Abstract
Morphological and molecular features were analyzed for a species of Phacus to better understand the phylogenetic relationships among them and establish the taxonomy. Phylogenetic analyses were based on nSSU rDNA and the research resulted in 55 new sequences. The study included species available in algal collections and those isolated directly from the environment in Poland and the Czech Republic. As a result, the obtained phylogenetic tree of Phacus includes 50 species, out of which 7 are represented on a tree for the first time (Phacus anacoelus, P. anomalus, P. curvicauda, P. elegans, P. lismorensis, P. minutus and P. stokesii) and many have been taxonomically verified. For all verified species, diagnostic descriptions were amended, the naming was reordered and epitypes were designated.
Keywords: Euglenida, environmental sampling, nSSU rDNA, Phacus, phylogeny, taxonomical revision
Abbreviations
- BI
Bayesian inference
- rbs
rapid bootstrap
- ML
maximum likelihood
- nt
nucleotide
- pp
posterior probability
Phacus was described in the 19th century (Dujardin 1841) and currently includes approximately 300 species names, of which 174 are taxonomically accepted (http://www.algaebase.org; Guiry and Guiry 2020). The number of species changes as taxonomic verifications (based on morphological and DNA sequence data) are ongoing (Kosmala et al. 2007, Karnkowska‐Ishikawa et al. 2010, Kim et al. 2010, Linton et al. 2010, Bennett and Triemer 2012, Kim and Shin 2014, Kim et al. 2015, Łukomska‐Kowalczyk et al. 2015). As a result, our understanding of the phylogenetic relationships among them is also constantly changing. The research cited above did not validate Phacus' separation into two subgenera, Chlorophacus (green forms) and Hyalophacus (colorless) as suggested by Pochmann (1942), or into four sections for the green forms (Proterophacus – cells flattened and leaf‐shaped; Pleuraspis – spherical or flat with ribbing instead of striae; Acanthochloris – slightly flattened with papillae on the surface; Kampylopter – round, triangular in cross‐section). First, it was shown that species from the Pleuraspis section are in fact representatives of Monomorphina (Marin et al. 2003). Later, when both Lepocinclis salina and Euglena limnophila, neither of which have flattened cells, were included in Phacus, the description of the genus underwent changes (Linton et al. 2010). Recently, Phacus horridus (=P. hispidulus from the Acanthochloris section) was reclassified as a member of Lepocinclis (Bennett and Triemer 2012, Łukomska‐Kowalczyk et al. 2020). As a result of all previous morphological‐molecular studies, descriptions were emended, epitypes were designated, many species were renamed, and nine new taxa were described (Kim et al. 2010, Łukomska‐Kowalczyk et al. 2015). Currently, Phacus is characterized by semi‐rigid to rigid cells, usually laterally compressed (with the exception of P. limnophilus and P. salinus), in most cases leaf‐shaped, sometimes twisted. They possess numerous chloroplasts of uniform shape (numerous, small, disc‐shaped, parietal, without pyrenoids). All species (with the exception of P. salinus) have dimorphic in size paramylon grains – in most cases the large grains are plate‐ or ring‐shaped. Colourless forms are known (e.g., Phacus ocellatus; Marin et al. 2003). Phacus is currently classified in the Phacaceae family, together with the representatives of the Lepocinclis and Discoplastis genera (Kim et al. 2010).
The most recently published phylogenetic trees of Phacus include 43 species (Kim et al. 2015, Łukomska‐Kowalczyk et al. 2015), some of which have been misidentified. We conducted this study to increase the number of species and strains representing Phacus in the phylogenetic trees and to perform comparative morphological and DNA sequence studies on new strains (=isolates). This will allow an estimation of morphological and genetic diversity and verification of morphological diagnostic features for particular taxa (well‐established clades) on a molecular phylogenetic tree; (b) the reconstruction of phylogenetic relationships and (c) taxonomic verification, amending diagnoses and designating epitypes for well‐distinguished taxa.
MATERIAL AND METHODS
Sampling and morphological study
During nine seasons (2011–2019), plankton samples were collected from 37 eutrophic water bodies located in Poland and one located in the Czech Republic using a plankton net with a mesh size of 10 µm (Fig. 1). Samples were screened in terms of their species diversity, and then a number of cells (7–300; see Table S1) exhibiting the same morphology were isolated from the sample using a micromanipulator (MM‐89 Narishige) with a micropipette installed on a Nikon Ni‐U microscope (Nikon, Tokyo, Japan). Morphological studies (descriptions and measurements) and documentation (photographs and video clips) of the isolated cells (isolates) of the Phacus morphotypes and those from six strains from algae collections (ACOI 1753, ACOI 1754, ACOI 1755, ACOI 1757, ACOI 3237, AICB 324) were executed with a NIKON Eclipse E‐600 microscope with differential interference contrast, equipped with the NIS Elements Br 3.1 software (Nikon) for image processing and recording. Photographs (and video clips) were taken using a NIKON DX‐1200 digital camera connected to the microscope. The NIS Elements Br measurement program was used for morphometric studies; three parameters were measured for the cells of each isolate (strain) – cell length, cell width, and tail length (which was defined as the hyaline projection); measurements were conducted from photographs of the isolates (strains). The data was analysed using the R v.3.2.0 software (R Development Core Team, 2008); means and standard deviations are given in Table 1. The isolates (i.e., morphologically identical cells) were transferred through multiple drops of sterile media (to clean the sample), and kept frozen at −80°C until DNA extraction.
Fig. 1.
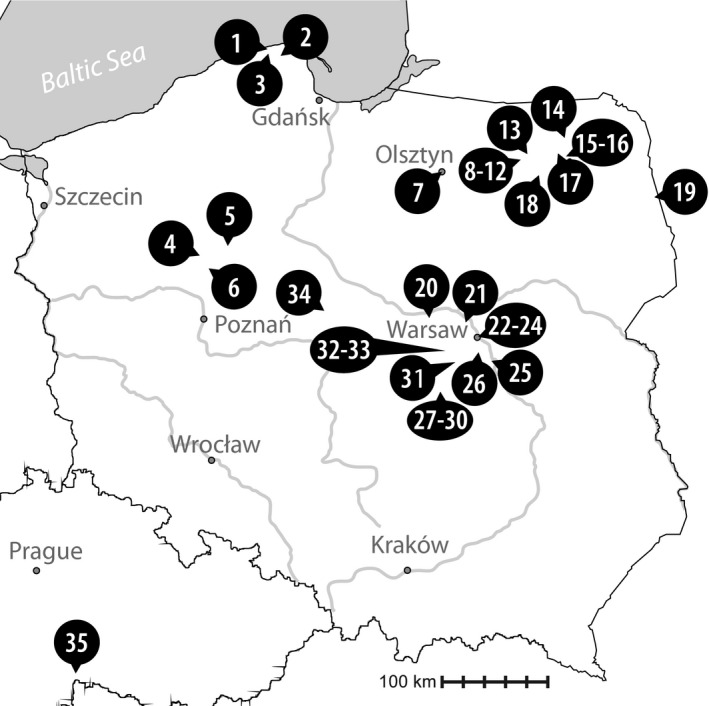
Map illustrating sampling locations in Poland and the Czech Republic. The names of lakes or towns in which the studied water bodies were located are marked with numbers: (1) Kopalino; (2) Strzebielinek; (3) Choczewo; (4) Chodzież; (5) Sokolec; (6) Budzyń 1 and 2; (7) Olsztyn; (8) Urwitałt 10; (9) Urwitałt 15; (10) Urwitałt 17; (11) Urwitałt 19; (12) Urwitałt 20; (13) Grabówek; (14) Doliwy; (15) Oracze; (16) Woszczele; (17) Jeziorowskie; (18) Pilchy; (19) Wojnowce; (20) Oracze; (21) Górki; (22) Łazienki; (23) Jelonki; (24) Żerań; (25) Baniocha; (26) Zgorzała; (27) Biała Rawska; (28) Regnów; (29) Cielądz; (30) Ossowice; (31) Nowa Bukówka; (32) Izdebno Nowe; (33) Izdebno Kościelne 1 and 2; (34) Mąkolno (35) Lásenický Stav.
Table 1.
Comparison of morphological traits among the study's isolates/strains of Phacus.
| Taxon | Isolate/strain | Number of measured cells |
Cell length (μm) Mean ± SD |
Cell width (μm)\Mean ± SD |
Tail length (μm) Mean ± SD |
Cell shape | Tail turning |
|---|---|---|---|---|---|---|---|
| P. acuminatus | ACOI 1753 | 107 | 31.2 ± 1.2 | 19.5 ± 1.8 | 2.4 ± 0.4 | Flat, leaf‐like, wide‐oval | Bent sideways |
| ACOI 1754 | 97 | 29.6 ± 0.9 | 23.3 ± 1.7 | 2.2 ± 0.3 | |||
| ACOI 3237 | 105 | 30.7 ± 1.2 | 22.8 ± 1.7 | 2.1 ± 0.4 | |||
| UTEX 1317 | 107 | 31.2 ± 1.5 | 18.5 ± 1.6 | 2.7 ± 0.6 | |||
| UW2269INo | 28 | 31.3 ± 1.2 | 22.6 ± 1.0 | 2.3 ± 0.4 | |||
| P. alatus | UW1898NBu | 43 | 42.8 ± 1.1 | 30.5 ± 1.4 | 6.5 ± 1.0 | Thick, wide‐oval, slightly flattened | Bent dorsally |
| UW2300Str | 15 | 42.5 ± 1.5 | 29.4 ± 0.9 | 7.8 ± 0.9 | |||
| P. anacoelus | UW1960Reg | 33 | 49.5 ± 2.4 | 34.9 ± 1.9 | 11.6 ± 1.7 | Thick, spherical, slightly flattened | Bent dorsally |
| UW2219IK1 | 30 | 47.4 ± 2.2 | 30.6 ± 2.5 | 10.9 ± 1.3 | |||
| P. ankylonoton | CCAC 0043 | 110 | 39.1 ± 3.1 | 24.6 ± 1.9 | 8.7 ± 1.7 | Thick, oval, triangular when cross‐sectioned | Straight |
| P. anomalus | UW1842LSt | 7 | 35.9 ± 1.2 | 32.4 ± 1.0 | 5.7 ± 1.2 | Thick, trapezoid‐shaped, slightly flattened | Bent dorsally |
| UW1936Wos | 2 | 41.1 ± 3.2 | 27.9 ± 0.5 | 7.8 ± 0.4 | |||
| UW1984Cie | 26 | 34.6 ± 1.7 | 26.3 ± 1.6 | ||||
| UW2193BRa | 42 | 42.9 ± 2.7 | 30.3 ± 1.7 | 6.7 ± 1.4 | |||
| P. applanatus | CCAC 2604 B | 92 | 39.5 ± 2.8 | 20.1 ± 2.4 | 6.8 ± 1.1 | Flat, elongated oval‐like | Straight |
| P. arnoldii | CCAC 2432 B | 10 | 57.8 ± 1.9 | 29.0 ± 1.9 | 12.8 ± 1.2 | Spherical, slightly twisted, triangular in cross section | Straight |
| UW1650Ur10 | 15 | 63.0 ± 2.5 | 38.0 ± 1.4 | 16.0 ± 2.0 | |||
| UW2313Pil | 30 | 60.4 ± 3.4 | 34.3 ± 1.3 | 12.8 ± 1.1 | |||
| P. caudatus | AICB 324 | 111 | 34.0 ± 1.1 | 20.5 ± 1.6 | 3.5 ± 0.6 | Flat, elongated oval‐like | Bent ventrally |
| ASW 08020 (CCAC 2415 B) | 108 | 39.3 ± 3.1 | 24.8 ± 2.2 | 8.4 ± 1.7 | |||
| CCAC 0034 | 109 | 34.6 ± 2.0 | 15.9 ± 1.7 | 5.5 ± 1.0 | |||
| UW2228INo | 11 | 35.0 ± 1.6 | 17.2 ± 0.9 | 4.3 ± 0.8 | |||
| UW2383Zer | 19 | 37.8 ± 1.6 | 20.8 ± 1.2 | 4.4 ± 0.6 | |||
| P. curvicauda | UW2163Wos | 2 | 28.5 ± 0.0 | 20.8 ± 0.5 | 2.7 ± 0.7 | Thick, wide‐oval | Bent dorsally |
| UW2262INo | 12 | 32.4 ± 1.8 | 21.3 ± 1.5 | 3.8 ± 0.7 | |||
| UW2461Mak | 34 | 31.2 ± 1.25 | 24.48 ± 1.26 | 3.3 ± 0.43 | |||
| UW2468Jel | 57 | 32.3 ± 1.1 | 25.1 ± 1.3 | 3.5 ± 0.5 | |||
| P. elegans | UW1837Ols | 1 | 142.3 | 39.0 | 51.2 | Flat, elongated oval‐like | Straight |
| UW2064Ur19 | 12 | 124.8 ± 4.9 | 47.4 ± 2.5 | 39.9 ± 3.6 | |||
| P. gigas | MSU | 1 | 123 | 71 | 28 | Flat, round | Bent sideways |
| UW1669Ora | 40 | 122.2 ± 2.5 | 76.7 ± 2.3 | 27.6 ± 2.0 | |||
| UW1823Ur19 | 16 | 125.2 ± 4.0 | 81.2 ± 2.0 | 24.7 ± 2.7 | |||
| P. hamatus | CCAC 2605 B | 1 | 52.3 | 28.9 | 10.3 | Flat, spoon‐shaped, convex | Bent ventrally |
| UW1900NBu | 23 | 63.8 ± 1.7 | 39.1 ± 1.7 | 12.7 ± 1.5 | |||
| UW1975Sok | 4 | 53.2 ± 1.0 | 33.2 ± 1.5 | 9.9 ± 0.9 | |||
| P. limnophilus | ACOI 1026 | 1 | 62 | 8.5 | Fusiform, not flattened | Straight | |
| UW1988Reg | 60 | 80.4 ± 2.6 | 9.4 ± 1.2 | ||||
| P. lismorensis | UW1665Ur10 | 2 | 124.5 ± 13.4 | 31.0 ± 1.4 | 45.5 ± 7.8 | Flat, long ovate, bent like a bow | Bent ventrally |
| UW1930Dol | 2 | 96.3 ± 4.8 | 26.4 ± 2.0 | 43.4 ± 1.3 | |||
| P. manginii | UW2082Zgo | 30 | 50.5 ± 1.9 | 28.4 ± 2.3 | 11.3 ± 1.5 | Flat, wide oval | Bent ventrally |
| UW2180Cie | 30 | 45.1 ± 2.0 | 27.3 ± 2.0 | 10.0 ± 1.2 | |||
| UW2225IK1 | 30 | 48.2 ± 0.0 | 25.6 ± 0.1 | 12.8 ± 0.1 | |||
| UW2264INo | 25 | 51.4 ± 2.1 | 29.1 ± 1.9 | 11.5 ± 1.1 | |||
| P. minutus | ACOI 1755 | 109 | 26.7 ± 1.2 | 19.9 ± 1.6 | 1.9 ± 0.4 | Flat, ovoid | Bent dorsally |
| P. orbicularis | UW2362Choc | 4 | 31.8 ± 2.3 | 23.2 ± 1.0 | 3.7 ± 0.2 | Flat, wide oval | Bent sideways |
| P. paraorbicularis | UW1977Bud2 | 28 | 76.6 ± 2.3 | 44.7 ± 2.3 | 16.7 ± 2.1 | Flat, wide oval | Bent ventrally |
| UW1980Oss | 3 | 77.7 ± 0.5 | 46.3 ± 1.1 | 14.9 ± 0.6 | |||
| P. pleuronectes | SAG 1261‐2b | 35 | 51.6 ± 4.8 | 25.5 ± 2.7 | 4.7 ± 0.9 | Flat, trapezoid‐like | Bent sideways |
| UW1981Oss | 11 | 34.5 ± 1.1 | 23.3 ± 1.1 | 5.9 ± 0.8 | |||
| UW2007Kro | 20 | 34.2 ± 0.9 | 21.2 ± 1.0 | 5.8 ± 0.8 | |||
| UW2202Ban | 29 | 32.4 ± 1.2 | 23.3 ± 1.2 | 4.5 ± 0.6 | |||
| UW2211Jel | 3 | 32.9 ± 0.4 | 23.4 ± 0.1 | 4.8 ± 0.0 | |||
| UW2230IK1 | 6 | 31.2 ± 0.7 | 22.9 ± 1.1 | 4.3 ± 0.4 | |||
| P. raciborskii | ACOI 1758 | 4** | 35.4 ± 1.9 | 10.5 ± 1.5 | 8.2 ± 1.5 | Flat, cylindrical, spirally twisted | Straight |
| UW1778Jez | 2 | 56.8 ± 4.0 | 13.3 ± 2.1 | 9.0 ± 0.7 | |||
| UW1815Ur20 | 8 | 56.6 ± 1.5 | 11.0 ± 1.7 | 11.1 ± 4.6 | |||
| UW2326Gra2 | 8 | 57.1 ± 2.3 | 15.1 ± 1.7 | 13.4 ± 1.8 | |||
| P. salinus | UW1914Bud | 10 | 50.9 ± 2.6 | 35.1 ± 2.6 | Egg‐like, not flattened | Lack | |
| UW1948Wos | 3 | 46.8 ± 4.6 | 34.0 ± 3.8 | ||||
| UW1970Chod | 39 | 47.0 ± 1.6 | 31.9 ± 2.1 | ||||
| UW1983Cie | 9 | 30.8 ± 1.2 | 21.2 ± 1.4 | ||||
| UW2392IK2 | 107 | 44.7 ± 3.9 | 32.7 ± 4.1 | ||||
| SAG 1244‐3 | 109 | 41.6 ± 1.8 | 26.7 ± 2.0 | ||||
| P. segretii | ACOI 1337 | 3 | 18.4 ± 0.6 | 12.9 ± 1.4 | Thick, wide‐oval | Lack | |
| P. stokesii | UW2399Ur19 | 11 | 41.3 ± 0.4 | 30.8 ± 0.4 | Thick, wide‐oval | Lack | |
| P. tenuis | ACOI 1757 | 111 | 32.4 ± 1.5 | 18.5 ± 1.5 | 4.4 ± 0.7 | Flat, oval | Bent ventrally |
*, Based on fig. 1c in Bennett and Triemer 2012; **, based on the photographs from the ACOI website: http://acoi.ci.uc.pt/spec_detail.php?cult_id=1442).
DNA isolation, amplification, and sequencing
Isolation of total DNA from samples (environmental samples and laboratory cultures of strains) was performed as described previously (Zakryś et al. 2002). An additional step of whole genome amplification was carried out in the case of environmental samples (according to Bennett and Triemer 2012). PCR amplification of the nSSU coding region, purification, and sequencing of the PCR products were performed by standard methods as previously described (Zakryś et al. 2002). To obtain some sequences, the nested PCR method was used as described previously (Łukomska‐Kowalczyk et al. 2020).
Sequence accession numbers, alignment, and sequence analyses
Fifty‐five new sequences (6 from strains and 49 from environmental isolates) used in the present study were submitted to GenBank with the following accession numbers: MN149548 – MN149602 and MN326770. Information about the accession numbers for the nSSU rDNA sequences reported here and those used for phylogenetic analyses (136 in total, of which 129 for Phacus) are shown in Table S1. Identical sequences from different strains/isolates were represented as single terminals in the phylogenetic analyses. The nSSU rDNA sequences were aligned using the MAFFT v.7.2 software (Katoh and Standley 2013) with E‐INS‐I strategy. Alignment was inspected by eye and corrected if necessary. Regions of doubtful homology between sites were removed from the alignments using TrimAl 1.2 with the option “automated1” (Capella‐Gutierrez et al. 2009). After trimming, 2,157 out of 4663 positions were left. Sequence diversity of nSSU rDNA was calculated using the Mega X software (Kumar et al. 2018) as pair‐wise distance based only on unambiguously aligned positions. The alignment and corresponding phylogenetic tree have been submitted to TreeBase (Study Accession URL: http://purl.org/phylo/treebase/phylows/study/TB2:S25743).
Phylogenetic analyses
The model of sequence evolution was selected using jModeltest 2.1.7 (Darriba et al. 2012) and the GTR+I+G model was chosen based on AIC, BIC or DT criteria (with ‐lnL = 48697.4; Lanave et al. 1984, Tavare 1986, Rodriguez et al. 1990) and was used to calculate maximum‐likelihood (ML) and Bayesian trees. The ML tree was inferred using RAxML 8.2.11 (Stamatakis 2014) using 1,000 rapid bootstrap inferences.
Bayesian inference (BI) with default priors was performed in MrBayes 3.2.6 software (Ronquist and Huelsenbeck 2003). A gamma correction with eight categories and proportion of invariable sites were used. Two independent runs with four Markov chains were performed. In each run, the chains lasted for 10,000,000 generations and trees were sampled every 1,000 generations. The first 25% of trees were discarded as burn‐in. Convergence among runs was assumed as the average standard deviation of split frequencies was below 0.01. The trees were visualized using FigTree v.1.4.2 (available at http://tree.bio.ed.ac.uk/software).
RESULTS
Phylogenetic analyses and morphological characteristics
Phylogenetic trees obtained by Bayesian (not shown herein) and maximum likelihood methods have a very similar topology, with 11 main clades and two branches with single sequences (Fig. 2, Fig. S1), although the relations between species are not always clearly defined.
Fig. 2.
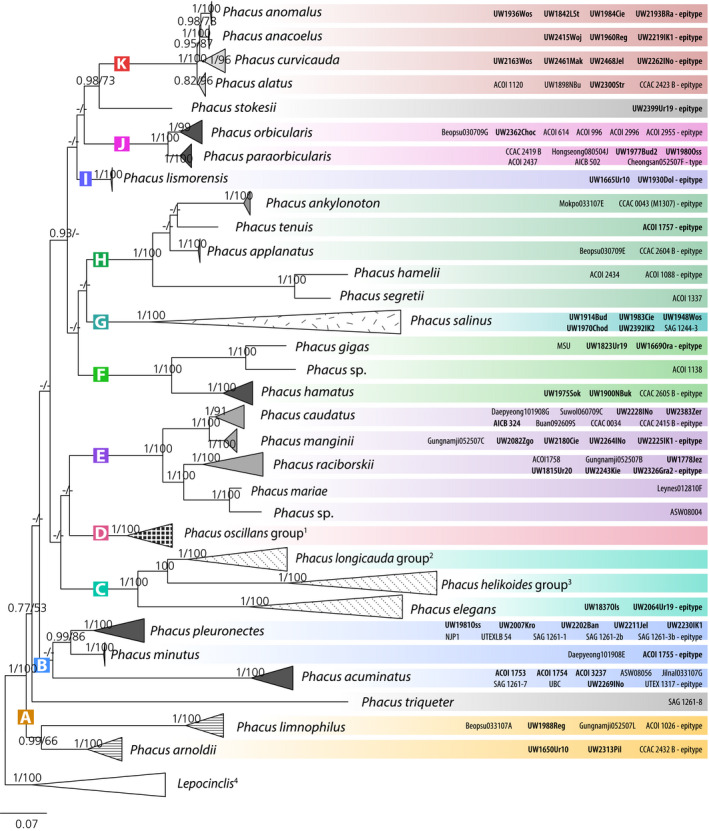
Consensus Maximum Likelihod tree based on 136 nSSU rDNA sequences (of which 129 represent Phacus). Isolates or strains with identical nSSU rDNA are represented by single tips. Sequences of the same species are represented by triangles of height proportional to the variability of sequences. Nodes are labeled with the Bayesian posterior probability (pp) values and rapid bootstrap (rbs) values. The pp values <0.75 and rbs values <50 and clades not present in the particular analysis are marked with a hyphen (‐). Scale bar represents number of substitutions per site. Sequences obtained in this study are indicated in bold type.
Groups of species:
1 P. brevisulcus Suwol060709A (type), P. oscillans ACOI 1339 (epitype), P. longisulcus Psurononuma100609J (type), P. smulkowskianus ACOI 1226 (epitype), UW2281Kop, P. granum AICB349, P. polytrophos CCAC2451B (epitype), P. minimus Buan092609I (type), P. claviformis Gungnamji052507F (type), P. hordeiformis Yongho092609A (type), P. inflexus ACOI 1336 (epitype), P. parvulus ACOI 1093 (epitype), P. viridioryza Sondang060709L (type), P. pusillus UTEX 1282 (epitype), P. skujae ACOI 1312 (epitype).
2 P. ranula Jigok090112, P. convexus UW1821Ora (epitype), P. tortus UW1845Jel (epitype), P. cordatus UW1808Ur17 (epitype), P. longicauda UW1896Laz (epitype), P. sp. Burni081809, P. circumflexus UW1844Jel (epitype).
3 P. crassus UW1566UR15 (type), P. cristatus UW1929Dol (type), P. helikoides UW1658Ur20 (epitype).
4 L. steinii Cheongsan052507K5, L. hispidula MSU, L. fusiformis ACOI 1025, L. spirogyroides ACOI 1027, L. acus ASW 08037, L. tripteris UWOB, L. fusca Saeraewool102007P. [Color figure can be viewed at wileyonlinelibrary.com]
The earliest branching clade (labelled as A), sister to all other Phacus sequences, is not strongly supported, but consists of two maximally supported subclades. It includes sequences of P. arnoldii and P. limnophilus originated from both laboratory cultures and environmental isolates. The genetic variability of nSSU rDNA does not exceed 4% for either species (Table S2). Representatives of both species morphologically resemble members of genus Lepocinclis – the three‐ridged cell shape of P. arnoldii makes it similar to L. tripteris, while the unflattened cells of P. limnophilus are typical for the majority of the representatives of the Lepocinclis genus (for details see Discussion). The clade corresponds to clade G (Kim and Shin 2014) and I6 (Kim et al. 2015).
The next clade (B), complementing clade F in Kim and Shin (2014) and clade I5 in Kim et al. (2015), includes three strongly supported subclades corresponding to the species: Phacus pleuronectes, P. minutus, and P. acuminatus. The cells of all three species are leaf‐like (flat), widely oval with a short, pointy tail. Sequences of P. pleuronectes are located in two groups: one with sequences of only environmental isolates and the second with all six strain sequences (out of which three are identical) and only one environmental sequence, sister to them. All isolates are characterized by a trapezoidal shape of cells in comparison to the ovoid cells of the cultivated strains. The genetic variability in the species does not exceed 3% and is lower than 0.5% among the strain sequences (Table S2). Phacus acuminatus and P. minutus are morphologically almost undistinguishable (see Taxonomic revisions), but they do not form a common clade and the genetic variability between them varies between 8.1% and 9.6% and is lower than 3% and 0.1% within the species respectively (Table S2).
Strongly supported clade C (1/100), that corresponds to clade D1 in Kim and Shin (2014) and to one subclade of the I3 clade in Kim et al. (2015), consists of sequences of ten species characterized by large cells (>75 µm) with long tails. In the clade there are three groups of sequences: (1) representatives of Phacus ranula, P. convexus, P. tortus, P. longicauda, P. cordatus, P. circumflexus with one sequence of the strain of unknown morphology (Phacus sp. Burni081809), (2) representatives of P. cristatus, P. crassus, P. helikoides, and (3) as sister clade to all other sequences, the subclade including two sequences of Polish environmental isolates of P. elegans – the species that is represented on the phylogenetic tree for the first time.
Maximally supported (1/100) clade D consists of sequences of 14 species closely related to Phacus oscillans, which are characterized by a small cell size (<40 µm long) and underwent taxonomic verification earlier (Karnkowska‐Ishikawa et al. 2010, Kim and Shin 2014). The clade corresponds to clade A in Kim and Shin (2014) and to clade I1 in Kim et al. (2015). The new environmental sequence of P. smulkowskianus UW2281Kop is located in this clade and it is identical with the sequence of the strain ACOI 1226 (epitype).
Clade E (1/100) includes four maximally supported subclades that correspond to four species: Phacus raciborskii and P. mariae branching together with one sequence of a strain of unknown morphology (Phacus sp. ASW 08004) and a pair of sister species: P. caudatus and P. manginii. Species located in this clade are easily distinguishable on the basis of morphology and their common diagnostic feature is flat cells with a conspicuous dorsal crest, which elongates into a sharp hyaline tail. The P. manginii subclade consists exclusively of sequences originated from environmental isolates (from Poland and South Korea), and the other subclades include sequences of both strains and isolates. Genetic diversity in P. caudatus and P. manginii does not exceed 1.8% and in the P. raciborskii clade there are five sequences with their genetic diversity below 0.3% and one (ACOI 1758) that is 4.5–5.0% divergent from all other (Table S2). The E clade corresponds to clade E in Kim and Shin 2014 and to clade I4 in Kim et al. 2015.
Strongly supported clades F, G, and H form a common clade in the maximum likelihood analyses only and in the bayesian analyses, relationships between them are not defined.
Clade F, absent in Kim and Shin (2014) and corresponding to a fragment of the I3 clade in Kim et al. (2015), consists of three identical sequences of Phacus gigas (one of the strain MSU and two Polish isolates), one sequence of a strain of unknown morphology (Phacus sp. ACOI 1138) and a subclade of three P. hamatus sequences. The P. gigas and P. hamatus individuals are flat, wide‐oval in general overview, and terminate with a sharp hyaline tail.
Clade G, not present in either Kim and Shin (2014) or in Kim et al. (2015), groups sequences of Phacus salinus: five from environmental isolates and one from a cultivated strain. Despite all of them being morphologically indistinguishable (spherical, egg‐like cells with both ends widely rounded), their genetic variability is very diverse (up to 11.3%) and the sequences vary considerably in length. The longest fragment of 18S rDNA belongs to the isolate UW1948Wos (3560 bp), and the sequences of other P. salinus isolates and strains are also fairly long: 2,124 bp for strain SAG 1244‐3, 2,151 bp for UW1970Chod, 2,778 bp for UW2392IK2. Usually the length of this fragment varies between 1,900 and 2,050 bp in most other Phacus species, and exceeds 2,100 bp only for P. helikoides, P. crassus, P. cristatus, and P. elegans.
Clade H, corresponding to the B clade in Kim and Shin (2014) and to clade I2 in Kim et al. (2015), consists of sequences of five species: Phacus ankylonoton, P. tenuis, P. applanatus, P. hamelii, and P. segretii, which can be easily distinguished based on morphology and molecular differences (see Figs. 3, q and r, 4f, and also fig. 1, a and b in Kosmala et al. 2007).
Fig. 3.
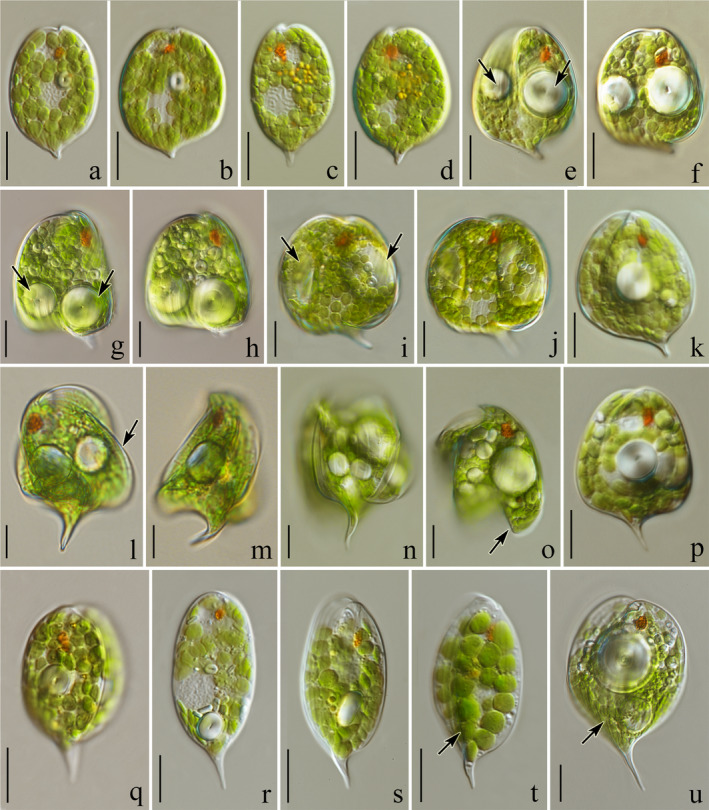
Light microscope photographs showing an overview of living cells of the studied Phacus strains (=isolates): (a, b) oval, leaf‐like flat cells of Phacus acuminatus (strain UTEX 1317) terminated with a short, sharp, wedge‐shaped tail; (c, d) cells of P. minutus (strain ACOI 1755) ending with a tail that is bent toward the dorsal side; (e, f) P. curvicauda (isolate UW2262INo), cells with centrally located, large, spherical paramylon grains (arrows); (g, h) trapezoid‐shaped cells of P. anomalus (isolate UW1984Cie) with two large, spherical paramylon grains located in the widest and thickest bottom part (arrows); (i, j) two large, parietal, ring‐like paramylon grains visible in the cell of P. alatus (strain CCAC 2423 B); (k) P. orbicularis (isolate UW2362 Choc); (l–o) spherical cells of P. anacoelus (isolate UW2219IK1) with visible crests (arrows) and large, spherical paramylon grains located in the center; (p) trapezoid‐shaped cell of P. pleuronectes (isolate UW2211Jel); (q) rotund (triangular when cross‐sectioned) cell of P. ankylonoton (strain CCAC 0043) terminated with a straight tail; (r) flat, elongated oval‐like cells of P. applanatus (strain CCAC 2604 B) ending with a straight tail; (s, t) cells of. P. caudatus (strain CCAC 0034) with a dorsal crest which elongates into a sharp hyaline tail slightly bent toward the ventral side; (u) wide‐oval cell of P. manginii (isolate UW2219IK1) with a low crest which elongates into a sharp hyaline tail bent slightly sideways. Scale bars 10 µm.
Fig. 4.

Light microscope photographs showing an overview of living cells of the studied Phacus strains (=isolates): (a) elongated oval‐like cell of Phacus elegans (isolate UW2064Ur19) elongated with a long, thorn‐like, straight tail; (b, c) spindle‐shaped cells of P. limnophilus with two large, rod‐like paramylon grains visible, (b) strain ACOI 1026, (c) isolate UW1988Reg; (d) wide‐oval cell of P. stokesii (isolate UW2399Ur19) rounded at both ends; (e) spherical cells of P. salinus (strain SAG 1244‐3); (f) P. tenuis (strain ACOI 1757), oval cell with a low crest elongated into a tail bent slightly toward the ventral side; (g–i) almost spherical, slightly twisted cells of P. arnoldii with three ridges (g, h) strain CCAC 2432B, (i) isolate UW2313Pil; (j) large, flat, almost round cell of P. gigas (isolate UW1669Ora) terminated with a sharp tail bent sideways; (k) spoon‐shaped (convex) cell of P. hamatus (strain CCAC 2605 B [=ASW 08032]) ending with sharp hyaline tail bent visibly toward the ventral side; (l–m) cylindrical, flat, slightly U‐bent and spirally twisted cells of P. raciborskii (isolate UW2326Gra2); (n) cell of P. lismorensis (isolate UW1930Dol) bent like a bow, terminated with a long, sharp, pointy tail. Scale bars 10 µm.
Two sequences of environmental isolates of Phacus lismorensis form a strongly supported clade I, however, that does not define their position. The species has a very characteristic morphology (cells bent like a bow; Fig. 4n) and its sequences were not hitherto present on any phylogenetic tree.
Clade J consists of two subclades corresponding to two sister species: Phacus orbicularis and P. paraorbicularis, which are characterized by wide, ovoid cells ending with a sharp curved tail; both had been previously taxonomically verified (Kosmala et al. 2007, Kim and Shin 2014). Two sequences of Polish isolates of P. paraorbicularis and one of P. orbicularis are included in the subclades. Variability in the whole clade does not exceed 1.5% (Table S2). The clade corresponds to clade C in Kim and Shin (2014) and to one subclade of the I3 clade in Kim et al. (2015).
Maximally supported clade K, corresponding to clade D2 in Kim and Shin (2014) and to one subclade of the I3 clade in Kim et al. (2015), includes sequences of four species. The genetic diversity in that clade is lower than 5% and in most cases (in spite of the variability of the sequence of Phacus curvicauda UW2163Wos) does not exceed 0.9% (Table S2). All representatives of the clade are characterized by thick cells with well‐developed ventral and dorsal sides, ending with a short tail curved toward the dorsal side. However, the four species are easily recognizable based on such features as the shape of the large paramylon grains and their location in the cell. Sister clades with the sequences of P. anacoelus and P. anomalus are maximally supported and the other two subclades have lower support – P. curvicauda: 1/96 and P. alatus: 0.82/96. All sequences in clade K (aside from the three of P. alatus) stem from environmental isolates from Poland.
The single sequence of Phacus stokesii has a sister position to clade K, although it is not strongly supported (0.98/73). It represents one out of two (along with P. segretii) tailless species (Fig. 4d).
Taxonomic revisions
Phylogenetic and morphological analyses and a review of the literature enabled the taxonomic identification of almost all clades present on the phylogenetic tree. For new or misidentified clades, as well as those represented by single sequences a taxonomic revision had been conducted. Its basis was to define a species as a group of singular morphotypes, which create a well‐supported clade on the phylogenetic tree. For such clades (morphologically well‐distinguished taxa) diagnostic descriptions were emended, epitypes designated and the nomenclature has been reordered.
Due to the lack of morphological data, the taxonomic affiliation of 5 strains (Jigok090112 as Phacus ranula on the phylogeny tree; Leynes012810F as P. mariae; Burni081809, ASW 08004 and ACOI1138 as Phacus sp.) was not verified.
Given that the isolated cells were destroyed for DNA extraction, the photographs are designated as epitypes (see International Code of Nomenclature for algae, fungi, and plants [Shenzhen Code]; chapter II, section 2, article 9.9; Turland et al. 2018).
The genus Phacus Dujardin 1841 . Emend. Linton and Karnkowska 2010 in Linton et al. 2010, p. 609.
Phacus acuminatus A.Stokes 1885a: 183, fig. 1 (Fig. 3a and b)
Emended diagnosis: Cells oval, wide‐oval or almost spherical (20–40 × 13–27 µm), leaf–like and flat (with very low crest); cells terminate with a short, sharp, wedge‐shaped, straight or bent sideways tail (on average 2–3 µm long).
Holotype: Stokes 1885a, fig. 1
Epitype: Figure 3a designated herein that supports the holotype (Stokes 1885a, fig. 1)
Representative DNA sequence: GenBank AF286209
Representative strain: UTEX 1317
Type locality: shallow ponds in Western New York, USA
Heterotypic synonyms: Phacus acuticauda Y.V. Roll 1925: 140, 148, pl. 5, fig. 17; P. acuminatus subspec. acuticauda (Y.V.Roll) Pochmann 1942: 143, fig. 32n; P. acuminatus var. acuticauda (Y.V.Roll) Huber‐Pestalozzi 1955: 194, fig. 230; P. acuminatus subspec. americana Pochmann 1942: 141, fig. 32 a‐c; P. acuminatus var. triangulatus (Y.V.Roll) Svirenko 1938: 56, fig. 60; P. acuminatus subspec. discifera Pochmann 1942: 143, figs. 32e and f; P. acuminatus var. discifera (Pochmann) Huber‐Pestalozzi 1955: 193, fig. 225; P. acuminatus subspec. indica Pochmann 1942: 144, fig. 32p; P. acuminatus var. indica (Pochmann) Huber‐Pestalozzi 1955: 194, fig. 229; P. acuminatus var. iowensis Allorge & Jahn 1943: 235, figs. 31, 32; P. acuminatus subspec. javana Pochmann 1942: 141, fig. 32d; P. acuminatus var. javana (Pochmann) Huber‐Pestalozzi 1955: 193, fig. 226; P. acuminatus subspec. variabilis (Lemmermann) Pochmann 1942: 143, figs. 32g, h; P. brachykentron Pochmann 1942: 145, fig. 33.
Comments: Following the nomenclatural priority rule, this morphological form (cells small, almost spherical, flat, and terminate with a short, sharp, straight or bent sideways, and wedge‐shaped tail) has been assigned the name Phacus acuminatus. The individual seen in Stokes' drawing (1885a, fig. 1) corresponds with the aforementioned characteristic, similarly to the cells from the UTEX 1317 strain (chosen as the representative strain), from which the epitype originates. However, due to the high morphological similarity to P. minutus, only molecular identification allows the differentiation of the two species. In this situation, designation of epitypes for both species seems justified (see Comments for P. minutus). The taxa that constitute P. acuminatus synonyms are those whose cells have a morphology similar to P. acuminatus and without proper diagnostic traits in their original descriptions.
Phacus alatus G.A.Klebs 1883: 312 (Fig. 3 i and j)
Emended diagnosis: Cells wide‐oval, almost round in general overview (35–46 × 27–34 µm); thick (slightly dorsally flattened); terminate with a short, sharp tail bent dorsally (on average 6–8 µm long). A wide furrow divides the cell in halves, which are slightly twisted longitudinally; two large, ring‐ or shield–like paramylon grains located parietally.
Basionym: Phacus triqueter in Stein 1878, Der Organismus Infusionsthiere 3, pl. 19 figs. 55‐57
Lectotype: designated herein, Stein 1878, pl. 19, fig. 56
Epitype: Figure 3i designated herein that supports the lectotype (Stein 1878, pl. 19, fig. 56)
Representative DNA sequence: GenBank AJ532474
Representative strain: CCAC 2423B (=ASW 08027)
Type locality: Germany
Heterotypic synonyms: Phacus alatus var. maximus Hübner 1886: 6, fig. 8; P. alatus var. lemmermanii Svirenko 1915a: 117, pl. 3 figs. 6, 7; P. alatus var. latviensis Skvortsov 1928: 112, pl. 2, fig. 19; P. lemmermannii (Svirenko) Skvortsov 1928: 114, pl. 2, figs. 30, 31; P. alatus var. incrassata Deflandre 1928: 215, figs. 14–17; P. alatus var. maior Drezepolski 1922: 4; P. alata var. indica Skvortsov 1937: 73, pl. 9, figs. 17, 18; P. angulatus Pochmann 1942: 171, figs. 70a–c; P. macrostigma Pochmann 1942: 170, fig. 68; P. moraviensis Pochmann 1942: 169, figs. 65, 66; P. platyaulax Pochmann 1942: 165, figs. 61–64.
Comments: The P. alatus individual seen in one of Stein's original drawings (fig. 56) – a thick cell (only slightly flattened, not leaf‐like flat) with a deep furrow and parietal paramylon grains – resembles the Polish representatives of P. alatus. Nonetheless, both this cell and others shown in the remaining drawings display a high similarity to P. curvicauda and P. anomalus due to the presence of a wide furrow dividing the cells, which are slightly twisted longitudinally, in halves. The morphological research presented herein demonstrates that the large paramylon grain location and shape are good diagnostic features for discriminating those three species (for more details see Discussion). The epitype designation for all three species will allow their proper identification. The taxa that constitute synonyms of P. alatus are those whose slightly flattened cells posses a furrow and two large, ring‐ or shield–like paramylon grains located parietally.
Phacus anacoelus A.Stokes 1885b: 19, fig. 2 (Fig. 3 l–o)
Emended diagnosis: Cells almost spherical (42–57 × 25–40 µm), slightly flattened, with the ventral and dorsal sides clearly visible; at the back cells terminate with a sharp, hyaline tail (10–12 µm long) bent toward the dorsal side; two high crests run along each side (four crests in sum), and as the cells are spirally twisted, when mobile it seems that the crests are evenly distributed; two large, spherical paramylon grains are located in the center of the cell.
Holotype: Stokes 1885b, fig. 2
Epitype: Figure 3l designated herein that supports the holotype (Stokes 1885b, fig. 2)
Representative DNA sequence: GenBank MN149556
Type locality: shallow ponds in Western New York, USA
Representative locality: Freshwater, Izdebno Kościelne village, field pond (52°08′21.0″ N, 20°32′03.2″ E)
Heterotypic synonym: Phacus anacoelus var. asiatica Skvortsov 1958: 164, pl. 3, fig. 27.
Comments: The cells of Polish populations are almost spherical with spiral ribs, what is consistent with the original description. However, the individual seen in Stokes's drawing (1885b, fig. 2) also highly resembles the species described later (P. asymmetricus, P. quinquemarginatus) (for more details see Discussion). Due to the aforementioned, emending the diagnostic description and designating an epitype will allow its proper identification.
Phacus ankylonoton Pochmann 1942: 148, fig. 37a–e (Fig. 3q ).
Emended diagnosis: Cells oval (30–50 × 20–30 µm), rotund (triangular when cross‐sectioned), terminate with a straight or slightly ventrally bent, sharp hyaline tail (on average 6‐9 µm long).
Lectotype: designated herein, Pochmann 1942, fig. 37d
Epitype: Figure 3q designated herein that supports the lectotype (Pochmann 1942, fig. 37d)
Representative DNA sequence: GenBank KF744064
Representative strain: CCAC 0043
Type locality: pond in small town Wolice, Eastern Poland (now Ukraine)
Comments: Due to its size and the general cell shape, P. ankylonoton is similar to P. caudatus, the only difference being the width of the oval cell, which in cross‐section is triangular. This particular trait is represented only in two of Pochmann's drawings (fig. 37d and e), which are in fact an original drawing by Dreżepolski (Dreżepolski 1925, fig. 112 – two cells). It is Dreżepolski's drawing that Pochmann refers to when describing the new taxon – one of them has been designated herein as the lectotype (fig. 37d). The remaining individuals in Pochmann's drawings (37a and b) resemble more P. caudatus due to the elongated oval‐like cell shape. The designation of an epitype will allow the proper identification of P. ankylonoton.
Phacus anomalus Fritsch & Rich 1929: 73, figs. 24h–n (Fig. 3 g and h)
Emended diagnosis: Cells wide‐egg or trapezoid‐shaped (30–50 × 20–35 µm) in general overview, thick, with a furrow running down the entire length of the cell and dividing it into halves, which are slightly twisted longitudinally; two large, spherical paramylon grains are located in the widest and thickest bottom part of the cell, which elongates into a sharp tail (on average 6–8 µm long) bent toward the dorsal side.
Lectotype: designated herein, Fritsch and Rich 1929, fig. 24h
Epitype: Figure 3g designated herein that supports the lectotype (Fritsch and Rich 1929, fig. 24h)
Representative DNA sequence: GenBank MN149560
Type locality: Griqualand West – the interior of South Africa
Representative locality: Freshwater, Cielądz village (51°42′50.9" N, 20°20′44.7" E)
Heterotypic synonyms: Phacus snitkovii Y.V. Roll 1938: 138, fig. 8; Phacus anomalus var. pullus‐gallinae Nygaard 1949: 168, fig. 102; P. drezepolskii Stawiński 1969: 39 and 50, fig. 78; P. curvicauda f. anomalus (F.E.Fritsch & M.F.Rich) Safonova in Popova and Safonova 1976: 58, pl. 12, figs. 1–22.
Comments: In most of the drawings by Fritsch and Rich (1929) the large, spherical paramylon grains are positioned centrally in the cell, which causes P. anomalus to be practically indistinguishable from P. curvicauda (both have thick cells with a furrow and the tail bent toward the dorsal side). Meanwhile, the study presented herein proved that in P. anomalus the large paramylon grains are located in the widest and thickest, bottom part of the cell. The lectotype designated herein – the cell visible in the drawing 24 h (Fritsch and Rich 1929) has the most antapical paramylon grains (located the closest to the posterior of the cell). Taxa that constitute P. anomalus synonyms are those whose cell morphology corresponds to the emended diagnostic description (for more details see Discussion).
Phacus applanatus Pochmann 1942: 152, fig. 42 (Fig. 3r )
Emended diagnosis: Cells elongated oval–like (40 × 20 µm on average), flat, terminate with a rather long, straight tail (6–7 µm on average).
Holotype: Pochmann 1942, fig. 42
Epitype: Figure 3r designated herein that supports the holotype (Pochmann 1942, fig. 42)
Representative DNA sequence: GenBank EU624031
Representative strain: CCAC 2604 B
Type locality: Sandberg pond near Teplitz‐Schönau, Germany
Comments: The morphological study of the CCAC 2604B strain (isolated in the Netherlands, channel in Leiden) shows that its representatives have cells terminated with a tail that is twice as long (6–7 µm) in comparison with the individual in the drawing by Pochmann (1942, fig. 42).
Phacus arnoldii Svirenko 1915a: 120 and 131, pl. 3, fig. 1 (Fig. 4 g‐i)
Emended diagnosis: The shape of the organism is particularly peculiar, due to the three wings (ridges) of the almost spherical and slightly twisted cells (46–95 × 25–69 µm); cells end with a hyaline tail (12–23 µm); in cross section the cells appear triangular with concave sides (similarly to L. tripteris); periplast spirally striated with numerous, perpendicular struts between longitudinal periplast strips; the longitudinal periplast strips are wide and sparsely arranged.
Holotype: Svirenko 1915a, pl. 3, fig. 1
Epitype: Figure 4g designated herein that supports the holotype (Svirenko 1915a, pl. 3, fig. 1).
Representative DNA sequence: GenBank GQ422793
Representative strain: CCAC 2432B (=ASW 08064)
Type locality: Kharkiv district, Zmiewsk, Krugloje Lake, Russia (currently Ukraine)
Heterotypic synonyms: Phacus warszewiczii Drezepolski 1925: 235, fig. 125; P. arnoldii var. ovatus T.G.Popova 1947: 57, pl. 2, fig. 12.
Comments: In the original description, “round, flat cells with a high, S‐shaped crest” and “wide, sparsely arranged periplast strips” are mentioned – thanks to these features the Polish strains could be identified. As our research has shown, cells appear triangular with concave sides in cross section that gives the impression of three wings (ridges) and is a diagnostic trait of P. arnoldii. Meantime, in Svirenko's drawing (1915a, pl. 3, fig. 1) only a high crest is visible that does not looks like one of the three equally sized wings. Furthermore, we have shown that a more or less round shape of the cells has no diagnostic meaning, which is why P. arnoldii var. ovatus has been included as a synonym of P. arnoldii.
Phacus brevisulcus Kim & Shin 2014: 955, fig. 2B
Type: (see Kim and Shin 2014), permanently preserved material from the strain Suwol060709A deposited at the Chungnam National University, as number CNU 025427. Figure 2B in Kim and Shin 2014 shows the type.
Representative DNA sequence: GenBank KF744067
Phacus caudatus Hübner 1886: 5, fig. 5 (Fig. 3 s and t)
Emended diagnosis: Cells elongated oval–like (31–47 × 15–30 µm), slightly asymmetrical, flat, with a high dorsal crest which elongates into a sharp hyaline tail (on average 4–8 µm), slightly bent toward the ventral side; periplast longitudinally striated.
Holotype: Hübner 1886, fig. 5
Epitype: Figure 3t designated herein that supports the holotype (Hübner 1886, fig. 5)
Representative DNA sequence: GenBank AJ532482
Representative strain: CCAC 2415B (=ASW 08020)
Type locality: Germany, Stralsund
Heterotypic synonyms: Phacus caudatus var. undulata Skvortsov 1922: 191, fig. 7; P. caudatus var. lata Allorge and Lefèvre 1925: 126, figs. 18–21; P. caudatus var. minor Drezepolski 1925: 230, fig. 107; P. caudatus var. ovalis Drezepolski 1925: 231, fig. 111; P. ovalis Skvortsov 1958: 165, pl. 3, fig. 33; P. caudatus var. volicensis Drezepolski 1925: 230, fig. 105.
Comments: Phacus caudatus is very similar to P. manginii and P. ankylonoton due to the presence of a crest that elongates into the hyaline tail. Meanwhile, both the diagnostic description as well as Hübner's drawing of P. caudatus (Hübner 1886, fig. 5) does not contain any diagnostic features that would allow the distinguishment of P. caudatus from the two species described below (for more details see Discussion). Due to the aforementioned, the designation of an epitype seems justified. The taxa that constitute P. caudatus synonyms are those whose cell morphology corresponds to the emended diagnosis.
Phacus circumflexus Pochmann 1942: 206, fig. 119a–f
Epitype: Łukomska‐Kowalczyk et al. 2015, fig. 3k
Representative DNA sequence: GenBank KP944083
Phacus claviformis Kim & Shin 2014: 955, fig. 2E
Type: (see Kim and Shin 2014), permanently preserved material from the strain Gungnamji052507F deposited at the Chungnam National University, as number CNU 025429. Figure 2E in Kim and Shin 2014 shows the type
Representative DNA sequence: GenBank KF744071
Phacus convexus (Pochmann) Zakryś & M.Łukomska, nom. nov.
Nomen novum: Phacus convexus [≡Phacus longicauda (Ehrenb.) subs P. rotundus Pochmann, Pochmann 1942, Arch. Protistenk. 95: 201, figs. 111a–e], non Phacus rotundus Brabez 1941, Beihefte zum Botanischen Centralblatt 61: 220, fig. 13b.
Lectotype: Stein 1878, pl. 20, fig. 2 (Łukomska‐Kowalczyk et al. 2015)
Epitype: Łukomska‐Kowalczyk et al. 2015, fig. 3g
Representative DNA sequence: GenBank KP944090 (Łukomska‐Kowalczyk et al. 2015)
Representative locality: Freshwater, pond in Oracze village (53°52′36.6" N, 22°20′41.2" E; Łukomska‐Kowalczyk et al. 2015)
Comments: The nomenclatural combination P. rotundus (Pochmann) Zakryś & M.Łukomska proposed by Łukomska‐Kowalczyk et al. (2015) for P. longicauda (Ehrenberg) subs. rotundus Pochmann (1942) is invalid, as a previously described taxon of the same name (though of a different morphology from P. longicauda subs. rotundus) exists (P. rotundus Brabez 1941). We propose P. convexus nomen novum for the morphological form known previously as P. longicauda subs. rotundus. The latin name convexus refers to the spoon‐shaped (convex) cells.
The proposed lectotype, Stein's drawing (1878 (pl. 20, fig. 2), was chosen not only because it is one of the few Pochmann (1942, p. 201) referred to when describing the subspecies rotundus, but also mainly because Stein (1878) was the first who noticed and documented the occurrence of various morphological forms of P. longicauda (pl. 20, figs. 1–3 in Stein 1878), including the form “rotunda” (pl. 20, fig. 2 in Stein 1878)” (Łukomska‐Kowalczyk et al. 2015). However, as we have no drawing, nor was the convex cell shape ever mentioned in Stein's diagnostic description for the “rotunda” form, we believe an epitype designation is justified.
Phacus cordatus (Pochmann) Zakryś & M.Łukomska 2015: 1151, fig. 3a–c.
Epitype: Łukomska‐Kowalczyk et al. 2015, fig. 3b
Representative DNA sequence: GenBank KP944102
Phacus crassus Zakryś & M.Łukomska 2015: 1153, fig. 3o and p
Type: Łukomska‐Kowalczyk et al. 2015, fig. 3p
Representative DNA sequence: GenBank KP944108
Phacus cristatus Zakryś & M.Łukomska 2015: 1153, fig. 3q
Type: Łukomska‐Kowalczyk et al. 2015, fig. 3q
Representative DNA sequence: GenBank KP944109
Phacus curvicauda Svirenko 1915a: 117 and 130, pl. 3 figs. 13–16 (Fig. 3 e and f)
Emended diagnosis: Cells wide‐oval or wide egg‐shaped (22–37 × 16–27 µm), thick, divided into two longitudinally twisted parts by a furrow; terminate with a sharp, short hyaline tail bent toward the dorsal side (on average 2.5–5 µm); two large, spherical paramylon grains positioned centrally in the cell.
Lectotype: designated herein, Svirenko 1915a, fig. 13.
Epitype: Figure 3e designated herein that supports the lectotype (Svirenko 1915a, fig. 13)
Representative DNA sequence: GenBank MN149567
Type locality: wetlands and small lakes near Kharkov, Russia (now Ukraine)
Representative locality: Freshwater, Izdebno Nowe village, pond (52°08′01.3″ N 20°32′52.9″ E)
Heterotypic synonym: Phacus curvicauda f. robusta Allorge & Lefèvre 1925: 127 figs. 50 and 51.
Comments: In Svirenko's drawings, the large paramylon grains are located centrally in the cells, which allowed us to identify the Polish strains of Phacus curvicauda. However, in cell shape, P. curvicauda is very similar to P. anomalus and P. alatus (they all have thick cells with a long furrow). The best diagnostic trait for differentiating the three species is cell shape, as well as the location and shape of the large paramylon grains (for more details see Discussion and Comments for P. anomalus and P. alatus). The drawing indicated as the lectotype (Svirenko 1915a, fig. 13) shows a long furrow and round paramylon grains – in the remaining images (figs. 14, 15, 16), the individuals either have short furrows, or ring‐like paramylon grains. The designation of an epitype will allow the proper identification of P. curvicauda.
Phacus elegans Pochmann 1942: 199, fig. 107 (Fig. 4a )
Emended diagnosis: Cells elongated oval‐like (112–147 × 38–51 µm), with a low crest which elongates into a long, thorn‐like, straight hyaline tail (on average 40–50 µm); the front of the cell visibly asymmetrical; cell straightened out both when immobile and swimming.
Holotype: Pochmann 1942, fig. 107.
Epitype: Figure 4a designated herein that supports the holotype (Pochmann 1942, fig. 107)
Representative DNA sequence: GenBank MN149571
Type locality: Southern Germany, peatbog Rauhen Wiese near Böhmenkirch (Schwarzwald)
Representative locality: Freshwater, Urwitałt, pond 19 (53°50′38.0" N, 21°38′06.5" E)
Comments: The Polish populations of this species have been identified based on the shape of the cells (elongated oval‐like with a visibly asymmetrical anterior end, ending with a long hyaline tail). According to Pochmann, Phacus elegans can be distinguished from P. lismorensis based on “a more elongated cell form and a shorter tail.” Furthermore, we have established that the presence of a crest that elongates into a long, straight hyaline tail is a diagnostic morphological feature; in P. lismorensis representatives the tail is ventrally bent (for more details see Discussion).
Phacus gigas A.M.Cunha 1913: 110, pl.10, fig. 3 (Fig. 4j ).
Emended diagnosis: Cells almost round (90–132 × 46–84 µm), flat, posterior visibly asymmetrical; cells terminate with a sharp hyaline tail (on average 20–28 µm long), which is bent sideways. Periplast longitudinally striated.
Holotype: Cunha 1913, pl.10, fig. 3
Epitype: Figure 4j designated herein that supports the holotype (Cunha 1913, pl.10, fig. 3)
Representative DNA sequence: GenBank MN149572
Type locality: Manguinhos, Rio de Janeiro
Representative locality: Freshwater, Oracze, farm pond (53°52′36.6" N, 22°20′41.2" E)
Comments: Phacus gigas has large, flat, almost round cells, and the Polish populations have been identified based on these traits. The study presented herein shows that the diagnostic feature allowing the proper identification of P. gigas (and therefore making the differentiation between other similar species, e.g., P. hamatus, P. paraorbicularis, or P. orbicularis possible) is the posterior end asymmetry as well as the tail bent sideways. As the cell asymmetry is not mentioned either in the diagnostic description or in Cunha's drawing (1913, pl.10, fig. 3), we believe that the designation of an epitype is justified.
Phacus granum Drezepolski 1925: 231 and 266, pl. 3, fig. 119.
Cells small (18–23 × 7.5–12 µm), cylindrical, slightly flattened, narrowing at both ends, terminate with a blunt papilla (see Karnkowska‐Ishikawa et al., 2010).
Representative DNA sequence: GenBank DQ249880
Representative strain: AICB 349
Phacus hamatus Pochmann 1942: 182, fig. 86 a–f (Fig. 4k )
Emended diagnosis: Wide egg–like, spoon‐shaped (convex) cells (35–66 × 25–43 µm) ending with a sharp hyaline tail (on average 7.5–15 µm long). The tail is visibly bent toward the ventral side, which causes the cells not to adhere to the substrate, but rather “lean” on the tail. Periplast longitudinally striated.
Lectotype: designated herein, Pochmann 1942, fig. 86b.
Epitype: Figure 4k designated herein that supports the lectotype (Pochmann 1942, fig. 86b)
Representative DNA sequence: GenBank AJ532473
Representative strain: CCAC 2605B (=ASW 08032)
Type locality: pond in small town Dobrostany, Eastern Poland (now Ukraine)
Comments: The characteristic shape of the Phacus hamatus cells (spoon‐shaped [convex]) visible in all of Pochmann's drawings (1942, fig. 86, a–f) allowed the identification of the species. On the other hand, the cell asymmetry visible in all drawings and stressed in the original diagnosis, is confusing, as our research has shown that it concerns only P. gigas. Phacus hamatus has symmetrical cells, but both species have so far been difficult to distinguish and often are confused (for more details see Discussion). The individual indicated here as the lectotype (fig. 86b in Pochmann 1942) has the least visible cell asymmetry.
Phacus hamelii Allorge & Lefèvre 1925: 128, figs. 55–57.
Epitype: designated herein, Kosmala et al. 2007, fig. 1a.
Representative DNA sequence: GenBank DQ397673 Comment: As the epitypification statement published in Kosmala et al. 2007 (p. 1078)) does not include the phase “designated here” (or on equivalent), thus the nomenclatural act has not been effected in accordance with ICN Art. 7.11 (Turland et al. 2018) and is conducted herein.
Phacus helikoides Pochmann 1942: 212, figs. 124, 125.
Epitype: Łukomska‐Kowalczyk et al. 2015, fig. 3r
Representative DNA sequence: GenBank KP944094
Phacus hordeiformis Kim & Shin 2014: 956, fig. 2, F and G
Type: (see Kim and Shin 2014), permanently preserved material from the strain Yongho092609A deposited at the Chungnam National University, as number CNU 025430. figures 2F and 2D in Kim and Shin 2014 show the type.
Representative DNA sequence: GenBank KF744073
Phacus inflexus (Kisselev) Pochmann 1942: 133, fig. 20 a–h
Epitype: designated herein, Karnkowska‐Ishikawa et al. 2010, fig. 1b.
Representative DNA sequence: GenBank FJ590503 Comment: As the epitypification statement published in Karnkowska‐Ishikawa et al. 2010 (p. 178) does not include the phrase “designated here” (or an equivalent), thus the nomenclatural act has not been effected in accordance with ICN Art. 7.11 (Turland et al. 2018) and is conducted herein.
Phacus limnophilus (Lemmermann) E.W.Linton & Karnkowska in Linton et al. 2010: 609 (Fig. 4 b and c)
Emended diagnosis: Cells spindle‐shaped or cylindrical (45–90 × 7–13 µm), not flattened (round in cross section); posterior end narrowed and sharply ended; two large, rod–like paramylon grains – one placed in front of the nucleus, the other behind it.
Basionym: Euglena limnophila Lemmermann, 1898, Botanisches Centralblatt 76: 152
Holotype: Lemmermann 1913 in Pascher's Süssw.‐Fl., fig. 205.
Epitype: Figure 4b designated herein that supports the holotype (Lemmermann 1913 in Pascher's Süssw.‐Fl., fig. 205)
Representative DNA sequence: GenBank DQ249877
Representative strain: ACOI 1026
Type locality: Germany, ponds in Düsseldorf, Grimma, Gohlis and Knautheim near Leipzig
Comments: Both the diagnosis and drawing (Lemmermann 1898, 1913, fig. 205) are confusing in terms of the number and location of the large paramylon grains – the description mentions one grain positioned at the back of the cell (behind the nucleus) or two grains placed at the nucleus's level on both sides (the left and the right). The second option is depicted in Lemmermann's drawing (1913, fig. 205). Meanwhile the rod‐like, large paramylon grains always exist as a pair – one in front of the nucleus, and the other behind; there is no room in the narrow, cylindrical cells of Phacus limnophilus for the paramylon grains to be located on nucleus level. Many euglenid researchers, including ourselves, have noted this (Pringsheim 1956, Popova and Safonova 1976, Tell and Conforti 1986 among others).
Phacus lismorensis Playfair 1921: 125, pl. 5 fig. 14 (Fig. 4n )
Emended diagnosis: Cells long ovate (70–134 × 24–40 µm), flat (no crest), bent like a bow; anterior end wide and rounded; posterior end gradually narrowed and terminated with a long, sharp, pointy tail (on average 40–50 µm long); the tail is set almost at a right angle and does not straighten even when the cells are immobile (not swimming).
Holotype: Playfair 1921, pl. 5 fig. 14.
Epitype: Figure 4n designated herein that supports the holotype (Playfair 1921, pl. 5 fig. 14)
Representative DNA sequence: GenBank MN149578
Type locality: Australia, neighbourhood of Lismore, freshwater pond
Representative locality: Freshwater, Doliwy, fish pond (54°01′55.5″ N, 22°18′02.0″ E)
Heterotypic synonyms: Phacus rostafińskii Drezepolski 1922: 13, fig. 3, a and u. Phacus pediformis Skvortsov 1958: 163, pl. 3, fig. 30.
Comments: Polish reresentatives of Phacus lismorensis have a long tail heavily bent toward the ventral side. In Playfair's drawing, it is only slightly bent which is why P. lismorensis might be mistaken with P. elegans, to which it is very similar both in cell shape and size (for more details see Discussion and Comments for P. elegans). The taxa that constitute P. lismorensis synonyms are those whose cells are long ovate, flat and terminate with a long tail heavily bent ventrally.
Phacus longicauda (Ehrenberg) Dujardin 1841: 337, pl. 5 fig. 6.
Epitype: Łukomska‐Kowalczyk et al. 2015, fig. 3d
Representative DNA sequence: GenBank KP944103
Phacus longisulcus Kim & Shin 2014: 955, fig. 2A
Type: (see Kim and Shin 2014), permanently preserved material from the strain Psurononuma100609J deposited at the Chungnam National University, as number CNU 025426. Figure 2A in Kim and Shin 2014 shows the type.
Representative DNA sequence: GenBank KF744079
Phacus manginii Léfévre 1931: 352, pl. 2, figs. 39–46 (Fig. 3u )
Emended diagnosis: Cells wide oval‐ or egg‐like (38–55 × 22–33 µm) with a well‐developed dorsal fold (crest) that elongates into a sharp hyaline tail (on average 10–13 µm long) slightly bent ventrally; periplast longitudinally striated.
Lectotype: designated herein, Léfévre 1931, fig. 40.
Epitype: Figure 3u designated herein that supports the lectotype (Léfévre 1931, fig. 40)
Representative DNA sequence: GenBank MN149581
Representative locality: Freshwater, Izdebno Kościelne village, field pond (52°08′21.0″ N, 20°32′03.2″ E)
Type locality: Indochina
Comments: Polish populations of Phacus manginii have been identified based on cell size as well as the presence of a well‐developed dorsal fold elongated into a sharp tail. The well‐developed dorsal fold makes P. manginii similar to P. caudatus. Moreover the differentiation of the two species is further complicated by the fact, that in two of Lefèvre's drawings (figs. 39 and 43) the cells of P. manginii are asymmetrical, while it is the representatives of P. caudatus that possess asymmetrical cells (for more details see Discussion). In such a situation, indicating a lectotype (fig. 40 in Léfévre 1931 displaying a symmetrical P. manginii cell) and designating an epitype will allow the proper identification of this species.
Phacus minimus Kim & Shin 2014: 955, fig. 2D
Type: (see Kim and Shin 2014), permanently preserved material from the strain Buan092609I deposited at the Chungnam National University, as number CNU 025428. figure 2D in Kim and Shin 2014 shows the type.
Representative DNA sequence: GenBank KF744081
Phacus minutus (Playfair) Pochmann 1942: 182, fig. 85 (Fig. 3 c and d)
Emended diagnosis: Cells flat, ovoid (20–30 × 11–28 µm), ending with an inconspicuous tail (on average 1‐2 long µm) that is bent toward the dorsal side. Periplast longitudinally striated.
Holotype: Pochmann 1942, fig. 85.
Epitype: Figure 3d designated herein that supports the holotype (Pochmann 1942, fig. 85)
Representative DNA sequence: GenBank MN149551
Representative strain: ACOI 1755
Type locality: Australia, Sydney, botanical garden
Comments: Strain ACOI 1755 has been identified as Phacus minutus due to its shape and cell size. Meanwhile, the morphological study presented has shown, that P. minutus has a tail bent toward the dorsal side that makes it different from P. acuminatus (bent sideways). Both species are very similar in size and shape, which is why the designation of their epitypes seems justified (for more details see Discussion).
Phacus orbicularis Hübner 1886: 5, fig. 1 (Fig. 3k )
Epitype: herein designated, Kosmala et al. 2007, fig. 1h.
Representative DNA sequence: GenBank DQ397671
Heterotypic synonym: Phacus heimii M.Lefèvre 1933: 261, figs. 11‐13.
Comments: Phacus heimii has been included as a synonym of P. orbicularis, as the sequence of the Polish isolate (UW2362Choc), the cells of which matched the morphological form described as P. heimii, was located in the orbicularis clade (for more details see Discussion). As the epitypification statement published in Kosmala et al. 2007 (p. 1077) does not include the phrase “designated here” (or an equivalent), thus the nomenclatural act has not been effected in accordance with ICN Art. 7.11 (Turland et al. 2018) and is conducted herein.
Phacus oscillans G.A.Klebs 1883: 313, pl. 3, fig. 6
Epitype: designated herein, Karnkowska‐Ishikawa et al. 2010, fig. 1h.
Representative DNA sequence: GenBank FJ590499 Comment: Because the epitypification statement published in Karnkowska‐Ishikawa et al. 2010 (p. 177) does not include the phrase “designated here” (or an equivalent), thus the nomenclatural act had been not effected in accordance with ICN Art. 7.11 (Turland et al. 2018) and that's why it's done now.
Phacus paraorbicularis Kim & Shin 2014: 956, fig. 2J
Type: (see Kim and Shin 2014), permanently preserved material from the strain Cheongsan052507F deposited at the Chungnam National University, as number CNU 025432. Figure 2J in Kim and Shin 2014 shows the type.
Representative DNA sequence: GenBank KF744083
Phacus parvulus G.A.Klebs 1883: 313, pl. 3, fig. 5
Epitype: designated herein, Karnkowska‐Ishikawa et al. 2010, fig. 1p.
Representative DNA sequence: GenBank AJ532472 Comment: Because the epitypification statement published in Karnkowska‐Ishikawa et al. 2010 (p. 177) does not include the phrase “designated here” (or an equivalent), thus the nomenclatural act had been not effected in accordance with ICN Art. 7.11 (Turland et al. 2018) and that's why it's done now.
Phacus pleuronectes (O.F.Müller) Nitzsch ex Dujardin 1841: 336, pl. 5, fig. 5a and b, 1841 (Fig. 3p )
Emended diagnosis: Cells ovoid or trapezoid‐like (30–55 × 20–28 µm).
Epitype: designted herein, Kosmala et al. 2007, fig. 1d.
Representative DNA sequence: GenBank AJ532475
Heterotypic synonyms: Phacus trapezoides Stawiński 1969: 39 and 50, fig. 76 a and b; P. trapezialis Shi et al., 1999: 264; pl. 69, figs. 3–5.
Comments: The names Phacus trapezoides and P. trapezialis have been included as synonyms of P. pleuronectes, as all five sequences of the Polish cells had a trapezoid–like shape and are in the pleuronectes clade (for more details see Discussion).
Phacus polytrophos Pochmann 1942: 128, fig. 15 a–d
Epitype: designated herein, Karnkowska‐Ishikawa et al. 2010, fig. 1m.
Representative DNA sequence: GenBank FJ590498 Comments: As the epitypification statement published in Karnkowska‐Ishikawa et al. 2010 (p. 178) does not include the phrase “designated here” (or an equivalent), thus the nomenclatural act has not been effected in accordance with ICN Art. 7.11 (Turland et al. 2018) and is conducted herein.
Phacus pusillus Lemmermann 1910: 514
Epitype: designated herein, Karnkowska‐Ishikawa et al. 2010, fig. 1r.
Representative DNA sequence: GenBank AF190815 Comment: As the epitypification statement published in Karnkowska‐Ishikawa et al. 2010 (p. 178) does not include the phrase “designated here” (or an equivalent), thus the nomenclatural act has not been effected in accordance with ICN Art. 7.11 (Turland et al. 2018) and is conducted herein.
Phacus raciborskii Drezepolski 1925: 234 and 266, fig. 113 (Fig. 4 l and m)
Emended diagnosis: Cells flat, cylindrical (35–60 × 8–15 µm), slightly U‐bent and spirally twisted; anterior rounded, narrowing toward the end in a wedge‐like manner and terminated with a sharp hyaline tail (on average 9–13 µm long); low crest running along the entire cell length; periplast longitudinally striated. Two large, ring‐like paramylon grains with one placed in front of the nucleus, and the other behind it.
Holotype: Dreżepolski 1925, fig. 113.
Epitype: Figure 4m designated herein that supports the holotype (Dreżepolski 1925, fig. 113)
Representative DNA sequence: GenBank MN149594
Type locality: pond in small town Dobrostany, Eastern Poland (now Ukraine)
Representative locality: Freshwater, Grabówek village, pond (53°50′52.9" N, 21°42′19.6" E)
Heterotypic synonyms: Phacus raciborskii var. triqueter Z.X.Shi 1987: 358, fig. 1n–p; P. trimarginatus Allorge & Jahn 1943: 242, figs. 33–35.
Comments: Following the priority rule, the representatives of Polish populations that displayed the characteristic cell shape (cylindrical, flat, with a low crest along the entire cell length, U‐bent and spirally twisted) are known as Phacus raciborskii, while the names of taxa with a morphology identical to P. raciborskii, but described later, have been included as its synonyms (for more details see Discussion).
Phacus salinus (F.E.Fritsch) E.W.Linton & Karnkowska in Linton et al. 2010: 609 (Fig. 4e )
Emended diagnosis: Cells wide egg‐like (32–57 × 25–48 µm), spherical (not flattened), widely rounded at the posterior end; lateral reservoir opening; wide periplast striation running spirally from right to left. Paramylon grains monomorphic (only small ones), oval and ring‐like.
Representative DNA sequence: GenBank EU624028
Representative strain: SAG 1244‐3
Comments: A new diagnostic trait – the lateral reservoir opening – has been included in the description. Moreover the measurements of the cells of the Polish populations have been taken into account, together with literature data. This seems justified, particularly that no such emendment had been done by Linton et al. (2010) when moving Phacus salinus from Lepocinclis to Phacus.
Phacus segretii Allorge & Lefèvre 1925: 128, figs. 58–60.
Cells wide‐oval (22–28 × 20–22 µm), thick, narrowed, and rounded on both sides, without a tail; the apical furrow almost reaches the posterior end; large, flat paramylon grain located in the center of the cell; periplast longitudinally striated (Pochmann 1942; ).
Representative DNA sequence: GenBank FJ719635
Representative strain: ACOI 1337
Heterotypic synomym: Phacus stokesii f. minor W.Conrad 1938: 12, figs. 45–47.
Comments: The small cells of Phacus stokesii f. minor indicate that it is a representative of P. segretii, which is why the name P. stokesii f. minor has been included as a synonym of P. segretii. The main difference between the two species is the size (P. stokesii: 40–46 × 30–35 µm; P. segretii: 22–28 × 20–22 µm).
Phacus skujae Skvortsov 1928: 116, pl. 2, fig. 42.
Epitype: designated herein, Karnkowska‐Ishikawa et al. 2010, fig. 1j.
Representative DNA sequence:FJ597146 Comment: As the epitypification statement published in Karnkowska‐Ishikawa et al. 2010 (p. 178) does not include the phrase “designated here” (or an equivalent), thus the nomenclatural act has not been effected in accordance with ICN Art. 7.11 (Turland et al. 2018) and is conducted herein.
Phacus smulkowskianus (Zakryś) W.H.Kusber 1998: 246.
Epitype: designated herein, Karnkowska‐Ishikawa et al. 2010, fig. 1e.
Representative DNA sequence: GenBank DQ249881 Comment: As the epitypification statement published in Karnkowska‐Ishikawa et al. 2010 (p. 179) does not include the phrase “designated here” (or an equivalent), thus the nomenclatural act has not been effected in accordance with ICN Art. 7.11 (Turland et al. 2018) and is conducted herein.
Phacus stokesii Lemmermann 1901:88, 1910: 518, fig. 9 (Fig. 4d )
Emended diagnosis: Cells wide‐oval (40–46 × 30–35 µm), thick, widely rounded at both ends, without a tail; apical furrow deep, running along the entire cell length; large, plate–like paramylon grain located in the center of the cell; periplast longitudinally striated.
Holotype: Lemmermann 1910, fig. 9
Epitype: Figure 4d designated herein that supports the lectotype (Lemmermann 1910, fig. 9)
Representative DNA sequence: GenBank MN149602
Type locality: North America, Teichen, ponds
Representative locality: Freshwater, Urwitałt village, field pond 19 (53°50'38.0" N, 21°38′06.5" E)
Heterotypic synonyms: Phacus aspidion Pochmann 1942: 193, fig. 99; P. balatonicus Hortobágyi 1943: 87, pl. 2, figs. 27–36; P. fominii Y.V. Roll 1938: 137, fig. 1; P. starmachii Stawiński 1969: 37 and 50, fig. 77; P. betkowski Stawiński 1969: 39 and 49, fig. 67a and b.
Comments: The morphology of the cells of Polish populations of Phacus stokesii (thick, widely rounded at both ends, without a tail) corresponds to the original diagnosis. The drawing by Lemmermann (1913, fig. 231), on the other hand, is confusing as it shows an almost round form, while in reality the shape of the cells of this species is wide‐oval. The taxa that constitute P. stokesii synonyms have wide‐oval cells (40–46 × 30–35 µm), thick and rounded at both ends (for more details see Discussion).
Phacus tenuis Svirenko 1915b: 336, pl. 2 figs. 17 and 18 (Fig. 4f )
Emended diagnosis: Cells oval (29–43 × 15–25 µm), flat, with a low crest that elongates into a sharp hyaline tail (3–6 µm), which is straight or slightly bent toward the ventral side.
Lectotype: designated herein, Svirenko 1915b, fig. 17.
Epitype: Figure 4f designated herein that supports the lectotype (Svirenko 1915b, fig. 17)
Representative DNA sequence: GenBank MN149552
Representative strain: ACOI 1757
Type locality: Republic of Armenia, Bogdan Lake and pond located close to city Echmiadzin
Homotypic synonym: Phacus caudata var. tenuis Svirenko 1915a: 121, pl. 3, figs. 17 and 18;
Heterotypic synonym: Phacus caudatus var. minor Drezepolski 1925: 230, fig. 107.
Comments: According to Svirenko (1915b) P. tenuis differs from P. caudatus by having more oval cells and a lower crest (the latin name “tenuis” means flat), and this has been confirmed by our research. Meanwhile the individuals in Svirenko's (1915b) drawings: P. caudatus (fig. 8) and P. tenuis (figs. 17, 18) are so similar, that the differentiation of the two species is almost impossible. figure 17 has been indicated as the lectotype, as it is in this drawing that the cell has the most oval shape and has the lowest crest.
Phacus tortus (Lemmermann) Skvortsov 1928: 110, pl.2, figs. 9, 10.
Epitype: Łukomska‐Kowalczyk et al. 2015, fig. 3n
Representative DNA sequence: GenBank KP944112
Phacus triqueter (Ehrenberg) Dujardin 1841: 338
Cells oval (37–68 × 30–45 µm) with a high crest, triangular when cross‐sectioned; terminate with a straight or diagonal tail (Pochmann 1942; ).
Representative DNA sequence: SAG 1261‐8
Representative strain: GenBank AJ532485
Phacus viridiozyra Kim & Shin 2014: 956, fig. 2H
Type: (see Kim and Shin 2014), permanently preserved material from the strain Sondang060709L deposited at the Chungnam National University, as number CNU 025431. Figure 2H in Kim and Shin 2014 shows the type.
Representative DNA sequence: GenBank KF744098
DISCUSSION
On the oldest molecular phylogenetic trees, Phacus was recognized as a monophyletic member of the family Phacaceae (Kim et al. 2010), but further investigation suggested that the genus was paraphyletic (Kim and Shin 2014, Karnkowska et al. 2015). In those two studies Phacus limnophilus and Phacus arnoldii (=P. warszewiczii; Karnkowska et al. 2015) or only P. limnophilus (Kim and Shin 2014) sequences formed a sister clade to the clade grouping other sequences of Phacus and Lepocinclis. Later, the increased number of species and utilization of multi‐marker analyses confirmed the monophyly of Phacus (Kim et al. 2015). Our research, based on nSSU rDNA only, but including more sequences and Phacus taxa (136 sequences, 50 species), yielded a phylogenetic tree with a topology similar to those published previously. Most clades present on our tree consisted of sequences that were also together on previous trees. This includes our clade D (P. oscillans group), that underwent taxonomic revisions earlier (Karnkowska‐Ishikawa et al. 2010, Kim and Shin 2014) and is always strongly supported (Kim et al. 2010, 2015), clade J with sequences of P. orbicularis and P. paraorbicularis (Kim et al. 2010, 2015, Kim and Shin 2014) and clade C (P. longicauda and P. helikoides group; Kim and Shin 2014, Kim et al. 2015, Łukomska‐Kowalczyk et al. 2015) that was extended by adding sequences of P. elegans. Clades F, B, E, and H were also present on previous trees (Kim and Shin 2014, Kim et al. 2015), but herein they were verified and new sequences were added.
Clade I (Phacus lismorensis) is represented for the first time, clade G (P. salinus) was previously represented only by one sequence, and clade K was represented at most by three sequences of only one species (Łukomska‐Kowalczyk et al. 2015), now includes sequences of four species.
In the following part, the details concerning nomenclature and history of particular species and groups of species are discussed.
Phacus acuminatus
Over 30 intraspecific taxa (subspecies, forms, and varieties) have been described for Phacus acuminatus (see: http://www.algaebase.org), mainly based on cell shape and size, tail length and shape as well as the number and morphology of the large paramylon grains. Many authors view such differences as intraspecific variability arising from either ontogenesis or environmental conditions (Huber‐Pestalozzi 1955, Popova and Safonova 1976 among others). This is further confirmed by our research. In regard to the shape and size of the cells, the obtained results (on average 30 × 20 µm) were compatible with the rich literature (e.g., Popova and Safonova 1976: 25–33 × 19–22 µm; short tail, no fold; Table 1) and the observed differences in each strain were the result of ontogenesis – the individuals tended to enlarge (particularly their width) right before cell division. Some dissimilarities were also found between strains, but they had no DNA sequence support – all strains occurred in a subclade of clade B (Fig. 2) with nSSU rDNA diversity below 3%. The UTEX 1317 strain (in the collection as P. brachykentron; Fig. 3a) was located among them. This morphological form, described by Pochmann (1942) as P. brachykentron, had been distinguished due to its slight cell asymmetry. For the aforementioned reasons, many taxa described in the literature as having a morphology similar to P. acuminatus and without proper diagnostic traits in their original descriptions, have been placed in synonymy.
Phacus alatus, P. anomalus, and P. curvicauda
These three species are closely related (clade K in Fig. 2) and morphologically similar. They all possess thick cells (not flat) divided into two parts by a furrow – the halves are also twisted in the longitudinal axis. Phacus alatus was the first to be described (Klebs 1883), when Klebs came to the conclusion that the morphological form identified by Stein (1878) as P. triqueter (pl. 19, figs. 55–57) represented a species new to science. In the original diagnosis, Klebs drew attention to the atypical, characteristic shape (“widened, wing–like sides of the cell which overlap; one from the ventral side, the other from the dorsal”) and the lateral placement (“in both wings”) of two large, discoidal paramylon grains. Since then, many taxa had been described at various taxonomical ranks (species, varieties and forms) of a similar morphology, the proper identification of which is practically impossible. It even came to the point, where Pochmann (1942) described P. platyaulax for the same exact morphological form previously described by Klebs as P. alatus, i.e., P. triqueter from Stein's original work (1878; see p. 165 in Pochmann 1942). As a result, based on the aforementioned reasons and the research presented herein, many of those forms have been placed in synonymy under P. alatus.
Two more species were described in the 20th century (Phacus curvicauda and P. anomalus) that differ from P. alatus by having spherical large paramylon grains positioned in the center of the cell (not parietal). As the criteria for their distinguishment were ambiguous, researchers tended to interpret their taxonomic rank in various ways (e.g., see Pochmann 1942; Popova and Safonova 1976, Starmach 1983). The species can be easily morphologically identified (Fig. 3, e–j); on the phylogenetic tree there is a clade with sequences of P. anacoelus sister to P. anomalus. Thus, distinguishing three separate taxa at the rank of species is justified. The shape and placement of the large paramylon grains are decent diagnostic traits that allow correct identification.
All three species are considered common and cosmopolitan (e.g., Popova and Safonova 1976, Starmach 1983). In Poland, Phacus curvicauda and P. anomalus are commonplace and are often found in large densities. Phacus alatus, on the other hand, is found rarely and only in low densities (B. Zakryś, pers. obs.). One other species – P. drezepolski (Stawiński 1969) had been described from Poland based on a single cell – however, as it is not different from P. anomalus, it has been listed as one of its synonyms.
Phacus anacoelus
This euglenid of a particularly characteristic shape (almost spherical, slightly flattened, with 4 spiral ribs; Fig. 3, l–o) was described from the USA at the end of the 19th century by Stokes (1885b). Forty years later, Sokoloff (1933) described a similar species from ponds in Mexico, but due to the asymmetry of the cell the species was named P. asymmetricus. Jahn and Shawhan (1942) described P. quinquemarginatus with 5 ribs from canals and ponds in Iowa (USA). All of the aforementioned species are of a similar size (on average around 40 × 35 µm; Table 1). Some authors of critical monographs recognized only P. anacoelus, deeming it a very rare species (Pochmann 1942; Popova and Safonova 1976). Phacus quinquemarginatus and P. asymmetricus had been reported from Argentina (Tell and Conforti 1986), while P. anacoelus had been recorded in Russia (Popova and Safonova 1976), Slovakia (Wołowski and Hindák 2005) and Poland (Dreżepolski 1925). We found two populations in Mazovia (Regnów and Izdebno Kościelne 1; see Fig. 1 and Table S1), one of which, in a small field pond in Izdebno Kościelne village, has been persisting for the past several years and appears regularly during summertime in high densities. The Polish populations have cells with bilateral symmetry visible in a cross section, but less so when observing mobile cells (Fig. 3, l–o). Thus, the term “asymmetrical” might be interpreted in various ways, and there is no basis to distinguish these species based on their imprecise, original diagnoses. However, we continue to recognize these three taxa pending the study of additional material. The sequences of P. anacoelus are located on the phylogenetic trees in a subclade within clade K (Fig. 2) along with P. curvicauda, P. anomalus and P. alatus, despite that “at first glance” these species appear morphologically very dissimilar (Fig. 3, e–j, l–o). However, they have a similar cell shape: slightly flattened, with well‐developed ventral and dorsal sides, terminating with a sharp, hyaline tail bent toward the dorsal side.
Phacus ankylonoton
This morphological form was described from Poland by Dreżepolski (1925) as Phacus caudata var. polonica, and later was recognized as separate species (P. ankylonoton) by Pochmann (1942). It differs from P. caudatus in the width of the cell, which in cross‐section is triangular (Fig. 3q, Table 1). The validity of distinguishing this species is further confirmed by two DNA sequences (strain CCAC 0043 from Germany and Mokpo033107E from South Korea), creating a common clade (H) on the phylogenetic tree at a distant position from the sequences of P. caudatus (clade E in Fig. 2). The species is rare – it has been noted only a few times in Europe, Asia and South America (Shi et al. 1999, Popova and Safonova 1976, Starmach 1983, Tell and Conforti 1986). In Poland it was previously reported (Stawiński 1969, Wołowski 1998), but we never found it ourselves.
Phacus applanatus
Two strains, CCAC 2604B (=ASW 08023) isolated in the Netherlands (channel in Leiden) and Beopsu030709E isolated in South Korea, occurred in clade H (Fig. 2 and Table S1). The morphological study of the CCAC 2604B strain demonstrates that it is an elongated‐oval, flat cell that terminates with a straight tail (Fig. 3r). These are the only reports of this species across the world. It is yet to be found in Poland.
Phacus arnoldii and P. limnophilus
These two taxa represent the basal branch of the Phacus phylogenetic tree (clade A in Fig. 2), despite being morphologically “more similar” to Lepocinclis. The greatest similarity is the three‐ridged cell shape of P. arnoldii, alike L. tripteris, while the spindle‐shaped (unflattened) cells of P. limnophilus are typical of the majority of Lepocinclis species (Fig. 4, b, c and g‐i). Increasing the number of DNA sequences only further supported this positioning. Phacus arnoldii had been described from Ukraine in 1915, but its presence is often reported in the literature as P. warszewiczii Drezepolski; named after Poland's capital city, Warsaw (“Warszawa” in Polish). In Poland P. arnoldii occurs rarely and always in small densities, in contrast to P. limnophilus, which is much more common and may create dense populations (B. Zakryś, pers. obs.). Both species are considered cosmopolitan (e.g., Popova and Safonova 1976, Starmach 1983, Tell and Conforti 1986, Shi et al. 1999).
The variety described from Russia (Phacus arnoldii var. ovatus; Popova 1947) has been included as a synonym of P. arnoldii, as minute differences in cell shape are not diagnostic – the sequence of the Polish strain (one with more rounded cells) is sister to a strain isolated in Austria (CCAC 2432E, =ASW 08064), the shape of which (less round, more elongated) seems to be a result of culturing conditions.
Phacus caudatus and P. manginii
These two species are closely related and morphologically similar due to their well‐developed (tall) dorsal fold that elongates into a sharp, hyaline, slightly ventrally bent tail, but they do differ in cell shape and size (subclades in clade E in Fig. 2; Fig. 3, s–u, Table 1). Phacus caudatus was described at the end of 19th century (Hübner 1886). Later, several varieties were distinguished (see http://www.algaebase.org) based on minute cell shape and size differences. We did not confirm the validity of several of these forms, which is why they are being synonymized under P. caudatus. The presence of Austrian, Korean, Romanian and Polish strains in the P. caudatus clade affirms the cosmopolitan nature of this species. Daepyeong101908G, as well as ASW 08020 (=CCAC 2415 B) strain, identified as P. carinatus in previous works, and likewise Suwol060709C strain, identified as P. swirenkoi (Kim and Shin 2014, Kim et al. 2015), have been placed in synonymy under P. caudatus based on their placement in the P. caudatus clade as well as their morphology (Table 1, Fig. 3, s and t). Moreover P. carinatus is a very dubious species. This morphological form was first identified in Australia by Playfair (1921, as P. triqueter) and later considered a new species (P. carinatus) by Pochmann (1942). It has not been found since. Moreover it is not present in any critical monograph on euglenids (Popova and Safonova 1976, Tell and Conforti 1986, Shi et al. 1999).
In Poland, Phacus caudatus is common and occasionally can be found in dense populations, similarly to P. manginii described from Indochina (Léfévre 1931). The latter has rarely been mentioned in the literature (from China, Shi et al. 1999; and Argentina, Tell and Conforti 1986 among others), most likely due to being mistaken for P. caudatus. However, its presence in Europe does affirm its cosmopolitan status. The Gungnamji052507C strain (identified as P. triqueter) is renamed as P. manginii due to the position of its sequence in the P. manginii clade.
The variety described by Dreżepolski from Poland as Phacus caudatus var. polonica was recognized by Pochmann (1942) as separate species (P. ankylonoton), which has been supported by this research – the sequence of CCAC 0043 strain (mistakenly identified in the collection as P. ranula) occurred in a clade with the Korean strain located far from the P. caudatus clade.
Phacus elegans and P. lismorensis
According to Pochmann these two very characteristic, yet still very similar morphological forms represent two species – Phacus elegans can be distinguished from P. lismorensis based on "a more elongated cell form and a shorter tail” (Pochmann 1942: 199). The Polish strains representing both morphological forms occur in different clades, Phacus elegans close to species with long tails (P. longicauda, P. helikoides among others), whereas P. lismorensis occurs in clade I in Figures 2 and S1. Furthermore, we have established that cell shape is a diagnostic morphological feature – with a bow–like arch and a visibly ventrally bent tail in P. lismorensis and straight (elongated) in P. elegans (Fig. 4, a and n). Additionally, P. elegans has a low crest that causes it to be slightly thicker than P. lismorensis. In Poland both species occur rarely and in minute densities. In the literature P. lismorensis is mentioned often and is considered to be cosmopolitan (e.g., Popova and Safonova 1976, Starmach 1983, Tell and Conforti 1986, Shi et al. 1999). On the other hand, P. elegans is rarely noted, which might be the result of the doubt over the validity of distinguishing the two species (Popova and Safonova 1976, Tell and Conforti 1986).
Phacus gigas and P. hamatus
These two species occur in the same clade (F in Fig. 2), despite being morphologically different both in cell shape and size – Phacus gigas is flat, slightly asymmetrical and large (the largest of all species in Phacus – on average 100 × 75 µm), whereas P. hamatus has symmetrical, much smaller (on average 50 × 30 µm) and spoon‐shaped (convex) cells (Table 1, Fig. 4, j and k). Despite such characteristic features, until recently some researchers questioned the validity of distinguishing P. gigas (e.g., Popova 1947, Popova and Safonova 1976). However both Bennett and Triemer (2012) and herein, in which the number of sequences for both species has been increased, unambiguously support two distinct species with interspecific nSSU rDNA variability above 7%. Phacus hamatus (as P. pleuronectes var. citriformis) has been described from Poland (Dreżepolski 1922), while P. gigas from Brazil (Cuncha 1913). Both species are cosmopolitan (Starmach 1983, Tell and Conforti 1986, Shi et al. 1999). In Poland P. hamatus is common, whereas P. gigas is far more rare, however both may occur in large densities (B. Zakryś, pers. obs.).
Phacus granum
This species had been described from Poland (Dreżepolski 1925) based on the presence of large paramylon grains in the cell that in the light of current knowledge is not a diagnostic trait, as it depends on the physiological state of the organism. Sadly, due to the small cell size and its morphological similarity to other representatives of the “small Phacus” group (P. brevisulcus, P. claviformis, P. hordeiformis, P. longisulcus, P. minimus, P. oscillans, P. parvulus, P. polytrophis, P. pusillus, and P. skujae – clade D in both Figs. 2; S1), we have failed to isolate material from environmental samples. Most likely, based on the same reasons, it is often omitted in the literature, despite being reported as cosmopolitan by some authors (Popova and Safonova 1976, Starmach 1983, Tell and Conforti 1986).
Phacus minutus
This morphological form has been described from Australia (a botanical garden in Sydney) as a variety of Phacus pleuronectes (P. pleuronectes var. minuta; Playfair 1921) due to their similar, ovoid cell shape, but of a smaller size (P. pleuronectes: 39–55 × 22–33 µm; P. minutus: 20–30 × 11–25 µm). Later, it was raised to the rank of species by Pochmann (1942). In his diagnosis, Pochmann stressed additional differences from P. pleuronectes, such as a more flattened cell shape. The validity of distinguishing this taxon is confirmed by our research. Although two sequences (Korean and Portuguese) form a sister clade with sequences of P. pleuronectes (Fig. 2, clade B subclades), the sequence divergence (2.4–4%) and morphological features (cell size and shape) allowed us to decide that P. pleuronectes and P. minutus should be accepted as distinct species (Fig. 3, c, d and p), at least pending the inclusion of new strains morphologically similar to P. minutus in the analyses. Moreover cells of the Portuguese strain (ACOI 1755) correspond morphologically to the original description by Pochmann with the exception of the tail, which is bent toward the dorsal side rather than sideways.
Phacus orbicularis and P. paraorbicularis
The two common, cosmopolitan and morphologically similar species are sister taxa (sequence divergence 0.7–1.4 %) and have already been verified taxonomically (Kosmala et al. 2007, Kim and Shin 2014). Phacus orbicularis was described in the 19th century (Hübner 1886), whereas P. paraorbicularis was described recently based on morphological and DNA sequence data (Kim and Shin 2014). In Poland, P. paraorbicularis occurs commonly and often in high densities. Phacus orbicularis is far rarer, and its populations are much more varied in cell shape and size (Kosmala et al. 2007). The form we collected in Poland is often known in the literature as P. heimii (symmetrical cells [31 × 23 µm] with a short tail [4 µm]; Fig. 3k). Its SSU sequence (MN149583 of the isolate UW2362Choc) places it in the P. orbicularis clade, confirming that P. heimii is a synonym of P. orbicularis (subclade in clade J in Figs. 2, S1).
Phacus pleuronectes
This species was taxonomically verified (Kosmala et al. 2007). After the addition of new sequences, it remains in a well‐established clade, in which two groups can be distinguished – one consists of the sequences from culture strains and one from Poland, while the other groups Polish strains only (Fig. S1). The cells from the Polish populations have a slightly more trapezoid–like shape and are slightly smaller (30–37 × 20–25 µm; Fig. 3p, Table 1) in comparison with cultured cells (38–55 × 26–28 µm – see table 2 in Kosmala et al. 2007). As genetic diversity in the clade is low, those morphological differences might be a result of different living conditions. The trapezoid–like form had been described from Poland under the name of P. trapezoides (Stawiński 1969) and from China as P. trapezialis (Shi 1987). Both names are now included in the list of synonyms for P. pleuronectes. It is cosmopolitan and a common species in Poland.
Phacus raciborskii
The species of Phacus with flat and permanently spirally twisted cells are located in different clades (see P. inflexus, P. smulkowskianus, P. tortus or P. helikoides – Figs. 2, S1). Another taxon of such morphology is P. raciborskii, described from Poland (Dreżepolski 1925). Four sequences of Polish strains have joined the Portuguese (strain ACOI 1758), two Korean (Gungnamji052507B as P. trimarginatus and Leynes012810F as P. mariae in Kim and Shin 2014, Kim et al. 2015) and the ASW 08004 (as Phacus sp.) strain sequences, creating a well‐supported clade (Fig. 2, clade E subclade, Fig. S1). The Portuguese sequence had been verified based on images posted on the website of the ACOI culture collection (http://acoi.ci.uc.pt/spec_detail.php?cult_id=1442). In the literature, we have found one more species (P. trimarginatus), described from ponds in Iowa (USA), that is morphologically indistinguishable from P. raciborskii and therefore has been included as a synonym. Phacus raciborskii is widely accepted as cosmopolitan and common (Pochmann 1942, Popova and Safonova 1976, Starmach 1983). It had been noted in Poland for many years (Dreżepolski 1925, Czosnowski 1948, Stawiński 1969) and is often found in dense populations (B. Zakryś, pers. obs.).
Phacus salinus
This very characteristic morphological form had been appearing in the literature since the 19th century – first under the name Crumenula texta (Dujardin 1836: 204, pl. 9, fig. M), later as Euglena texta (Hübner 1886) or Lepocinclis texta (Lemmermann 1901). None of the aforementioned descriptions refer to the striation direction. Only Fritsch (1914) drew attention to this feature and described a species new to science (L. salina) due to its periplast striation (striae running from right to left). Popova (1955) however considered it to be a variant of L. texta (L. texta var. salina). Unexpectedly, based on DNA sequence data, L. salina is transferred to Phacus (P. salinus), in spite of being morphologically (oblong, rigid cells ; Fig. 4e) more similar to Lepocinclis (Linton et al. 2010). Until now only one sequence had been available (EU624028 of the strain of SAG 1244‐3), and its position was not well supported (Linton et al. 2010). Five additional sequences form a common, strongly supported clade with the previous sequence, but the position remains uncertain (clade G in Fig. 2, Fig. S1). Phacus salinus occurs commonly in Poland, often in large densities. Both forms (texta and salina) are universally accepted as cosmopolitan and common (e.g., Popova and Safonova 1976, Starmach 1983, Tell and Conforti 1986, Shi et al. 1999). In spite of that, we have never found texta (with periplast striation running from left to right), despite having inspected over 100 populations.
Variability of nSSU rDNA fragments (lengths and sequences) of Phacus salinus is much higher than the observed morphological diversity. There are also no morphological features that could be used to distinguish the species. This occurs in several common euglenid species with very high intraspecific genetic diversity, e.g., Lepocinclis fusiformis up to 7.8%, L. hispidulus up to 5.1% (Łukomska‐Kowalczyk et al. 2020), and P. circumflexus up to 4.9% (Łukomska‐Kowalczyk et al. 2015).
The sequence MN149599 of Phacus salinus UW1948Wos (3,560 bp) is almost the longest known nSSU rDNA in autotrophic euglenids, only that of Euglena sanguinea is longer‐the fragment exceeds 6,000 bp (Karnkowska‐Ishikawa et al. 2013).
Phacus stokesii and P. segretii
Despite the two species being very similar morpologically (wide‐oval cells without a tail and with a long apical furrow (Fig. 4d and the photo of Phacus segretii strain ACOI 1337 on the collection's website: http://acoi.ci.uc.pt/spec_detail.php?cult_id=815), their sequences are found in separate clades (in clade H and as a sister to clade K in Fig. 2, Fig. S1). The most significant difference is cell size – P. stokesii is almost twice as big (40‐46 × 30‐35 µm) as P. segretii (22–28 × 20–22 µm; Table 1). In the literature, there are several other species of a similar morphology described, out of which five (P. fominii, P. aspidion, P. balatonicus, P. starmachii, and P. betkowski) are herein designated as synonyms of P. stokesii; one other (P. stokesii f. minor) has been synonymized under P. segretii due to a similar size and a lack of any other well‐defining feature. Popova and Safonova (1976: 62‐63) listed the same synonyms, except for P. fominii. Both species (P. stokesii and P. segretii) are considered cosmopolitan, but P. stokesii is far more common (Popova and Safonova 1976, Starmach 1983, Tell and Conforti 1986, Shi et al. 1999). Historically, only P. stokesii has been reported in Poland (Dreżepolski 1925, Stawiński 1969, Wołowski 1998), which currently is very uncommon and never found in large densities (B. Zakryś, personal observation)
Phacus tenuis
The Portuguese strain ACOI 1757 (as Phacus caudatus in ACOI) occurs outside of the “caudatus” clade (clade H in Fig. 2), despite being morphologically very similar to P. caudatus (cells flat, elongated‐oval in shape; Figs. 3, s and t, 4f). It does however differ in terms of having a lower crest and a straight tail, which in turn makes it similar to P. applanatus (Fig. 3r). A detailed literature study shows, that such a morphological form was described from Ukraine by Svirenko (1915a), first as P. caudatus var. tenuis, and in the same year was raised to the rank of species (P. tenuis) by Svirenko (1915b). The research presented herein supports such a decision. Phacus caudatus var. tenuis is reported from various countries: Russia (Popova and Safonova 1976), the Czech Republic (Wołowski 1992), Slovakia (Wołowski and Hindák 1996) and Poland (Stawiński 1969).
Phacus triqueter
The diagnostic trait for this species is its high crest that causes the cell's triangular appearance when cross‐sectioned. This three‐ridged cell shape of Phacus triqueter makes it similar to Lepocinclis tripteris or P. arnoldii. The very characteristic morphological form was described by Ehrenberg (1838) as Euglena triquetra and later moved to Phacus as P. triqueter (Dujardin 1841). Many authors considered it to be cosmopolitan (e.g., Pochmann 1942, Popova and Safonova 1976, Starmach 1983, Tell and Conforti 1986, Shi et al. 1999). Meanwhile, it has yet to be found in Poland by us, despite previous reports of its presence (Czosnowski 1948, Stawiński 1969, Wołowski 1998). Only one sequence of P. triqueter (from strain SAG 1261‐8) was included in the phylogenetic analysis (Fig. 2). Based on the image posted on the Culture Collection of Algae and Protozoa (CCAP) website, it appears that the strain had been verified correctly (https://www.ccap.ac.uk/results2014.php?mode=attr&Environment=All&Country=All&Pathogen=All&Type_Culture=All&Genus_Name=Phacus&Strain_Name=Phacus+triqueter&Strain_No=1261%2F8).
conclusion
As a result of the presented studies the phylogenetic trees of Phacus now includes 50 species represented by 129 sequences of SSU rDNA, of which seven species (Phacus anacoelus, P. anomalus, P. curvicauda, P. elegans, P. lismorensis, P. minutus, and P. stokesii) and 55 sequences are new for the tree. The new sequences were obtained from laboratory strains (6) and those isolated directly from the environment (49). The environmental isolates came from 37 eutrophic water bodies located in Poland and one located in the Czech Republic. Comparative morphological and molecular studies on new laboratory strains and environmental isolates as well as research in the literature, have allowed (a) an estimation of morphological and genetic diversity and verification of morphological diagnostic features for particular taxa (well‐established clades on a molecular phylogenetic tree) and as a result their proper identification; due to the lack of morphological data, the taxonomic affiliation of five strains (Jigok090112 as P. ranula on the phylogeny tree; Leynes012810F as P. mariae; Burni081809, ASW 08004 and ACOI1138 as Phacus sp.) was not verified; (b) the reconstruction of phylogenetic relationships among 50 species of Phacus (c) taxonomic verification, emending diagnoses, and designating epitypes for well‐distinguished taxa (19 species).
Supporting information
Figure S1. The Maximum Likelihood phylogenetic tree based on 136 nSSU rDNA sequences (of which 129 represent Phacus). The Bayesian posterior probability (pp) and the bootstrap (bs) values obtained by maximum likelihood analysis are marked at the nodes. The pp <0.75, bs values <50 and clades not present in the particular analysis are marked with a hyphen (‐). The sequences obtained in this study are indicated in bold type. Scale bar represents number of substitutions per site.
Table S1. List of species and sampling data of isolates/strains used in this study. GenBank accession numbers and their nuclear SSU rDNA gene sequenced are given, with new sequences indicated in bold type.
Table S2. The nuclear SSU rDNA pair‐wise sequence distances (%) among strains and isolates.
Acknowledgments
This work was supported by the OPUS 2016/23/B/NZ8/00919 grant from the National Science Centre, Poland. The Open access for this publication was partially funded by the University of Warsaw. The authors thank Prof. Richard Triemer, MI, USA, for providing the photos of P. limnophilus (strain ACOI 1026). The authors thank the three anonymous reviewers for the input and Prof. Paul Gabrielson, NCU, USA, who has been a great help in improving the manuscript.
References
- Allorge, C. F. & Jahn, T. L. 1943. Survey of the genus Phacus Dujard. Trans. Amer. Microsc. Soc. 62:233–44. [Google Scholar]
- Allorge, P. & Lefèvre, M. 1925. Algues de Sologne. Fascicule speciale de la session extraordinaire tenue dans la Sologne en Juillet. Bull. Soc. Bot. Fr. 72:122–50. [Google Scholar]
- Bennett, M. S. & Triemer, R. E. 2012. A new method for obtaining nuclear gene sequences from field samples and taxonomic revisions of the photosynthetic euglenoids Lepocinclis (Euglena) helicoideus and Lepocinclis (Phacus) horridus (Euglenophyta). J. Phycol. 48:254–60. [DOI] [PubMed] [Google Scholar]
- Brabez, R. 1941. Zur Kenntnis der Algen des Franzensbader und Sooser thermenbereicches. Beih. Bot. Zbl. 61:13–236. [Google Scholar]
- Capella‐Gutierrez, S. , Silla‐Martinez, J. M. & Gabaldon, T. 2009. TrimAl: a tool for automated alignment trimming in large‐scale phylogenetic analyses. Bioinformatics 25:972–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad, W. 1938. Flagellates des îles de la Sonde (Euglénacées). Bull. Mus. R. Hist. Nat. Belgique 14:1–20. [Google Scholar]
- Cunha, A. M. da 1913. Contribuiçâo para o conhecimento da fauna de Protozoários do Brasil. Mem. Inst. Oswaldo Cruz 5:101–22. [Google Scholar]
- Czosnowski, J. 1948. Materiały do flory wiciowców Polski. [Materials for flagellated flora of Poland.] Pozn. Tow. Przyj. Nauk Wydz. Mat.‐Przyr. Prace Kom. Biol. 11:41–5. [Google Scholar]
- Darriba, D. , Taboada, G. L. , Doallo, R. & Posada, D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9:772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deflandre, G. 1928. Algues d'eau douce du Vénézuéla. Rev. Algol. 3:212–41. [Google Scholar]
- Dreżepolski, R. 1922. Eugleniny wolnożyjące ze zbioru glonów podlaskich i litewskich dr J. Grochmalickiego [De Eugleninis se ipsis sustinentibus ex collectione facta a dr. J. Grochmalicki in Podlachia et Lithuania]. Rozpr. Wiad. Muz. Dzieduszyckich 7 ⁄ 8:1–19 (in Polish with Latin summary).
- Dreżepolski, R. 1925. Przyczynek do znajomości polskich Euglenin [Supplément à la connaissance des Eugléniens de la Pologne]. Kosmos 50:173–270 (in Polish with French summary). [Google Scholar]
- Dujardin, F. 1836. Recherches sur les organismes inférieurs IV. Ann. Sci. Nat. Zool, ser. 2, tom 5: 193–205. [Google Scholar]
- Dujardin, F. 1841. Histoire Naturelle des Zoophytes‐Infusoires. Libraire encyclopedique de Roret, Paris, 684 pp. [Google Scholar]
- Ehrenberg, C. G. 1838. Die Infusionsthierchen als vollkommene Organismen. Ein Blick in das tiefere organische Leben der Natur. Nebst einem Atlas von vierundsechszig colorirten Kupfertafeln, gezeichnet vom Verfasser, Leipzig, 547 pp. [Google Scholar]
- Fritsch, F. E. 1914. Notes on British Flagellates I‐IV. New Phytol. 13:341–52. [Google Scholar]
- Fritsch, F. E. & Rich, M. F. 1929. Contributions to our knowledge of the freshwater algae of Africa. 7, Freshwater algae from Griqualand West. Trans. Roy. Soc. S. Africa 18:1–92. [Google Scholar]
- Guiry, M. D. & Guiry, G. M. 2020. AlgaeBase. World‐wide electronic publication, National University of Ireland, Galway: https://www.algaebase.org; searched on 30 April 2020. [Google Scholar]
- Hortobágyi, T. 1943. Adatok a Balatonboglári sestnjában, psammonjában és lasionjában élö moszatok ismeretéhez. Magyar Biol. Kutzt . Munk, Tihany 15:75–127. [Google Scholar]
- Huber‐Pestalozzi, G. 1955. Das Phytoplankton des Süsswassers; Systematik und Biologie: 4 Teil; Euglenophyceen. E. Schweizerbartsche Verlagsbuchhandlung, Stuttgart, Germany, 606 pp. [Google Scholar]
- Hübner, E. F. W. 1886. Euglenaceenflora von Stralsund. Program d. Realgymnasiums zu Stralsund. K. Regierungs‐buchdruckerei, Stralsund, Germany, pp. 1–20. [Google Scholar]
- Jahn, T. L. & Shawhan, F. M. 1942. Phacus quinquemarginatus sp. nova ( Protozoa, Mastigophora, Euglenoidina). Trans. Amer. Micr. Soc. 56:48–54. [Google Scholar]
- Karnkowska, A. , Bennett, M. S. , Watza, D. , Kim, J. I. , Zakryś, B. & Triemer, R. E. 2015. Phylogenetic relationships and morphological character evolution of photosynthetic euglenids (Excavata) inferred from taxon‐rich analyses of five genes. J. Eukaryot. Microbiol. 62:362–73. [DOI] [PubMed] [Google Scholar]
- Karnkowska‐Ishikawa, A. , Milanowski, R. , Kwiatowski, J. & Zakryś, B. 2010. Taxonomy of the Phacus oscillans (Euglenaceae) and its close relatives – balancing morphological and molecular features. J. Phycol. 46:172–82. [Google Scholar]
- Karnkowska‐Ishikawa, A. , Milanowski, R. , Triemer, R. E. & Zakryś, B. 2013. A redescription of morphologically similar species from the genus Euglena: E. laciniata, E. sanguinea, E. sociabilis, and E. splendens . J. Phycol. 49:616–26. [DOI] [PubMed] [Google Scholar]
- Katoh, K. & Standley, D. M. 2013. MAFFT multiple sequence Alignment software version 7: improvements in performance and usability. J. Mol. Biol. Evol. 30:772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J. I. , Linton, E. W. & Shin, W. 2015. Taxon‐rich multigene phylogeny of the photosynthetic euglenoids (Euglenophyceae) . Front. Ecol. Evol. 3:1–11. [Google Scholar]
- Kim, J. I. & Shin, W. 2014. Molecular phylogeny and cryptic diversity of the genus Phacus (Phacaceae, Euglenophyceae) and the descriptions of seven new species. J. Phycol. 50:948–59. [DOI] [PubMed] [Google Scholar]
- Kim, J. I. , Shin, W. & Triemer, R. E. 2010. Multigene analyses of photosynthetic euglenoids and new family, Phacaceae (Euglenales). J. Phycol. 46:1278–87. [Google Scholar]
- Klebs, G. A. 1883. Über die Organisation einiger Flagellaten‐Gruppen und ihre Beziehungen zu Algen und Infusorien. Untersuchung Botan. Inst. Tübingen 1:233–362. [Google Scholar]
- Kosmala, S. , Bereza, M. , Milanowski, R. , Kwiatowski, J. & Zakryś, B. 2007. Morphological and molecular examination of relationships and epitype establishment of Phacus pleuronectes, Phacus orbicularis and Phacus hamelii . J. Phycol. 43:1071–82. [Google Scholar]
- Kumar, S. , Stecher, G. , Li, M. , Knyaz, C. & Tamura, K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35:1547–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusber, W. H. 1998. A study on Phacus smulkowskianus (Euglenophyceae) – a rarely reported taxon found in waters of the Botanic Garden Berlin‐Dahlem. Willdenowia 28:239–47. [Google Scholar]
- Lanave, C. , Preparata, G. , Saccone, C. & Serio, G. 1984. A new method of calculating evolutionary substitution rate. J. Mol. Evol. 20:86–93. [DOI] [PubMed] [Google Scholar]
- Léfévre, M. 1931. De la valuer des caractéres chez quelques Eugléniens. Trav. Cryptogamiques dédis á L. Mangin, Paris, pp. 343–54. [Google Scholar]
- Lefèvre, M. 1933. Contribution à la connaissance des Flagelles d'Indochine. Ann. Cryptogam. Exotique 6:58–264. [Google Scholar]
- Lemmermann, E. 1898. Beiträge zur Kenntnis der Planktonalgen. II. Beschreibung neuer Formen. Botanisches Centralblatt 76:150–6. [Google Scholar]
- Lemmermann, E. 1901. Beiträge zur Kenntniss der Planktonalgen, XII: Notizen über einige Schwebealgen. Ber. Deutsch. Bot. Ges. 19:85–95. [Google Scholar]
- Lemmermann, E. 1910. Kryptogamenflora der Mark Brandenburg und angrenzender Gebiete III. Verlag von Gebrüder Borntraeger, Leipzig, Algen, 712 pp. [Google Scholar]
- Lemmermann, E. 1913. Euglenineae In Pascher A. [Ed.] Die Süsswasserflora Deutschlands, Österreichs und der Schweiz, Flagellatae II. Verlog von Gustav Fisher, Jena, pp. 115–74. [Google Scholar]
- Linton, E. W. , Karnkowska‐Ishikawa, A. , Kim, J. I. , Shin, W. , Bennett, M. S. , Kwiatowski, J. , Zakryś, B. & Triemer, R. E. 2010. Reconstructing euglenoid evolutionary relationships using three genes: nuclear SSU and LSU, and chloroplast SSU rDNA sequences and the description of Euglenaria gen. nov. (Euglenophyta). Protist 161:603–19. [DOI] [PubMed] [Google Scholar]
- Łukomska‐Kowalczyk, M. , Karnkowska, A. , Łach, Ł. , Milanowski, R. & Zakryś, B. 2015. Delimiting species boundaries within the Phacus longicauda complex (Euglenida) through morphological and molecular analyses. J. Phycol. 51:1147–57. [DOI] [PubMed] [Google Scholar]
- Łukomska‐Kowalczyk, M. , Chaber, K. , Fells, A. , Milanowski, R. & Zakryś, B. 2020. Molecular and morphological delimitation of species in the group of Lepocinclis ovum‐like taxa (Euglenida). J. Phycol. 56:283‐99. [DOI] [PubMed] [Google Scholar]
- Marin, B. , Palm, A. , Klingberg, M. & Melkonian, M. 2003. Phylogeny and taxonomic revision of plastid‐containing euglenophytes based on SSU rDNA sequence comparisons and synapomorphic signatures in the SSU rRNA secondary structure. Protist 154:99–145. [DOI] [PubMed] [Google Scholar]
- Nygaard, G. 1949. Hydrobiological studies on some Danish ponds and lakes; Part II: the quotient hypothesis and some new or little known phytoplankton organisms. Det. Kon. Danska Vid. Selsk. Biol. Skr. 7:1–293. [Google Scholar]
- Playfair, G. J. 1921. Australian freshwater flagellates. Proc. Linn. Soc. (N. S. Wales) 46:99–146. [Google Scholar]
- Pochmann, A. 1942. Synopsis der Gattung Phacus . Arch. Protistenk. 95:81–252. [Google Scholar]
- Popova, T. G. 1947. Sistematiceskije zametki po evglenovym [Taxonomical note about euglenophytes]. Izv. Zap. Sib. Fil. AN SSSR Ser. Biol. (Novosibirsk) 2:47–71 (in Russian). [Google Scholar]
- Popova, T. G. 1955. Euglenovyje vodorosli. Opredelitel prosnovodnych vodoroslej SSSR, 7 [Euglenophyta. The handbook of freshwater algae]. Sov. Nauka, Moscow, 267 pp (in Russian). [Google Scholar]
- Popova, T. G. & Safonova, T. A. 1976. Flora Sporovych Rastenij SSSR, 9. [Flora plantarum cryptogamarum URSS, 9]. Euglenophyta, vol. 2 Izd. Nauka, Leningrad. 286 pp. (in Russian). [Google Scholar]
- Pringsheim, E. G. 1956. Contribution towards a monograph of the genus Euglena . Nova Acta Leopold. 18:1–168. [Google Scholar]
- R Core Team 2008. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.R‐project.org/. [Google Scholar]
- Rodriguez, F. , Oliver, J. L. , Marin, A. & Medina, J. R. 1990. The general stochastic model of nucleotide substitution. J. Theor. Biol. 142:485–501. [DOI] [PubMed] [Google Scholar]
- Roll, Y. V. 1925. Novye vidy vodoroslej najdemyje w okrestnostjach Sew. Doneckoj biologiceskoj stancji [Les nouvelles éspèces des algues trouvées aus environs de la station biologique du Donetz du Nord]. Russk. Arch. Protistol. 4:137–52. [Google Scholar]
- Roll, Y. V. 1938. Algologiczni notatki. III. Niekatoryje i redkije vodorosli [Algological notes. III. Some and rare algae]. In: Symposium dedicated to the memory of A. V. Fomin Ukrainian SSR Akademy of Sciences Press, Kiev, pp. 136–45 (in Ukrainian). [Google Scholar]
- Ronquist, F. & Huelsenbeck, J. P. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–4. [DOI] [PubMed] [Google Scholar]
- Shi, Z. X. 1987. New taxa of Euglenophyta from Western Hubei. Acta Hydrobiol. Sin. 11:357–66. [Google Scholar]
- Shi, Z. X. , Wang, Q. , Xie, S. & Dai, I. 1999. Flora Algarum Sinicarum Aquae Dulcis. Tomus VI. Euglenophyta. Science Press, Beijing, 414 pp. (in Chinese). [Google Scholar]
- Skvortsov, B. W. 1917. Über Flagellata aus Mandschurei. I Teil. J. Microbiol. 4:57–77. [Google Scholar]
- Skvortsov, B. W. 1922. On the phytoplankton from the ponds of Tiensin. J. R. Asiatic Society Shanghai 53:189–95. [Google Scholar]
- Skvortsov, B. W. 1928. Die Euglenaceengattung Phacus Dujardin. Eine systematische Übersicht. Ber. Deutsch. Bot. Ges. 46:105–25. [Google Scholar]
- Skvortsov, B. W. 1937. Contributions to our knowledge of the freshwater algae of Rangoon, Burma, India. Arch. Protist. 90:73. [Google Scholar]
- Skvortsov, B. W. 1958. New and rare Flagellatae from Manchuria, eastern Asia. Philipp. J. Sci. 86:139–202. [Google Scholar]
- Sokoloff, D. 1933. Algunas nuevas formas de flagellados del Valle de Mexico. Ann. Inst. Biol. Mex. 4:197–206. [Google Scholar]
- Stamatakis, A. 2014. RAxML version 8: a tool for phylogenetic analysis and post‐analysis of large phylogenies. Bioinformatics 30:1312–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starmach, K. 1983. Euglenophyta – eugleniny In Starmach K. & Sieminńska J. [Eds.] Flora Słodkowodna Polski 3. P. W. N, Warsaw, 594 pp. [Google Scholar]
- Stawiński, W. 1969. Arten der Gattung Phacus, welche in verschiedenen Biotopen der Umgebung von Bielsko‐Biała vorkommen. Acta Hydrobiol. 11:1–55. [Google Scholar]
- Stein, F. R. 1878. Der organismus der infusionsthiere. III. Abt. Der Organismus der Flagellaten. Verlag von Wilhelm Engelmann, Leipzig, 154 pp. [Google Scholar]
- Stokes, A. C. 1885a. Notices of new fresh‐water Infusoria. IV. Am. Mon. Microsc. J. 6: 183–90. [Google Scholar]
- Stokes, A. C. 1885b. Some apparently undescribed Infusoria . Am. Nat. 19:18–27. [Google Scholar]
- Svirenko, D. O. 1915a. Matérial pour servir à l'étude des algues de la Russie. Étude systématique et géographique sur les Euglénacées. Trav. Inst. Bot. Univ. Charkov 48:67–143 (in Russian). [Google Scholar]
- Svirenko, D. O. 1915b. Zur Kenntnis der russischen Algenflora. 2. Euglenophyceae (excl. Trachelomonas). Arch. Hydrobiol. Planktonk 10:321–40. [Google Scholar]
- Svirenko, D. O. 1938. Vyznachnyk prisnovodnykh vodorostey URSR [Identification book of freshwater algae of the URSR (Ukrainian Soviet Socialist Republic)]. 2. Euglenineae. Publ. House of the Acad. Sci. USSR, Kyiv, 176pp (in Ukrainian). [Google Scholar]
- Tavare, S. 1986. Some probabilistic and statistical problems on the analysis of DNA sequences. Lec. Math. Life Sci. 17:57–86. [Google Scholar]
- Tell, G. & Conforti, V. 1986. Euglenophyta pigmentadas de la Argentina. Bibliotheca Phycologica. J. CRAMER, Berlin, Stuttgart, 301 pp. [Google Scholar]
- Turland, N. J. , Wiersema, J. H. , Barrie, F. R. , Greuter, W. , Hawksworth, D. L. , Herendeen, P. S. , Knapp, S. et al. 2018. International Code of Nomenclature for Algae, Fungi, and Plants (Shenzhen Code) Regnum Vegetabile 159. Koeltz Botanical Books, Glashütten. [Google Scholar]
- Wołowski, K. 1992. Occurrence of Euglenophyta in the Třebon Biosphere Reserve (Czechoslovakia). Algol. Stud. 66:73–98. [Google Scholar]
- Wołowski, K. 1998. Taxonomic and environmental studies on Euglenophytes of the Kraków‐Częstochowa upland (Southern Poland). Fragm. Florist. Geobot. 6:3–192. [Google Scholar]
- Wołowski, K. & Hindák, F. 1996. Contribution to the knowledge of euglenophytes from Western Slovakia. Biologia 51:1–11. [Google Scholar]
- Wołowski, K. & Hindák, F. 2005. Atlas of Euglenophytes. VEDA, Publishing House of the Slovak Academy of Science, Bratislava, 136 pp. [Google Scholar]
- Zakryś, B. , Milanowski, R. , Empel, J. , Borsuk, P. , Gromadka, R. & Kwiatowski, J. 2002. Two different species of Euglena, E. geniculata and E. myxocylindracea (Euglenophyceae), are virtually genetically and morphologically identical. J. Phycol. 38:1190–9. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. The Maximum Likelihood phylogenetic tree based on 136 nSSU rDNA sequences (of which 129 represent Phacus). The Bayesian posterior probability (pp) and the bootstrap (bs) values obtained by maximum likelihood analysis are marked at the nodes. The pp <0.75, bs values <50 and clades not present in the particular analysis are marked with a hyphen (‐). The sequences obtained in this study are indicated in bold type. Scale bar represents number of substitutions per site.
Table S1. List of species and sampling data of isolates/strains used in this study. GenBank accession numbers and their nuclear SSU rDNA gene sequenced are given, with new sequences indicated in bold type.
Table S2. The nuclear SSU rDNA pair‐wise sequence distances (%) among strains and isolates.


