J.; Radu C.; Matesoi D.; Medesan C.; Ober R. J.; Ward E. S. Increasing the Serum Persistence of an IgG Fragment by Random Mutagenesis. Nat. Biotechnol. 1997, 15, 637–640. 10.1038/nbt0797-637. [DOI] [PubMed] [Google Scholar]
- Kim J.-K.; Firan M.; Radu C. G.; Kim C.-H.; Ghetie V.; Ward E. S. Mapping the Site on Human IgG for Binding of the MHC Class I-Related Receptor, FcRn. Eur. J. Immunol. 1999, 29, 2819–2825. . [DOI] [PubMed] [Google Scholar]
- Wang L.; Cheung N.-K. V.; Hoseini S. S.; Xu H.; Ponomarev V. Silencing Fc Domains in T Cell-Engaging Bispecific Antibodies Improves T-Cell Trafficking and Antitumor Potency. Cancer Immunol. Res. 2019, 7, 2013–2024. 10.1158/2326-6066.CIR-19-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vita R.; Mahajan S.; Overton J. A.; Dhanda S. K.; Martini S.; Cantrell J. R.; Wheeler D. K.; Sette A.; Peters B. The Immune Epitope Database (IEDB): 2018 Update. Nucleic Acids Res. 2019, 47, D339–D343. 10.1093/nar/gky1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan A. C.; Jacobson D.; Yatsenko K.; Sritharan D.; Weinreich T. M.; Shaw D. E. Atomic-Level Characterization of Protein–Protein Association. Proc. Natl. Acad. Sci. U. S. A. 2019, 116, 4244–4249. 10.1073/pnas.1815431116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.