Abstract
Aims
The emergence of carbapenem‐resistant Klebsiella pneumoniae (CRKP) strains has led to increased mortality and morbidity rates. Tigecycline, a new class of broad‐spectrum glycyl‐tetracycline antibiotics, has been used to target multi‐ and pan‐drug‐resistant bacterial infections. This study aimed to assess the molecular characteristics of CRKP in a tertiary hospital, and its susceptibility to tigecycline, to create a reference for hospital infection control and clinical drug use.
Methods and Results
We retrieved patient clinical information and CRKP characterization from medical records and detected the MIC of tigecycline using the micro‐broth dilution method. Multi‐locus sequence typing was performed, and antibiotic resistance genes associated with CRKP were detected by qPCR. A total of 166 CRKP strains were detected in the sputum, urine and blood among intensive care unit patients (average age, 69·6 years). The most infrequently observed resistance genes were amikacin resistance genes, followed by tobramycin resistance genes. KPC ‐2, CTX‐M9 and CTX‐M1 were the most frequently detected resistance genes.
Conclusions
No strain was resistant to tigecycline (MIC ≥ 8 µg ml−1). Twenty‐four sequence types were identified, with ST11 being the most common type.
Significance and Impact of the Study
Clinicians and infection control experts should be aware of CRKP prevalence to facilitate clinical treatment and improve nosocomial infection control.
Keywords: carbapenem resistance, Klebsiella pneumoniae, KPC‐2, nosocomial infection, tigecycline
Introduction
In recent years, Klebsiella pneumoniae drug resistance, pressured by antibiotic treatment, has been continuously increasing. Carbapenem antibiotics were effective against multi‐drug‐resistant and extended‐spectrum beta‐lactamase‐producing strains; however, with the emergence of carbapenem‐resistant bacteria, clinical treatment faces a new dilemma. In 2017, both Chinese and European antibiotic resistance monitoring networks showed an increase in the detection rate of carbapenem‐resistant K. pneumoniae (CRKP). Specifically, an increase from 4·9% in 2013 to 9·0% in 2017 was reported in China, with an alarming increase of 26·9% in Shanghai. Meanwhile, the rate of European CRKP remained relatively unchanged, from 7·3% in 2014 to 7·2% in 2017, with the greatest change being from 62·3 to 64·7% in Greece (Simonsen 2018). According to the 2018 CHINET Resistance Monitoring Network, K. pneumoniae resistance to imipenem has risen rapidly from 16·1% in 2016 to 20·9% in 2017 and 26·1% in 2018. Therefore, CRKP resistance in China is increasing at an alarming rate, which has led to increased attention being paid to the CRKP drug resistance mechanism. Presently, the focus is on carbapenemase, the efflux pump, outer membrane pore proteins and the formation of biofilms.
However, the abundance of CRKP, its sensitivity to drugs and the percentage of strains carrying drug‐resistant genes in our hospital remain unknown. Moreover, multi‐locus sequence typing (MLST) and phylogenetic analysis have not been performed at our hospital. Therefore, the results of our research are expected to provide data that may be of clinical use in the treatment and control of these nosocomial infections.
Materials and methods
Patient characteristics and CRKP characterization
The clinical characteristics of the patients, including age, sex, inpatient ward and disease, were obtained from digitally stored medical records. Klebsiella pneumoniae strains were identified using the Vitek 2 system (BioMérieux, Marcy‐l'Étoile, France) between January 2013 and December 2017. Antibiotic susceptibility tests were performed using a Vitek‐2 Compact instrument. The ethics committee of Fujian Provincial Hospital (approval number: K2018‐01‐001) approved this study.
Susceptibility to tigecycline
Susceptibility to tigecycline was determined using the micro‐broth dilution method. Tigecycline was purchased from Selleck Chemicals (Houston, TX; Lot: S140303) and was used at a concentration ranging from 0·06 to 128 μg ml−1. Cation‐adjusted Mueller–Hinton broth was purchased from Bio‐Kont Co., Ltd (Wenzhou, China; Lot: HB6231‐1). The ratio of the strain to the cation‐adjusted Mueller–Hinton broth was 1 : 200, with 100 μl added to each well of a drug sensitivity plate. The plate was placed in an incubator with 5% carbon dioxide at 35°C overnight, and the results were observed on the second day.
Multi‐locus sequence typing
Bacterial DNA was extracted using the Bacteria Genomic DNA Kit (CWBio Co., Beijing, China). MLST for K. pneumoniae was performed following methods previously described (Diancourt et al. 2005). The allelic profiles and the sequence types (STs) of each strain were determined using online databases (https://pubmlst.org/bigsdb?db=pubmlst_mlst_seqdef).
Detection of antibiotic resistance genes
Primers for the detection of resistance genes (CTX‐M1, CTX‐M9, KPC‐2, Ompk35, Ompk36, Ompk37, IPM‐4, NDM‐1 and OXA‐48) were designed using Primer Premier 5 (ver. 5.00) software and are presented in Table 1. Bacterial DNA was obtained as described above. qPCR was performed with an UltraSYBR mixture kit (CWBIO Co., Ltd) on a Cobas z 480 analyzer (LightCycler 480; Roche, Basel, Switzerland) with an initial incubation at 95°C for 10 min, followed by 40 cycles of 15 s at 95°C and 60 s at 60°C. Melting curve fluorescence was evaluated five times per degree Celsius from 60 to 95°C. Each reaction was carried out in triplicate.
Table 1.
Primers of resistance genes associated with carbapenem‐resistant Klebsiella pneumoniae
| Genes | Primer sequence (5′→3′) | Product length (bp) | Annealing temperature (°C) |
|---|---|---|---|
| CTX‐M1 | AGGAAGTGTGCCGCTGTAT | 216 | 55 |
| AGATTCGGTTCGCTTTCAC | |||
| CTX‐M9 | ACGCAGGTGCTTTATCGC | 183 | 57 |
| TGTGCCGTTGACGTGTTTT | |||
| IMP‐4 | GCAGAGCCTTTGCCAGATT | 288 | 57 |
| CGTGGGGATGGATTGAGA | |||
| KPC‐2 | GCGGCTCCATCGGTGTGTA | 282 | 60 |
| TGGCGGCGGCGTTATCA | |||
| NDM‐1 | ATGTCTGGCAGCACACTTCC | 300 | 58 |
| CCGCAACCATCCCCTCT | |||
| OmpK35 | CAGGTCCTTGCCTTTGGTCT | 283 | 58 |
| CAACGGTATCGCACTGTCTG | |||
| OmpK36 | GCAAAGCCCAGGGAACC | 254 | 57 |
| CGTACCGCCTTGAAACAGA | |||
| OmpK37 | GGCGATTACGGCTCCTT | 265 | 55 |
| TGCTGCGGTTATTGGTG | |||
| OXA‐48 | CCATAAGGCAACCACCACA | 183 | 57 |
| CCATAACCAACACGCTTCACT |
Results
Demographic and clinical characteristics
The demographic and clinical characteristics of patients and specimens are shown in Table 2. The patients’ ages ranged from 1 to 98 years with an average age of 69·57 ± 17·89, and the proportion of patients older than 60 years of age was 77·10%. Moreover, the number of male patients was 2·77 times that of female patients. The primary sources of specimens were sputum, blood, urine, alveolar lavage fluid, secretions, pus, pleural effusion, ascites and bile. The three most significant sources were sputum, which accounted for 42·17% of the specimens; urine, accounting for 27·10%; and blood, accounting for 15·67%. The diseases identified in our study primarily included pulmonary infection, cerebral infarction, gastrointestinal bleeding, nephrotic syndrome, bile duct stones, urinary tract infection and bloodstream infection. The four most common diseases were brain diseases (30·12%), based on samples from neurology, neurological surgery and brain‐related diseases in the intensive care unit (ICU); respiratory diseases (21·08%); digestive tract diseases (16·87%); and urinary system diseases (6·02%). Patients in the hospital were mainly admitted in the ICU, gastroenterology, neurology, neurosurgery, cardiovascular medicine, hepatobiliary surgery and senior officials’ inpatient wards. The four most common wards were the ICU (63·25%), neurosurgery (7·83%), neurology (3·01%) and senior officials’ inpatient wards (3·01%). The 7‐day, 30‐day and total mortality numbers of CRKP‐infected patients were as follows: 1 (0·6%), 5 (3·4%) and 8 (5·4%) respectively.
Table 2.
Demographic and clinical characteristics of patients and specimens
| Characteristics (unit) | Numerical data, n (%) |
|---|---|
| Median age (years) | 73 |
| Interquartile range (years) | 61–83 |
| Category by age (years), n (%) | |
| <20 | 1 (0·6) |
| 20–40 | 13 (7·8) |
| 41–60 | 24 (14·5) |
| 61–80 | 74 (44·6) |
| 81–100 | 54 (32·5) |
| Gender, no. males (%) | 121 (72·9) |
| Source, n (%) | |
| Sputum | 70 (42·2) |
| Urine | 45 (27·1) |
| Blood | 26 (15·7) |
| Others | 25 (15·0) |
| Comorbidities, n (%) | |
| Brain diseases | 50 (30·1) |
| Pulmonary infections | 35 (21·1) |
| Diseases of the digestive system | 28 (16·9) |
| Diseases of the urinary system | 10 (6·0) |
| Heart diseases | 9 (5·4) |
| Others | 34 (20·5) |
| Origin, n (%) | |
| Intensive care unit | 105 (63·3) |
| Neurosurgery | 13 (7·8) |
| Neurology | 5 (3·0) |
| Senior officials’ inpatient ward | 5 (3·0) |
| Others | 38 (22·89) |
| Average hospital stay (days) | 50 |
| Interquartile range (days) | 22–58 |
| Outcomes, n (%) | |
| Crude 7‐day mortality | 1 (0·6) |
| Crude 30‐day mortality | 5 (3·4) |
| Total mortality | 8 (5·4) |
Susceptibility of CRKP to antibiotics
The detection rate of CRKP strains collected in this study was 15·74% (166/1055). All strains showed multi‐drug resistance. Resistance rates to amikacin, tobramycin, minocycline, gentamicin, sulfamethoxazole and cefoperazone/sulbactam were 41·60, 61·60, 64·00, 68·80, 69·20 and 69·60% respectively. Cefotetan, levofloxacin, ciprofloxacin, cefepime, ertapenem, aztreonam, piperacillin/tazobactam, ampicillin/sulbactam, ceftazidime and imipenem resistance rates were >90%. The resistance rate against IPM, according to the Clinical & Laboratory Standards Institute (CLSI) M100‐S27 standard (IPM ≥4 μg ml−1 or inhibition zone diameter ≤19 mm), was 100%, as shown in Fig. 1. Tigecycline MIC values are shown in Fig. 2. MICs were mainly concentrated at 0·25 and 0·5 μg ml−1 (73·5%), and no tigecycline‐resistant strains were found, according to the Food and Drug Administration standard (MIC ≥8 μg ml−1).
Figure 1.
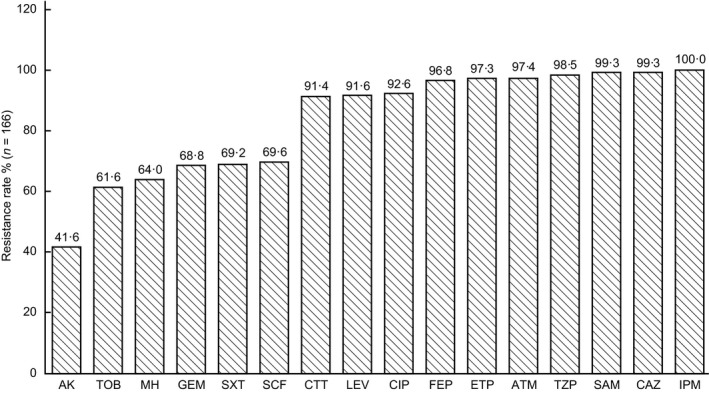
Susceptibility of CRKP to antibiotics.
Figure 2.
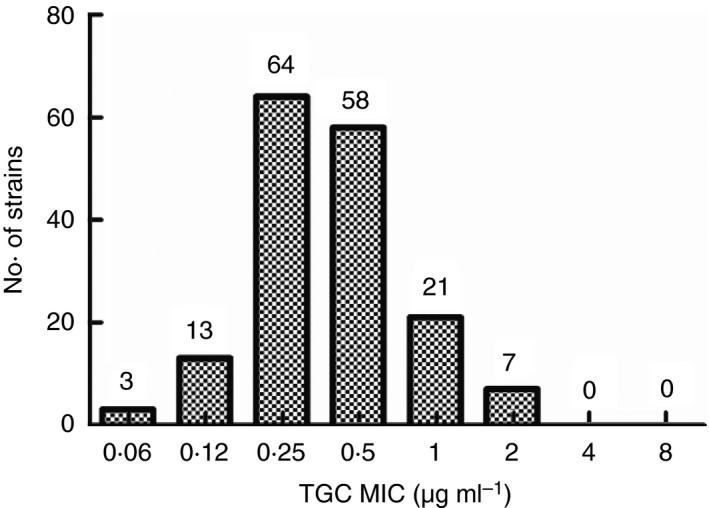
MICs of tigecycline against CRKP.
Sequence alignment and phylogenetic analysis
We compared our sequence analysis to the MLST database and identified a total of 24 ST strains (ST1, ST11, ST15, ST23, ST34, ST36, ST37, ST147, ST231, ST313, ST369, ST550, ST571, ST859, ST873, ST1224, ST1517, ST1647, ST2230, ST2235, ST2370, ST2894, ST3034 and ST3087). These strains were divided into two distinct groups: group 1 contained 18 strains (ST1, ST11, ST15, ST23, ST34, ST36, ST37, ST147, ST231, ST313, ST550, ST859, ST873, ST1517, ST2370, ST2894, ST3034 and ST2230), as shown in the phylogenetic analysis in Fig. 3; whereas group 2 contained six strains (ST369, ST571, ST1224, ST1647, ST2235 and ST3087). The most abundant ST strains of carbapenem‐resistant K. pneumoniae in our hospital were as follows: ST11, 80·12% (133/166); ST15, 3·01% (5/166); ST147, 2·41% (4/166); and ST23 1·81% (3/166). Moreover, ST258 was determined to be the network download strain (BAA1705), and ST37 was the outbreak strain in the neonatal ICU (NICU). (Chen et al. 2019).
Figure 3.
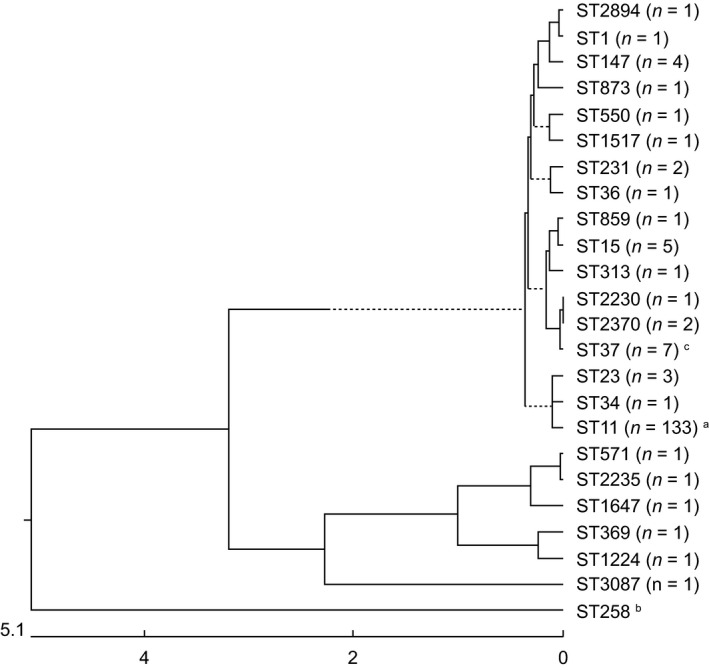
MLST and phylogenetic trees of CRKP. a, ST11 is the most common type; b, ST258 is the network download strain BAA1705; c, outbreak strains in the NICU.
CRKP resistance genes
The results of drug resistance gene analysis are shown in Fig. 4. KPC‐2 was identified in 75·90% (126/166), CTX‐M9 was identified in 68·07% (113/166), CTX‐M1 was identified in 7·83% (13/166), OmpK35 deletion was identified in 4·82% (8/166), OmpK36 deletion was identified in 93·98% (156/166) and OmpK37 deletion was identified in 5·42% (9/166) of all strains. IPM‐4, NDM‐1 and OXA‐48 were not detected.
Figure 4.
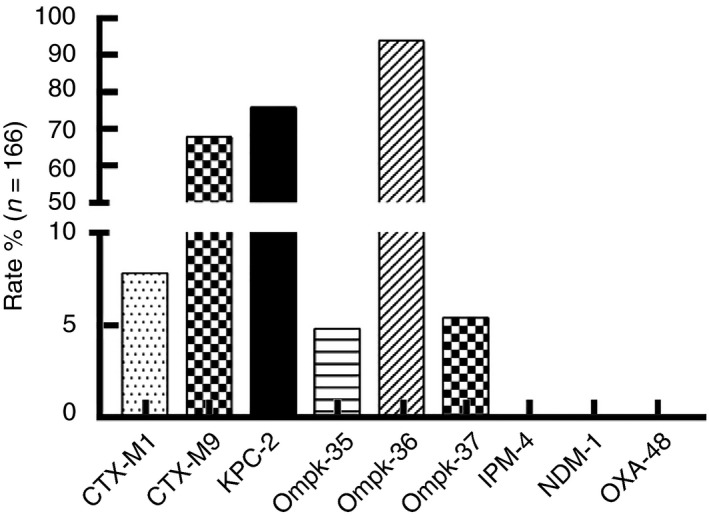
CRKP resistance genes.
Discussion
Klebsiella pneumoniae is the leading cause of nosocomial infections worldwide, and its incidence is on the rise (Saidel‐Odes and Borer 2013). Although its prevalence is second only to that of Escherichia coli, its drug resistance is broader and more problematic than that of E. coli. Treating CRKP is thus exceedingly difficult. Xu et al. (2017) retrieved reports published before December 22, 2015, that included terms such as ‘Klebsiella pneumoniae’, ‘drug resistance’, and ‘carbapenemase’, ‘imipenem’, ‘meropenem’, or ‘ertapenem’. Their systematic review and meta‐analysis of the mortality rates of infected patients unveiled that the mortality rate for 2462 patients with CRKP was 42·14%, whereas that for patients with carbapenem‐sensitive K. pneumoniae infection was 21·16%. Moreover, the mortality rates of patients with bloodstream or urinary tract infections were 54·30 and 13·52% respectively. The mortality rates of patients admitted to the ICU or receiving solid organ transplantation were 48·9 and 43·13% respectively. Geographical differences have also been observed: North American, South American, European and Asian studies have reported mortality rates of 33·24, 46·71, 50·06 and 44·82% respectively. Therefore, it is crucial to monitor drug sensitivity, as well as drug resistance genes and the molecular epidemiological characteristics of CRKP.
A total of 166 CRKP strains were collected in this study, primarily from patients over 60 years of age, which is consistent with previous reports (van Duin et al. 2014; Meng et al. 2019). Han et al. (2017) reported a CRKP prevalence of 24·6% (946/3846); among these, 507 (53·6%) were from respiratory specimens, 350 (37·0%) from the urinary system and 9 (9·4%) from blood, which is also consistent with our research. The detection rate of CRKP in our hospital was 15·74%, which was higher than the averages in the Fujian province (10·7%) and all of China in 2017 (9·0%). There were significant differences between regions in our study; Shanghai had the highest resistance rate (26·9%), whereas Qinghai had the lowest (0·3%). Our results were quite different from those of the sensitivity analysis that included 244 CRKP strains performed by Li et al. (2019), who concluded that, because of its high sensitivity rate (95·5%), tigecycline is still the best choice for CRKP. The rate of sensitivity to the remaining antibiotics was only slightly higher than 30%, except for fosfomycin, which had a sensitivity rate of 35·2%. The differences in sensitivity could be related to the different CRKP strains that are prevalent in each region. Tigecycline‐resistant K. pneumoniae strains were not detected, which may be because the period of initial use of tigecycline is short, and it is a specific antibiotic in our hospital. In addition, to detect the MIC of tigecycline, the appropriate detection method should be selected. When the method shows mediation or drug resistance, the micro‐broth dilution method or E‐Test strip is required to determine the exact MIC.
Globally, the most common CRKP strains are ST11 and ST258; ST11 is the main ST in China (64·2%), and ST258 is the most common in the United States (70·0%) (Kitchel et al. 2009; Andrade et al. 2011; Qi et al. 2011). Previous studies (Chen et al. 2014) have shown that ST258 is a hybrid clone: 80% of the genome originates from an ST11‐like strain, and 20% originates from an ST442‐like strain. Lu et al. (2018) studied 174 CRKP strains from hospitalized patients at the Affiliated Hospital of Sun Yassin University Medical Sciences, and approximately 98·0% of CRKP strains belonged to ST11. The proportion of the ST11 strain in our hospital was similar to that described by Shu et al. (2019), who reported a proportion of 78·0%. MLST and evolutionary tree typing showed that the strains were mainly divided into two branches. No other ST has caused an epidemic, except for a NICU outbreak of ST37 observed in our previous study (Chen et al. 2019).
In our hospital, the KPC‐2 gene was most commonly detected (75·9%), followed by the CTX‐M9 gene (68·07%). OmpK36 deletion was observed at a rate of 93·98%; however, IPM‐4, NDM‐1 and OXA‐48 were not detected. Galani et al. (2019) detected 300 CRKP strains in hospitals across Greece and found KPC‐2 (66·7%), NDM (16·7 %), VIM (7%) and OXA‐48 (4·0%); 14 strains carried both KPC and VIM (4·7%), two strains carried both NDM and OXA (0·7%) and one strain carried both KPC and OXA (0·3%) resistance genes. Tian et al. (2018) retrospectively analysed the drug‐resistant phenotypes and clinical molecular epidemiology of 170 CRKP strains in Shanghai from January 2016 to December 2017 and found that the most abundant gene was bla OXA‐232 (42·35%), followed by bla NDM‐1 (20·59%), bla KPC‐2 (17·65%), bla NDM‐5 (16·47%) and bla IMP‐4 (1·18%). Furthermore, they found that the genes present were related to different ages and epidemic strains in different regions. The major drug resistance gene in China and the United States is KPC, found in >90% of strains, with KPC‐2 being the most common. The NDM‐1 carrier rate is <10% in China and <5% in America (Deleo et al. 2014; Li et al. 2014). We suggest that screening be performed to detect other drug resistance genes to better understand the epidemiological trends of drug‐resistant strains and provide a theoretical basis for infection control and clinical treatment.
Here common drug resistance genes were detected. However, the expression of efflux pump genes and biofilm formation among the 166 CRKP strains were not analysed. The drug resistance gene carrier vehicles, such as plasmid vectors, insert sequences, integrins or other drug resistance gene mobile elements, were not assessed in this study. Therefore, the present investigation of CRKP strain carbapenem resistance mechanisms must be complemented by further studies. Future work will aim to develop clinical solutions to treat and prevent multi‐ and pan‐drug‐resistant bacterial infections.
Funding details
This work was supported by the National Major Science and Technology Project for the Control and Prevention of Major Infectious Diseases of China (grant no. 2017ZX10103004), Fujian Medical Science and Technology Innovation Project (grant no. 2016Y9005), Fujian Natural Science Foundation (grant no. 2019J01178), Youth Scientific Research Project of Fujian Provincial Health and Family Planning Commission (grant no. 2017‐1‐9) and High‐level hospital foster grants from Fujian Provincial Hospital (grant no. 2019HSJJ11).
Conflict of Interest
No conflict of interest declared.
Acknowledgements
We are grateful to all those who have participated in our research and to those who have funded our work. Thanks to the language editing company, Editage (www.editage.com).
Dongjie Chen, Hongru Li and Yunan Zhao are co‐first authors.
Contributor Information
Y. Chen, Email: falinchen@126.com.
F. Chen, Email: falinchen@126.com.
References
- Andrade, L.N. , Curiao, T. , Ferreira, J.C. , Longo, J.M. , Clímaco, E.C. , Martinez, R. , Bellissimo‐Rodrigues, F. , Basile‐Filho, A. et al (2011) Dissemination of blaKPC‐2 by the spread of Klebsiella pneumoniae clonal complex 258 clones (ST258, ST11, ST437) and plasmids (IncFII, IncN, IncL/M) among Enterobacteriaceae species in Brazil. Antimicrob Agents Chemother 55, 3579–3583. 10.1128/AAC.01783-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D. , Hu, X. , Chen, F. , Li, H. , Wang, D. , Li, X. , Wu, C. , Li, N. et al (2019) Co‐outbreak of multidrug resistance and a novel ST3006 Klebsiella pneumoniae in a neonatal intensive care unit: a retrospective study. Medicine (Baltimore) 98, e14285 10.1097/MD.0000000000014285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L. , Mathema, B. , Pitout, J.D. , DeLeo, F.R. and Kreiswirth, B.N. (2014) Epidemic Klebsiella pneumoniae ST258 is a hybrid strain. MBio 5, 10.1128/mBio.01355-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleo, F.R. , Chen, L. , Porcella, S.F. , Martens, C.A. , Kobayashi, S.D. , Porter, A.R. , Chavda, K.D. , Jacobs, M.R. et al (2014) Molecular dissection of the evolution of carbapenem‐resistant multilocus sequence type 258 Klebsiella pneumoniae . Proc Natl Acad Sci USA 111, 4988–4993. 10.1073/pnas.1321364111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diancourt, L. , Passet, V. , Verhoef, J. , Grimont, P.A. and Brisse, S. (2005) Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol 43, 4178–4182. 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galani, I. , Nafplioti, K. , Adamou, P. , Karaiskos, I. , Giamarellou, H. and Souli, M. (2019) Nationwide epidemiology of carbapenem resistant Klebsiella pneumoniae isolates from Greek hospitals, with regards to plazomicin and aminoglycoside resistance. BMC Infect Dis 19, 167 10.1186/s12879-019-3801-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, J.H. , Goldstein, E.J. , Wise, J. , Bilker, W.B. , Tolomeo, P. and Lautenbach, E. (2017) Epidemiology of carbapenem‐resistant Klebsiella pneumoniae in a network of long‐term acute care hospitals. Clin Infect Dis 64, 839–844. 10.1093/cid/ciw856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitchel, B. , Rasheed, J.K. , Patel, J.B. , Srinivasan, A. , Navon‐Venezia, S. , Carmeli, Y. , Brolund, A. and Giske, C.G. (2009) Molecular epidemiology of KPC‐producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob Agents Chemother 53, 3365–3370. 10.1128/AAC.00126-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B. , Xu, X.H. , Zhao, Z.C. , Wang, M.H. and Cao, Y.P. (2014) High prevalence of metallo‐β‐lactamase among carbapenem‐resistant Klebsiella pneumoniae in a teaching hospital in China. Can J Microbiol 60, 691–695. 10.1139/cjm-2014-0291. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Shen, H. , Zhu, C. and Yu, Y. (2019) Carbapenem‐resistant Klebsiella pneumoniae infections among ICU admission patients in Central China: prevalence and prediction model. Biomed Res Int 2019 10.1155/2019/9767313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, M.C. , Tang, H.L. , Chiou, C.S. , Wang, Y.C. , Chiang, M.K. and Lai, Y.C. (2018) Clonal dissemination of carbapenemase‐producing Klebsiella pneumoniae: two distinct sub‐lineages of sequence type 11 carrying blaKPC‐2 and blaOXA‐48. Int J Antimicrob Agents 52, 658–662. 10.1016/j.ijantimicag.2018.04.023. [DOI] [PubMed] [Google Scholar]
- Meng, X. , Yang, J. , Duan, J. , Liu, S. , Huang, X. , Wen, X. , Huang, X. , Fu, C. et al (2019) Assessing molecular epidemiology of carbapenem‐resistant Klebsiella pneumoniae (CR‐KP) with MLST and MALDI‐TOF in Central China. Sci Rep 9, 2271 10.1038/s41598-018-38295-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, Y. , Wei, Z. , Ji, S. , Du, X. , Shen, P. and Yu, Y. (2011) ST11, the dominant clone of KPC‐producing Klebsiella pneumoniae in China. J Antimicrob Chemother 66, 307–312. 10.1093/jac/dkq431. [DOI] [PubMed] [Google Scholar]
- Saidel‐Odes, L. and Borer, A. (2013) Limiting and controlling carbapenem‐resistant Klebsiella pneumoniae . Infect Drug Resist 7, 9–14. 10.2147/IDR.S44358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu, L.B. , Lu, Q. , Sun, R.H. , Lin, L.Q. , Sun, Q.L. , Hu, J. , Zhou, H.W. , Chan, E.W. et al (2019) Prevalence and phenotypic characterization of carbapenem‐resistant Klebsiella pneumoniae strains recovered from sputum and fecal samples of ICU patients in Zhejiang Province, China. Infect Drug Resist 12, 11–18. 10.2147/IDR.S175823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen, G.S. (2018) Antimicrobial resistance surveillance in Europe and beyond. Euro Surveill 23, 10.2807/1560-7917.ES.2018.23.42.1800560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, D. , Pan, F. , Wang, C. , Sun, Y. and Zhang, H. (2018) Resistance phenotype and clinical molecular epidemiology of carbapenem‐resistant Klebsiella pneumoniae among pediatric patients in Shanghai. Infect Drug Resist 11, 1935–1943. 10.2147/IDR.S175584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Duin, D. , Perez, F. , Rudin, S.D. , Cober, E. , Hanrahan, J. , Ziegler, J. , Webber, R. , Fox, J. et al (2014) Surveillance of carbapenem‐resistant Klebsiella pneumoniae: tracking molecular epidemiology and outcomes through a regional network. Antimicrob Agents Chemother 58, 4035–4041. 10.1128/AAC.02636-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, L. , Sun, X. and Ma, X. (2017) Systematic review and meta‐analysis of mortality of patients infected with carbapenem‐resistant Klebsiella pneumoniae . Ann Clin Microbiol Antimicrob 16, 18 10.1186/s12941-017-0191-3. [DOI] [PMC free article] [PubMed] [Google Scholar]


