Summary
Fabeae legumes such as pea and faba bean form symbiotic nodules with a large diversity of soil Rhizobium leguminosarum symbiovar viciae (Rlv) bacteria. However, bacteria competitive to form root nodules (CFN) are generally not the most efficient to fix dinitrogen, resulting in a decrease in legume crop yields. Here, we investigate differential selection by host plants on the diversity of Rlv.
A large collection of Rlv was collected by nodule trapping with pea and faba bean from soils at five European sites. Representative genomes were sequenced. In parallel, diversity and abundance of Rlv were estimated directly in these soils using metabarcoding. The CFN of isolates was measured with both legume hosts. Pea/faba bean CFN were associated to Rlv genomic regions.
Variations of bacterial pea and/or faba bean CFN explained the differential abundance of Rlv genotypes in pea and faba bean nodules. No evidence was found for genetic association between CFN and variations in the core genome, but variations in specific regions of the nod locus, as well as in other plasmid loci, were associated with differences in CFN.
These findings shed light on the genetic control of CFN in Rlv and emphasise the importance of host plants in controlling Rhizobium diversity.
Keywords: competitiveness, genospecies, nod genes, nodules, Pisum sativum, Rhizobium leguminosarum symbiovar viciae, symbiosis, Vicia faba
Introduction
Rhizobia are soil bacteria that have the ability to form root nodules with legumes. These symbiotic organs fix atmospheric dinitrogen (N2) into organic forms that relieve the plant of nitrogen limitation. The importance of biological nitrogen fixation (BNF) for agriculture and the nitrogen cycle in the biosphere is well recognised. Rhizobium leguminosarum symbiovar viciae (Rlv) is the specific symbiont of legumes of the tribe Fabeae (formerly Vicieae) that include crops of agronomic interest such as pea (Pisum sativum L.) and faba bean (Vicia faba L.). Rlv bacteria belong to the same species complex as Rhizobium leguminosarum symbiovars trifolii (Rlt) and phaseoli (Rlp), which nodulate clover (Trifolium species) and Phaseolus bean (Phaseolus vulgaris L.), respectively. Rlv genome sequences belong to at least five distinct genospecies that are not symbiovar specific (Kumar et al., 2015). The genetic determinants of the symbiosis are plasmid encoded (Young, 2016; Andrews et al., 2018), so symbiovars reflect the symbiosis plasmid rather than the chromosome diversity. The nodD gene, encoding a regulator of symbiosis genes, has frequently been used as a marker to discriminate Rlv bacteria based on their symbiotic capacities (Zézé et al., 2001; Laguerre et al., 2003). Indigenous populations of Rlv are frequent in soils (Laguerre et al., 2003; Mutch & Young, 2004). Plant benefits rely on both legume and Rhizobium traits that operate at different stages of interaction. Early stages of symbiotic association require Rhizobium and host plant compatibility to form root nodules together (ability to form nodules, AFN) that is generally assessed by inoculating the bacteria alone on its host. Nevertheless, in soil there are generally several compatible Rlv bacteria for a host plant and the bacteria which finally form nodules vary among multiple possible associations. Compatible combinations display various levels of competitive to form root nodules (CFN). Pioneer studies showed that CFN was not associated with the ability to fix nitrogen in Sinorhizobium/Medicago sativa associations (Amarger, 1981). Co‐inoculation of a mixture of diverse pea‐nodulating Rlv strains on a panel of 104 pea genotypes, representative of the variability of the genus Pisum, revealed that the CFN varied greatly depending on both pea and Rlv genotypes and it was not associated with BNF efficiency (Bourion et al., 2018). Plants may sanction BNF‐inefficient partners by reducing the number of cultivable cells present in nodules (Kiers et al., 2003) and preferentially stimulate the growth of symbiotic organs formed with most BNF‐efficient rhizobia (Laguerre et al., 2012). Nevertheless, BNF may be suboptimal because of the presence of poorly effective but highly competitive Rlv that outcompete BNF‐efficient compatible bacteria (Laguerre et al., 2003). Despite the potential interest for inoculation of pea with effective Rlv strains (Bremer et al., 1988; Fesenko et al., 1995; McKenzie et al., 2001), inoculant strains are frequently outcompeted by naturally occurring ineffective rhizobia (Meade et al., 1985). Understanding the determinants of CFN will allow the selection of bacteria for improved inoculation strategies (Triplett & Sadowsky, 1992; Laguerre et al., 2003).
The mechanisms controlling CFN have not been fully elucidated. There is evidence for microbe–microbe interactions such as antibiosis and quorum sensing that are potentially involved in CFN (Robleto et al., 1998; McAnulla et al., 2007; Naamala et al., 2016). It is likely that differential proliferation of competitive bacterial genotypes in the host rhizosphere contributes to CFN, but the association between symbiosis plasmid diversity and partner choice supports the hypothesis that plant–Rhizobium interaction mechanisms are major drivers of CFN (Moawad, Ellis, & Schmidt, 1984; Laguerre et al., 2003). They involve the synthesis and secretion by rhizobia of lipo‐oligosaccharides called Nod Factors (NFs). NFs are recognised by plasma membrane localised plant receptors and trigger pathways that activate nodule formation. This interaction can confer species specificity (Dénarié et al., 1992; Mergaert et al., 1997; Broughton et al., 2000; Radutoiu et al., 2007). In rhizobia transcription of the nod genes is controlled by NodD, a transcriptional regulator, activated by flavonoids secreted by the plant (Broughton et al., 2000). The common rhizobial genes nodABC are responsible for biosynthesis of the core NF, and nodIJ are involved in their secretion from the bacteria (Mergaert et al., 1997). NFs from different rhizobia share the same chitin‐like N‐acetyl glucosamine backbone with a fatty acyl chain at the nonreducing end, but differ in the backbone length, the size and the saturation of the fatty acyl chain, as well as substitutions such as glycosylation, acetylation and sulfation (Dénarié et al., 1992). These differences are encoded by accessory nod genes and may result in variations of the interactions among rhizobial and plant species (Debellé et al., 1988; Surin & Downie, 1988; Downie & Surin, 1990; Surin et al., 1990; Lewis‐Henderson & Djordjevic, 1991a,b; Spaink et al., 1991; Bloemberg et al., 1995). The nodX gene of some Rlv, such as the strain TOM, is responsible for the acetylation of the NFs at the reducing terminus, and this modification allows nodulation with specific pea genotypes from the Middle East which have a SYM2 allele encoding a LysM‐RLK able to recognise this modified NF (Firmin et al., 1993; Hogg et al., 2002; Sulima et al., 2017). In addition, bacterial surface polysaccharide recognition by specific LysM receptors modulates plant‐bacterial recognition and potentially CFN (Kawaharada et al., 2015). Secretion of bacterial effectors by the type I and type III bacterial secretion systems have been implicated in modulating the partner choice (Devine et al., 1980; Sutton et al., 1994; Deakin & Broughton, 2009; Linhartová et al., 2010; Yang et al., 2010).
In this study, we focus on early partner choice and investigate the genetic basis of CFN of Rlv with pea and faba bean (the most cultivated Fabeae crops in Europe) in natural populations of bacteria. The strategy was to characterise extensively the large diversity of Rlv from various geographic sites (directly in soil or using trapping in pea and faba bean nodules), to measure CFN with both hosts, and to associate Rlv genomic variation with pea/faba bean CFN phenotype in order to identify candidate genes and/or markers controlling this early symbiotic trait.
Materials and Methods
Bacterial collection
Rhizobia were isolated from an agricultural site in each of five European countries (France, Spain, Sweden, Serbia and the Czech Republic; Supporting Information Table S1). Soils were collected from each site at the beginning of the growing season by collecting five subsamples of top soil (1–10 cm) from 6 to 15 plots belonging to diverse culture systems (Table S2). Pooled samples were formed by combining soils from the different plots at each site. Pool samples have relatively homogenous chemical compositions, except the Spanish soil that was more clayey than others. Rlv bacteria were trapped by growing pea or faba bean (Table S3) in 1 l pots filled with a mixture of soil and sterile siliceous sand. For each pool, 100 nodules were sampled individually from 32‐d‐old plants, surface sterilised in 3% sodium hypochlorite for 3 min, washed four times in sterile water, crushed in 12.5% glycerol and plated on yeast‐extract–mannitol (YEM) broth. Bacteria were isolated from all pooled soil samples and both hosts except the Spanish pool (unsuccessful trapping with pea). After three cycles of single colony purification, 210 isolates were collected (130 from faba bean and 80 from pea nodules). A part of the coding sequence of the nodD gene was amplified by PCR using specific primers (Table S4) and sequenced (Genoscreen, Lille, France; https://www.genoscreen.fr/).
The diversity and abundance of nodD sequences in soil
Soil subsamples were taken from each of the plots at each European site, and of the pooled soil from each site that was used for plant nodulation trapping. Soil was well mixed and 100 000 copies of the QQstd‐nodD artificial template (Table S4) were added to 250 mg soil. This template was based on the nodD sequence of R. leguminosarum sv. viciae strain 3841, retaining matches to the primers, but with each base of the sequence between them replaced with its complement, in order to ensure that the base composition of the amplicon was not altered but the actual sequence resembled no natural sequence. Soil DNA was extracted with the MoBio Powerlyzer Powersoil extraction kit (Quiagen, Manchester, UK). The relative abundance of symbiovar viciae nodD genes in soil samples was estimated by PCR amplification and high‐throughput DNA sequencing using the MAUI‐seq protocol (Fields et al., 2019). The NodD136fwd and NodD136rev primers (Table S4) were designed to amplify an informative sequence 136 bp long (excluding the primers) at the start of the coding sequences of all known nodD genes of symbiovar viciae (based on published and our unpublished data), but not of the related symbiovar trifolii or any other published DNA sequences. These sequences were incorporated in the extended primers (including linkers and a 12‐nt random unique molecular identifier) used in the MAUI‐seq method. After Illumina sequencing, the reads were processed according to the MAUI‐seq protocol (Fields et al., 2019) to estimate the relative abundance of NodD sequence variants, and their absolute abundance was estimated by comparison with the counts for the spiked artificial template.
Effectiveness and competitiveness measurement
Plant varieties regularly cultivated at the five European sites were used (Table S3). Seeds were surface sterilised in 3% Ca‐hypochlorite solution for 10 min. Plants were grown in a glasshouse (16 h : 8 h, 22°C: 18°C, day : night cycle) in 2 l pots filled with sterilised siliceous sand. High‐pressure sodium lamps with a mean photosynthetically active radiation of 250 µmol photons m−2 s−1 were used to complement natural light at morning and evening. Pots were supplied with N‐free nutrient solution (K2HPO4 0.8 mM, MgSO4 1 mM, K2SO4 0.6 mM, CaCl2 2 mM, NaCl 0.2 mM adjusted to pH 6.5) twice a week. Plants were inoculated at sowing with 400 ml suspension of bacteria of interest (107 CFU ml−1) grown in YEM broth. For effectiveness experiments, shoots were collected from 6‐wk‐old plants inoculated with single Rlv isolates or well characterised control strains (P221 for pea and 3841 for faba bean). Relative normalised biomass was calculated by dividing shoot biomass by the shoot biomass obtained with the control strain in a parallel experiment on the same plant variety. For competitiveness experiments, the strain of interest was co‐inoculated in equal proportion with a reference strain carrying antibiotic resistance. P1NP3CSt (Laguerre et al., 2007) and P1NP2HSp (spontaneous mutant of P1NP2H; Laguerre et al., 2003), resistant respectively to streptomycin (100 µg ml−1) and spectinomycin (100 µg ml−1), were used as reference with, respectively, pea and faba bean. Nodules (50–96 from three individual plants) were surface sterilised as mentioned above, crushed individually in 12.5% glycerol and the suspension streaked onto YEM agar plates. The bacteria present in each nodule are generally clonal and multiple infections (mixed nodules) are below 1% (Amarger, 1981). The number of nodules occupied by the reference strain was scored using antibiotic resistance. The CFN was defined as the ratio of nodules formed with the strain of interest to the reference strain. For relative comparison on a given host, CFN distribution was then centred on the median of the host dataset (i.e. 24 CFN tests for faba bean, 26 for pea).
Genome sequencing, genomic and genetic association analysis
Bacterial genomes were sequenced by MicrobesNG (Birmingham, UK, https://microbesng.uk/) on an Illumina HiSeq platform using a 250 bp paired‐end protocol. Genomic DNA libraries were prepared using the Nextera XT Library Prep Kit (Illumina, San Diego, CA, USA). High‐quality paired‐end reads were assembled using the Galaxy/BBRIC pipeline (https://bbric-pipelines.toulouse.inra.fr/) and genome annotations were performed using EuGene‐PP (Sallet et al., 2014) and Rast (http://rast.nmpdr.org/; Tables S8–S10). Pairwise average nucleotide identity (ANI) values were calculated using jspecies software (http://jspecies.ribohost.com/jspeciesws), and dDDH values were calculated using the Ggdc2.1 webtool (Meier‐Kolthoff et al., 2013). Heatmaps were built using the pheatmap R package. Core and accessory genomes of Rlv bacteria were generated using the SPINE/AGEnt webtools (http://vfsmspineagent.fsm.northwestern.edu/index_age.html; Ozer et al., 2014) using the default parameters (except minimum core genome segment size to output = 200 bp). Genomic regions specific to strains of interest were selected and their presence/absence checked by a Blast search in their genomes. The nucleotide sequences of the nod genes were aligned using ClustalOmega webtool (https://www.ebi.ac.uk/services) and biallelic single nucleotide polymorphisms (SNP) were selected. A Mixed Linear Model implemented in the Emmax software (Kang et al., 2010) was used to detect associations between CFN values and SNPs of the nod region. Briefly, the model, which is routinely used in plant genome‐wide association studies, estimates allelic effect at each SNP accounting for the genetic relationships between individuals which are described by a kinship matrix calculated using the whole SNP dataset. Each allelic effect is then tested for significance using an F‐test, producing then a P‐value for each SNP tested.
Results
Diversity of nodD gene sequences
A nodD sequence was successfully amplified from each of the 210 strains that were isolated from root nodules on pea and faba bean plants grown in soils from five European countries (Table S5). The 31 distinct sequences (alleles) were more similar to symbiovar viciae than to trifolii (strain WSM1689) or phaseoli (strain 4292; Fig. 1). They fall into seven clades or ‘nodD groups’ (bootstraps > 82%; Figs 1, S1; Table S5). The two main nodD groups are B1 (89 isolates) and A1 (91 isolates). The other five nodD groups (31 isolates) are closely related to these two groups. The strains isolated in the present study represent all known nodD groups and most of the known nodD alleles (23/31). They covered and enriched the Rlv diversity previously characterised on a limited number of bacteria. Although there were differences in their relative abundance, the two main nodD groups were found at all the European locations (Fig. 2; Tables S5, S6).
Figure 1.
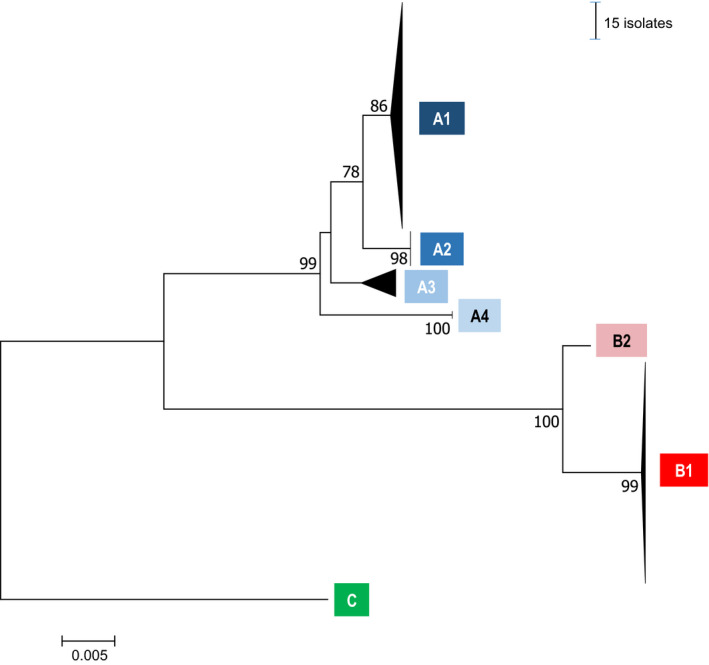
Phylogenetic tree based on nodD gene sequences of the 210 pea and faba bean Rlv isolates. Blue, red and green colour boxes define the A, B and C nodD groups respectively. Horizontal scale indicate the number of base substitutions per site. Vertical scale is related to the number of isolates present in nodD groups. Detailed phylogenetic tree is provided in the Supporting Information Fig. S1.
Figure 2.
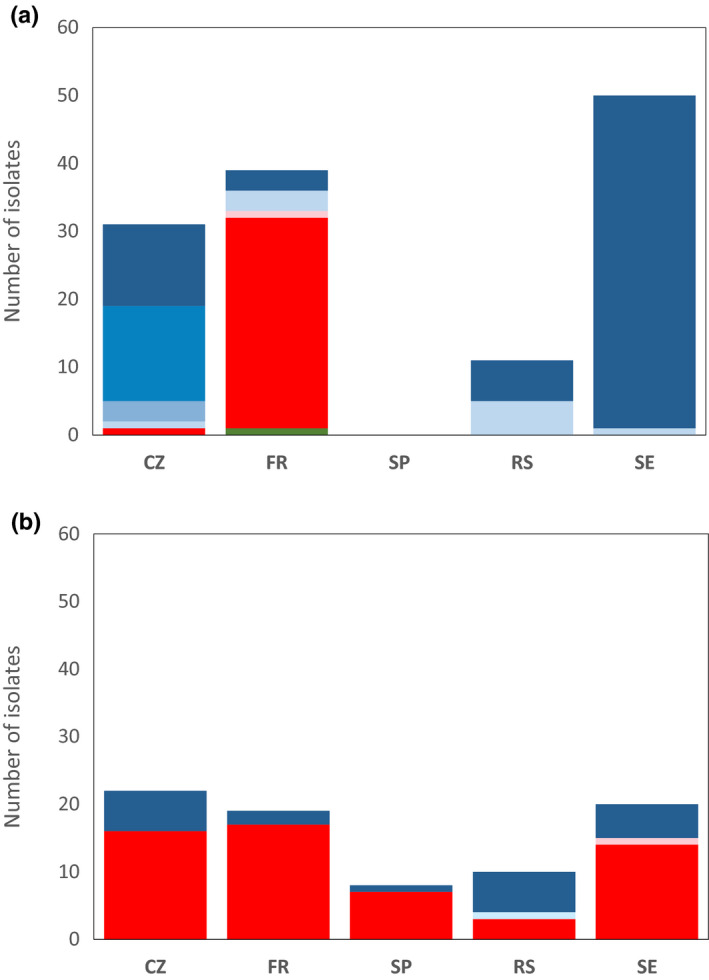
Diversity of the Rlv isolates as a function of their host and their geographic origin. (a) Isolates from pea. (b) Isolates from faba bean. Colours represent the nodD groups. A1–A4: dark to pale blue; B1 and B2: dark and pale red; C: green (see Fig. 1). Origins are Sweden (SE), France (FR), Spain (SP), Czech Republic (CZ) and Serbia (RS). More details are available in Supporting Information Tables S5 and S6.
Plant hosts select different nodD genotypes from the soil population
The relative frequency of nodD groups among rhizobia isolated from pea and faba bean root nodules was very different, even though the plants were exposed to the same set of soil populations. From all soils, the frequency of isolates belonging to the group B1 was higher in faba bean (72%) than in pea nodules (24%). An opposite result was observed for isolates belonging to the group A1 (Fig. 2; Table S6). To understand whether pea or faba bean, or both, are being selective in their choice of rhizobia, the composition of the soil populations from which they trapped symbionts is required. We used specific PCR primers for symbiovar viciae to amplify a part of the nodD sequence from DNA extracted directly from soil samples. The most abundant sequences recovered from the soil samples were in the nodD group A1, which was also the most abundant in the isolates from these soils (Fig. 3). The second group in abundance was B1, which was the second most abundant type among all isolates. Together, these two nodD groups represent 89.5% of all nodD sequences recovered from the soil. Only three other sequences exceeded 1% in overall frequency, and all of these were also found among the isolates. This demonstrates that direct amplification from the soil does recover relevant sequences, and that isolates from nodules include most of the Rlv diversity available in the soil. Similar sequences were found in all European soils, but their relative abundance varied widely even among soils sampled from different plots at the same location (Fig. 3a), although replicate analyses of individual soil samples were reproducible (Fig. 3b). Quantitation using an artificial template added to soils immediately before DNA extraction did not work perfectly, as this sequence was not recovered from a few of the samples, perhaps because the DNA was rapidly immobilised or lost. However, results for the remaining samples (Fig. S2a) indicated that soils with a high abundance of nodD sequences were overwhelmingly dominated by group A1 (Fig. S2b), which is consistent with the observation that the soil mixes all had a high frequency of A1 (Fig. 3c). Group B sequences were in the minority in most soils, indicating that the high frequency of group B among faba bean nodule isolates (Fig. 2b) was the result of strong discrimination by this host.
Figure 3.
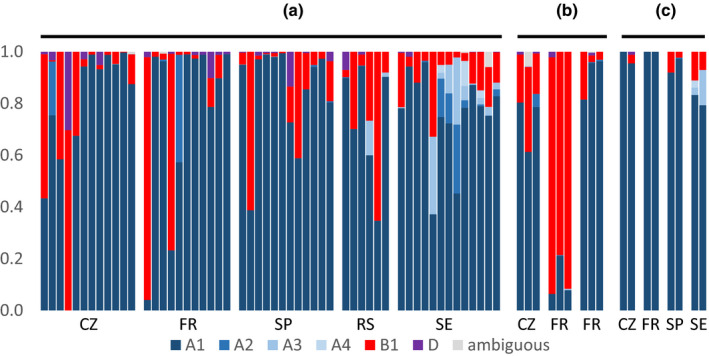
Frequencies of nodD sequences amplified directly from soils. (a) Samples from individual plots of five sites located in Sweden (SE), France (FR), Spain (SP), Czech Republic (CZ) and Serbia (RS). In each site, soils samples were collected from plots belonging to diverse culture systems (Supporting Information Table S2). (b) Triplicate analyses of three additional plots. (c) Duplicate samples of the mixed soils (pooled samples of the different plots of each site) used for nodulation of host plants. Group D is a clade of sequences that have not been found in nodule isolates. Ambiguous sequences are close to either A or B groups but cannot be placed in a specific group.
Symbiotic effectiveness is not related to nodD genotype
Symbiotic effectiveness was judged by plant shoot biomass production on N‐free growth medium for a representative subset of strains inoculated onto their host of origin. Large variations of biomass were observed between isolates on both pea and faba bean but this variation was not significantly explained either by nodD groups or by geographic origins (ANOVA P < 0.05, Table S7).
Genome diversity of the Rlv isolates
Fifty isolates representative of the nodD sequence diversity as well as geographical origins were sequenced (Table S8). Based on ANI (Fig. 4) and digital DNA–DNA hybridisation (dDDH; Fig. S3) comparisons, these isolates are mainly related to three genospecies of R. leguminosarum, gsE (27 strains), gsC (10 strains) and gsB (3 strains), that have been previously identified in European isolates (Kumar et al., 2015; Jain et al., 2018; Cavassim et al., 2019). Eight strains belonged to two other genospecies that were not observed in these previous studies. These two new genospecies gsF‐1 and gsF‐2 include respectively the R. leguminosarum strain TOM (Firmin et al., 1993; 5 strains) and the strain R. laguerreae FB206 (Saïdi et al., 2014; 3 strains). The last two strains (CZP3G4 and SEF4G12) display only low genomic similarities with R. leguminosarum bacteria (88% < ANI < 89%) but are further distinct from other Rhizobium species (González et al., 2019).
Figure 4.
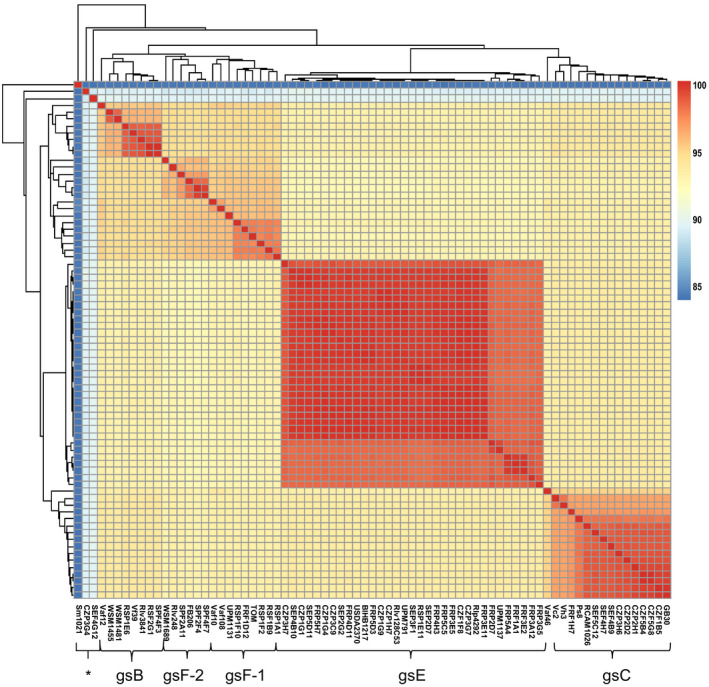
Genomic diversity of Rhizobium leguminosarum sv. viciae isolates. Hierarchical clustering and heatmap based on average nucleotide identity (ANI) values. Genospecies gsF‐1 and gsF‐2 have been defined based on an ANI threshold of 95%, and gsB, gsC and gsE correspond to those previously defined by Kumar et al. (2015). Already published genomes of R. leguminosarum sv. viciae are included, as well as examples of sv. trifolii (WSM1689) and sv. phaseoli (4292). Sinorhizobium meliloti 1021 is used as outgroup. Asterisk indicate isolates putative new genospecies, outside the R. leguminosarum species complex (ANI < 90%).
All the Rlv genomes had the canonical cluster of nodulation genes, arranged in several different operons: nodABCIJ(X), nodD, nodFEL nodMN(T) and nodO. A phylogeny based on the concatenated sequences of these genes was very similar to that based on nodD alone, and defined similar clusters, except that group C is split (Fig. S4). It is notable that there is no association between the nodD groups and the genospecies (Table S8).
Competitiveness for nodulation is host specific and associated with nod genotype
Although it is generally expected that Rlv bacteria form nodules with all legumes within the Fabeae tribe (including pea and faba bean), isolates may display differences in their CFN, depending on hosts. To test this hypothesis, we selected a subset of 33 pea or faba bean isolates that efficiently fix dinitrogen with their initial host when inoculated alone (Table S7), and that are representative of the genomic diversity of Rlv (Fig. 4). Each was mixed individually with a reference strain before inoculation on its original plant host species. Bacterial densities were equal and saturating (> 107 cfu ml−1) to prevent the potential effects of differential bacterial growth on nodulation performance. CFN was estimated from the ratio of nodules formed (Table 1). A wide range of CFN index values was found, especially on pea where the CFN index ranged from 0 to 3.37 (Table 1). There were host‐dependent CFN differences between the nodD groups A1 and B1. On faba bean, the median CFN for nodD group B1 strains was 1.06 and for A1 strains it was 0.55 (Wilcoxon test; P‐value = 0.0378). By contrast, the median CFN of B1 strains on pea was only 0.14, whereas for A1 strains it was 1.28 (Wilcoxon test; P‐value = 0.0218). The results were consistent with the higher proportion of B1 strains isolated from faba bean nodules than from pea nodules (Figs 1, 2). The trade‐off between competitive performance on pea and on faba bean was confirmed by focusing on 16 of these Rlv isolates that were tested on both host species. None of these was highly competitive on both hosts. A negative relationship between CFN on pea versus on faba bean was systematically found (Table 1; Fig. 5). Nevertheless, there was considerable variation in the CFN of strains within a nodD group, and even those with identical nodD alleles (Table 1). Some strains were uncompetitive on pea (or faba bean), as they formed no nodules in competition with the reference strain (CFN = 0). However, in these cases, their ability to form nodules when inoculated alone was confirmed (except SPFP2A11, which did not nodulate Pisum sativum cv. Lucy, although it was very competitive on faba bean; Fig. S5). Globally, the data indicated an association of pea and faba bean CFN with the nodD groups A1 and B1 respectively (Table 1). However, although all Rlv competitive with faba bean were found in B1 and most of the Rlv competitive with pea belong to A1, some A1 strains were more competitive on faba bean.
Table 1.
Competitive to form root nodules (CFN) of diverse Rlv isolates.
| Strain | Isolated from | Genospecies | nodD group | nodD allele | Normalised CFN | |||
|---|---|---|---|---|---|---|---|---|
| Faba bean | Median | Pea | Median | |||||
| SEF5G12 | faba | a | A1 | 18 | 0.31 | 0.55 | nd | 1.28* |
| SPF4F3 | faba | B | A1 | 18 | 0.6 | 2.41 | ||
| RSF2G1 | faba | B | A1 | 17 | 0.00 | 2.17 | ||
| FRF1H7 | faba | C | A1 | 21 | 0.2 | 1.78 | ||
| CZF5G8 | faba | C | A1 | 26 | 1.38 | 0.38 | ||
| CZF5B4 | faba | C | A1 | 18 | 1.03 | 0.16 | ||
| SEF4H7 | faba | C | A1 | 21 | 0.69 | nd | ||
| RSP1E11 | pea | E | A1 | 26 | 0.49 | 3.37 | ||
| SEP3F1 | pea | E | A1 | 26 | 0.51 | 3.02 | ||
| FRP5H7 | pea | E | A1 | 26 | 1.4 | 0.78 | ||
| RSP1F2 | pea | F‐1 | A1 | 21 | nd | 0.77 | ||
| CZP2D1 | pea | nd | A1 | 22 | nd | 0.06 | ||
| RSP1E6 | pea | B | A4 | 15 | nd | 0.43 | ||
| CZP3C9 | pea | E | A2 | 15 | 1.29 | 0.89 | ||
| CZP1G4 | pea | E | A2 | 14 | 1.69 | 0.5 | ||
| CZP1G9 | pea | E | A3 | 10 | nd | 2.31 | ||
| SEP2G2 | pea | E | A4 | 11 | nd | 1.88 | ||
| FRP5D3 | pea | E | A4 | 10 | nd | 0.00 | ||
| RSP1F10 | pea | F‐1 | A4 | 10 | nd | 1.34 | ||
| SEF4G12 | faba | a | B1 | 6 | 1.45 | 1.06* | 0.79 | 0.14 |
| CZF1B5 | faba | C | B1 | 4 | 0.74 | nd | ||
| SEF4B9 | faba | C | B1 | 4 | 0.71 | nd | ||
| CZF1F8 | faba | E | B1 | 6 | 1.16 | nd | ||
| FRF1A1 | faba | E | B1 | 6 | 0.96 | nd | ||
| FRF3E2 | faba | E | B1 | 4 | 0.42 | nd | ||
| FRP3E5 | pea | E | B1 | 6 | nd | 0.23 | ||
| FRP5C5 | pea | E | B1 | 6 | 1.48 | 0.05 | ||
| FRP5A4 | pea | E | B1 | 6 | 1.33 | 0.00 | ||
| SPF2F4 | faba | F‐2 | B1 | 4 | 1.67 | 0.00 | ||
| SPF2A11b | faba | F‐2 | B1 | 5 | 1.25 | 0.00 | ||
| RSF1B9 | faba | F‐1 | B1 | 6 | 0.96 | 0.27 | ||
| FRF1D12 | faba | F‐1 | B1 | 4 | 0.82 | 0.36 | ||
| FRP3E11 | pea | E | C | 1 | nd | 0.00 | ||
Isolates were co‐inoculated with reference strains P1NP3CSt (pea) and P1NP2HSp (faba bean) onto host plants. CFN is expressed as the ration of nodules formed with the strain of interest to the reference strain. CFN values in bold are for strains on their original host species. nd, not determined.
Genospecies displaying < 90% ANI with all known R. leguminosarum symbiovar viciae (Rlv) sequences.
SPF2A11 was unable to nodulate Pisum sativum cultivar Lucy.
P < 0.05.
Figure 5.
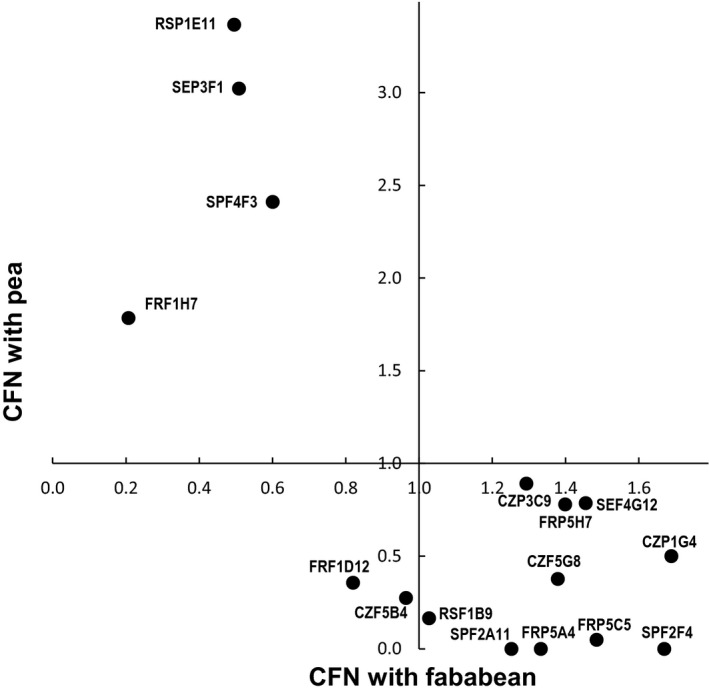
Competitive to form root nodules (CFN) of the Rlv isolates tested with both pea and faba bean. Data were extracted from the Table 1.
Genetic variation associated with differences in the CFN
In order to identify genetic differences that might be responsible for the CFN variation, we first examined structural variation within the nod gene region between bacteria of nodD groups A1 and B1. All the strains had the equivalent set of nod genes, arranged in the same order, except those in nodD group B1 that have the additional gene nodT together with a small gene of unknown function inserted downstream of nodT. Five strains of the group A1 have the nodX gene located downstream of the nodABCIJ operon but this insertion was not associated with pea/faba bean CFN in our experimental conditions (data not shown). The N‐terminal end of NodO in B1 bacteria is also substantially divergent compared with A1 bacteria (only 57% identity is observed in the first 100 amino acids). Both the presence of the nodT gene and the nodO allele variation in B1 strains, compared with A1, is significantly associated with low CFN on pea, but there is no effect on faba bean (P = 0.00553 and P = 0.3694 respectively, Kruskal–Wallis test). It is possible that these structural differences may contribute to the contrasted CFN phenotypes on pea.
The second approach aimed to identify SNPs within the conserved nod genes region, which may be associated with pea/faba bean CFN phenotypes. The two hosts were first considered independently; 24 isolates have been tested on faba bean and 26 on pea. The nod regions of these isolates were aligned to identify the SNPs associated with pea or faba bean CFN values. In total, 1449 and 1785 biallelic SNPs (with faba bean or pea respectively) were identified. Following association tests with faba bean and pea CFN phenotypes, two clusters of 53 and 76 significant SNPs, respectively, were detected (P‐value threshold of 5%; Fig. 6). In order to account for multiple tests, we calculated a chromosome‐wide 5% P‐value threshold for each dataset by using permutations (see Methods S1). The empirical P‐value thresholds were 0.004 and 0.0015 for the faba bean and pea dataset, respectively. Using these thresholds, no SNP was detected as significant, although the smallest SNP P‐values were 0.01 and 0.0068 in the faba bean and pea association analyses, respectively, thus close to the thresholds. However, 92% (49/53) and 76% (58/76) of the top significant SNPs (i.e. with P‐value < 0.05) associated with CFNs in faba bean and pea respectively, were clustered within the nodMNO genes zone. Using a procedure of spatial permutations of each P‐value dataset (see Methods S1), the probabilities to observe in the permuted datasets > 92% (faba bean) or 76% (pea) of SNPs with a P‐value threshold < 0.05 and lying within the 3300 bp nodMNO zone, were null. This indicated that the physical clustering of these significant SNPs within the nodMNO zone is unlikely to be due to random chance.
Figure 6.
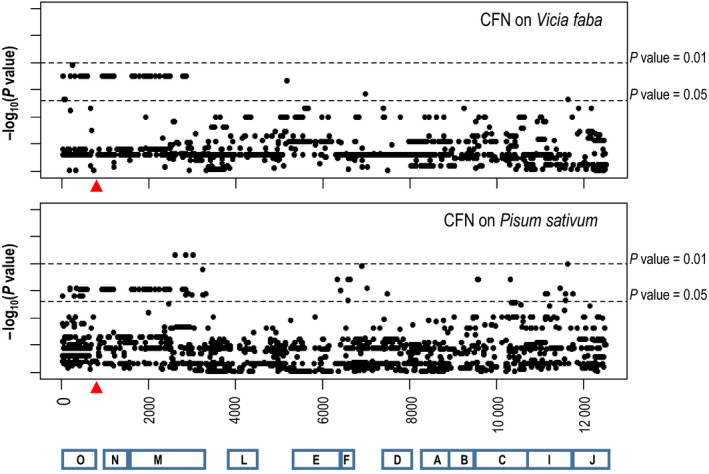
Association between pea or faba bean competitive to form root nodules (CFN) and single nucleotide polymorphism (SNP) within the nod genes sequence. Black circles represent biallelic SNPs along nod gene cluster. The location of the nod genes is indicated. Red arrow correspond to the 3 kb region between nodN and nodO genes (containing nodT in some strains), too variable to be included in this analysis.
To identify loci outside of the nod region potentially associated with CFN phenotypes, we compared bacteria with contrasted CFN phenotypes sharing close or contrasted nod genotypes (i.e. A vs A or A vs B). In the first approach, nod type A bacteria displaying contrasted pea/faba bean CFN phenotypes were compared. Five significant genomic sequences were identified and annotated by reference to the genome of the strain 3841 (Tables 2, S9). Three of these regions were found on pRL9 and pRL12, but it was not possible to assign the two others regions to a replicon because they were absent from the reference. Presence/absence of these specific genomic regions was then characterised in all sequenced genomes of Nod type A isolates, and the association with the CFN phenotype was estimated (Table S9). These five regions were significantly associated with faba bean CFN but only two of these with pea CFN (Tables 2, S9). In a second approach, the genomes of eight bacteria displaying the most contrasted CFN phenotypes (i.e. a high CFN with pea and a low CFN with faba bean, or vice versa), and belonging to the Nod type A or B were compared using the same strategy. This identified 32 specific genomic sequences (Tables 2, S10). Except for one region located on the chromosome and five regions absent in the reference strain 3841, all others clustered within 14 regions of the plasmids pRL8, pRL9, pRL10 (symbiosis plasmid) and pRL12. One of these regions included the nodO gene and the insertion of the nodT gene in the pRL10, as expected. The presence/absence of these specific genomic sequences was characterised in all genomes of isolates, and the association with the CFN phenotype was estimated (Table S10). Finally, 14 regions were significantly associated with both pea and faba bean CFN phenotypes, whereas six were solely associated with pea CFN (Table 2). Among these genomic regions, only one (encoding an acetyl‐CoA hydrolase transferase) was identified in both analyses (i.e. A vs A and A vs B).
Table 2.
Specific genomic regions associated with CFN with Pisum sativum or Vicia faba.
| Genomic comparison (nod types) | Repliconsa | Region/CDS positionsb | Annotationc | Association with pea CFN | Association with faba bean CFN | ||||
|---|---|---|---|---|---|---|---|---|---|
| Total (n = 20) | A1 (n = 12) | B1 (n = 8) | Total (n = 23) | A1 (n = 11) | B1 (n = 12) | ||||
| A vs B | chromosome | 833874–838499 | Hypothetical protein | * | 0.10 | (*) | * | * | 0.35 |
| A vs B | pRL7 | 51839–52045 | Hypothetical protein | * | 1.00 | 1.00 | 0.12 | 1.00 | (*) |
| A vs B | pRL8 | 77684–78604 | LysR transcriptional regulator | * | * | 0.26 | * | 0.22 | * |
| A vs B | 78784–79167 | Endoribonuclease L‐PSP | * | * | 0.26 | * | 0.22 | * | |
| A vs B | 79231–80253 | Nitrilase | * | * | 0.26 | * | 0.22 | * | |
| A vs B | pRL9 | 19902–20633 | Transcriptional regulator Crp‐Fnr family | * | 0.37 | 1.00 | * | * | * |
| A vs B | 20661–22586 | fixL | * | 0.37 | 1.00 | * | * | * | |
| A vs B | 24138–25490 | Coproporphyrinogen III oxidase, oxygen‐independent | * | 0.37 | 1.00 | * | * | (*) | |
| A vs B | 26312–27064 | Transcriptional regulator Crp‐Fnr family | * | 0.37 | 1.00 | * | * | (*) | |
| A vs B | 27191–27916 | Truncated response regulator | * | 0.37 | 1.00 | * | * | (*) | |
| A vs B | 28190–29263 | Alcohol dehydrogenase | * | 0.37 | 1.00 | * | * | (*) | |
| A vs B, A vs A | 30916–32436 | Acetyl‐CoA hydrolase | * | * | 1.00 | * | * | (*) | |
| A vs B | 32449–33273 | Universal stress protein family, tandem domain | * | * | 1.00 | * | * | (*) | |
| A vs B | 33432–33740 | hypothetical protein, miscellaneous | * | * | 1.00 | * | * | (*) | |
| A vs B | pRL9 | 49741–50607 | Universal stress protein family | * | 0.37 | 1.00 | * | * | (*) |
| A vs B | pRL9 | 8920–9131 | no annotated gene | * | * | 1.00 | * | * | 1.00 |
| A vs B | pRL10 | 174948–175802 | nodO | * | 1.00 | 1.00 | 0.27 | 1.00 | 1.00 |
| A vs B | 176905–177522 | Hypothetical protein | * | 1.00 | 1.00 | 0.27 | 1.00 | 1.00 | |
| A vs B | 177479–178927 | nodT | * | 1.00 | 1.00 | 0.27 | 1.00 | 1.00 | |
| A vs B | pRL10 | 205375–205767 | Transposase | * | 1.00 | 1.00 | 0.12 | 1.00 | (*) |
| A vs B | pRL10 | 217295–216726 | Transposase | * | 1.00 | 1.00 | 0.12 | 1.00 | (*) |
| A vs B | pRL10 | 153549–153184 | Hypothetical protein | * | 1.00 | 1.00 | 0.12 | 1.00 | (*) |
| A vs B | pRL10 | 215237–215476 | IS481 transposase | * | 1.00 | 1.00 | 0.32 | 1.00 | 0.31 |
| A vs B | pRL10 | 182899–182656 | no annotated gene | * | 1.00 | 1.00 | 0.27 | 1.00 | 1.00 |
| A vs A | pRL12 | 57481–57729 | VapC toxin protein antagonist | 1.00 | * | 0.45 | * | 0.25 | |
| A vs A | pRL12 | 58177–58587 | Hypothetical protein | 1.00 | * | 0.45 | * | 0.25 | |
| A vs B | pRL12 | 99541–100443 | Transcriptional regulator | * | 0.31 | 0.17 | * | * | * |
| A vs B | 100563–101459 | UDP‐glucose 4‐epimerase | * | 0.31 | 0.17 | * | * | * | |
| A vs B | 101512–102480 | Hypothetical protein | * | 0.31 | 0.17 | * | * | * | |
| A vs B | 102545–103369 | Aldo/keto reductase family | * | 0.31 | 0.17 | * | * | * | |
| A vs B | pRL12 | 371983–372822 | Hypothetical protein | * | 0.17 | 0.12 | (*) | 0.53 | * |
| A vs B | 372947–373888 | LysR transcriptional regulator | * | 0.17 | 0.12 | (*) | 0.53 | * | |
| A vs B | pRL12 | 598629–598466 | No annotated gene | * | * | 1.00 | * | * | 1.00 |
| A vs B | pRL12 | 868438–869751 | Hypothetical protein | * | 0.28 | * | * | * | 0.19 |
| A vs B | scaffold#4d | 255964–256491 | ECF family sigma factor | * | 0.66 | * | * | 0.21 | * |
| A vs A | scaffold#12d | 37450–40707 | DNA polymerase III subunit alpha | 1.00 | * | 0.82 | * | 0.52 | |
| A vs B | scaffold#14d | 157714–158751 | Aspartate‐semialdehyde dehydrogenase | * | 0.39 | 1.00 | * | 0.10 | (*) |
| A vs B | scaffold#32d | 52588–528333 | Hypothetical protein | * | 0.39 | 1.00 | * | 0.10 | (*) |
| A vs A | scaffold#39d | 21998–22116 | no annotated gene | 1.00 | * | 1.00 | * | 1.00 | |
| A vs A | scaffold#39d | 28925–29324 | no annotated gene | 1.00 | * | 1.00 | * | (*) | |
| A vs B | scaffold#42d | 16775–17494 | Methyl‐accepting chemotaxis protein | * | 1.00 | 1.00 | 0.12 | 1.00 | (*) |
| A vs B | scaffold#42d | 19264–20550 | DegT/DnrJ/EryC1/StrS aminotransferase | * | 1.00 | 1.00 | 0.12 | 1.00 | (*) |
| A vs B | 20691–21362 | HAD family hydrolase | * | 1.00 | 1.00 | 0.12 | 1.00 | (*) | |
| A vs B | 21391–22038 | GnaT‐family acetyltransferase | * | 1.00 | 1.00 | 0.12 | 1.00 | (*) | |
| A vs B | 22343–23551 | Glycerophosphoryl diester phosphodiesterase | * | 1.00 | 1.00 | 0.12 | 1.00 | (*) | |
| A vs B | 23588–24670 | Hypothetical protein | * | 1.00 | 1.00 | 0.12 | 1.00 | (*) | |
Genomic regions are listed. Associations of presence/absence of these genomic regions with pea or faba bean CFN were estimated by a Kruskal–Wallis test using the data of the Table 1. Association was tested in global populations of bacterial isolates as well as subpopulations belonging to the nod type A or B (the size of the population used for the test is indicated). *, P < 0.05; (*), P < 0.1; no label, P‐value > 0.1.
Based on sequence homology to the Rlv 3841 reference genome.
Positions of the homologous sequences in the Rlv 3841 reference genome.
Annotations of RAST databases.
Positions of the homologous sequences in the Rlv RSP1E11 genome.
Discussion
A widespread nod gene polymorphism in Rlv is associated with host preference
The diversity of R. leguminosarum sv. viciae populations was characterised either by trapping bacteria in pea or faba bean root nodules or by quantifying them directly in the soils. This study confirmed that bacteria of the same symbiovar may belong to different genospecies and a genospecies may include bacteria from different symbiovars (Kumar et al., 2015). The phylogeny of nod genes located on the symbiosis plasmid is independent of chromosome variations. Lateral gene and plasmid transfers are the major drivers of the evolution of the symbiotic phenotype (at least at the level of R. leguminosarum) and therefore genospecies are not ecologically relevant for symbiotic traits (Kumar et al., 2015; Andrews et al., 2018). However this does not exclude that some genes present on the chromosome may also contribute to the symbiotic phenotype and its variation.
Most of the Rlv nodule isolates (87%) and sequences identified directly in the soil (90%) belong to the two main nodD groups A1 and B1. These groups include and extend those already defined by RFLP analysis in previous studies on bacteria trapped from European soils with pea and faba bean (Laguerre et al., 2003; Mutch & Young, 2004) or from Chinese soils with faba bean (Tian et al., 2010). Few examples of specific nodD alleles in particular geographic location were found (even when bacteria isolated outside of Europe and present in GenBank were included in the analysis), giving little support for strong phylogeographic structuring of the Rlv diversity. This study confirmed previous observations, made on a limited number of soils, that pea and faba bean select differentially A1 and B1 nod groups of Rlv from the same soil populations to form nodules (Laguerre et al., 2003; Mutch & Young, 2004; Jorrin & Imperial, 2015).
Characterising the diversity of the ‘hidden’ soil population of rhizobia
As evaluated either by the number of collected isolates or by the level of nodD sequences estimated in the soil, the relative sizes of nodD groups may vary greatly between geographic sites. Indeed, local variation of the relative composition of Rlv communities between soil plots in each site is large and in the same range as the variation between geographic locations. Systematic analyses have already revealed high local variation of Rlt population sizes and their symbiotic potentials (Wakelin et al., 2018). Whether these variations are the consequences of independent evolution trends of the bacteria, agronomic practices, the presence of different hosts, environmental parameters or soil composition favouring specific groups of isolates remain to be investigated. Nevertheless, it is noteworthy that the main nodD groups are maintained in all sites. This suggests that nodD groups of Rlv bacteria are nonsubstitutable. They are likely under positive selection in the various sites probably because of specific ecological functions. Whether these functions are related to their interactions with Fabeae legumes, other plant species or to factors related to their free‐living lifestyle remains to be discovered.
We have no explanation: (1) why pea trapped many nodD group B1 bacteria on the French pool of soil, whereas it generally trapped nodD group A1 bacteria on other pools; and (2) why we were repeatedly unable to trap bacteria with pea from the Spanish pool despite the presence of nodD group A1 bacteria in these soils. We can only speculate that CFN is interacting with abiotic and biotic environmental factors such as soil composition, pH or culture system that may contribute to modulate nodulation and may result in CFN variation. Response of CFN to environment is an open question that requires further investigation beyond the objective of this study.
The amplification of nodD sequences directly from soil DNA, followed by high‐throughput sequencing, is a novel approach to address the challenge of describing the diversity of potential symbionts in the soil. On the whole, the sequences found and their relative abundance are consistent with the genotypes of the nodule isolates, but there is one clear anomaly. Most of the isolates from the French soil were nodD group B1 (Fig. 2), but all the sequences amplified from the pooled French soil were nodD group A1 (Fig. 3c). We can speculate that this was the result of inadequate mixing of the soil before the small subsample was taken for DNA amplification. The various French soil samples that were pooled for the nodulation tests showed extreme variation in allele frequency (Fig. 3a,b) and, in retrospect, we can see that extremely thorough mixing is needed before subsampling of pooled soils.
Competitiveness for nodulation is host specific and associated with the nod genotypes
Both microbe–microbe and plant–microbe interactions are likely to affect competitiveness for nodulation. As our main interest was in plant–microbe interactions, we chose to minimise the opportunity for microbe–microbe interactions by inoculating with a mixture of Rlv at high densities. Although most Rlv strains can nodulate both pea and faba bean when inoculated alone, in mixture they display highly contrasted CFN with these hosts. As this trait operates only in mixture, it explains possible discrepancies between the capacity of individual strains tested alone and their performance in competition with large Rlv communities in the soil. We found that Nod types A and B are associated with pea and faba bean CFN, respectively, although additional genetic determinants not linked to nod genes can be also involved. Whether equivalent associations of CFN with specific nod types are also found with other Fabeae legumes (lentil, for example) remains to be investigated. The negative relationship between pea and faba bean CFN in natural isolates suggests a trade‐off between these two traits. More generally, this implies that the diversity and evolution of rhizobial populations cannot be understood without considering interactions with all the potential hosts that it has encountered.
Genetic factors underlying differences in CFN
Comparative genomic analyses and genetic association studies are a powerful approach for identifying genetic determinants of important traits in symbiotic bacteria (Lipuma et al., 2014; Porter et al., 2017). In this study, we identified genetic markers (specific genomic regions or SNPs) directly associated with pea/faba bean CFN. As expected, most of these genomic markers belong to Rlv plasmids and only a few to the chromosome. The association between specific SNP clusters and the pea/faba bean CFN phenotype revealed that the nodMNO zone within the nod gene island is likely to be one genetic component of CFN. A pool‐seq analysis of rhizobia populations trapped with pea, faba bean, lentil or Vicia sativa in a single soil has already identified this region as highly polymorphic (Jorrin & Imperial, 2015), but the potential association with CFN was not directly documented. Although global nodMNO sequence variation (including the insertion of nodT within this zone) is associated with the nod group, SNPs specifically related to pea and faba bean CFN were identified. The nodM, N, T and O genes encode, respectively, glucosamine synthases (likely involved in the synthesis of the GlcNAc Nod factor moiety), a putative outer membrane translocator, and a cation‐specific channel. These are not essential for nodulation but may modulate the process (Economou et al., 1990; Baev et al., 1992; Wakelin et al., 2018). Consistent with a role in modulating nodulation capacities in a host‐specific manner, the nodM and nodT genes have been implicated in cultivar‐specific nodulation of Rlt (Lewis‐Henderson & Djordjevic, 1991a,b), and nodO was found to complement loss of nodE, an essential determinant of host specificity in Rlv, which is required for the synthesis of NFs with unsaturated acyl moieties (Downie & Surin, 1990; Demont et al., 1993).
Presence/absence of additional genomic regions that potentially contribute to the pea or/and faba bean CFN phenotypes was identified, suggesting that the control of CFN may involve other genetic determinants in addition to nod genes. Again, these genomic regions are mainly on plasmids and therefore are potentially horizontally transferred. Interestingly, these regions are associated with both pea and faba bean CFN, with several exceptions associated only with pea CFN (including nodT). The association of the presence/absence of these genomic regions with pea or faba bean CFN may vary depending on the Nod type context, arguing for interactions between nod genes and additional genetic loci. Altogether, the data suggest that pea and faba bean CFN may be associated with a relatively limited number of specific genetic loci. Functions of the candidate genes are consistent with a role in competitiveness for nodulation. For example a putative methyl‐accepting chemotaxis protein (Rivilla et al., 1995; Yost et al., 1998), or a putative protein involved in a type II toxin–antitoxin system (Yost et al., 2004; Lipuma et al., 2014) may be implicated either in plant–microbe or microbe–microbe interactions and possibly modulate CFN. However, it would be too speculative at this stage to propose specific roles for these loci in pea/faba bean CFN. Indeed, even if the corresponding sequences are statistically associated with the CFN phenotype, this could be the result of genetic associations with variations in neighbouring genes of interest and not to direct roles in CFN. Reverse genetics studies with several combinations of various alleles will be required to confirm the biological impact of candidate genes on pea/faba bean CFN. Nevertheless, these loci represent valuable markers to select Rlv strains competitive for nodulation with pea and/or faba bean and open the way to the identification of genes controlling competitiveness for nodulation.
Author contributions
ML and PY designed research. GC, EJ, E‐PJ, RL‐B, MS and JM collected soils. NAL, KH‐G and BB performed trapping experiments. SB, MP, DD, AL‐Q, SC and MT performed phylogenetic analysis and genome sequencing. DS and PY performed metabarcoding experiments from soils. SB and MB performed genetic association and comparative genomic studies. ML, PY and SB wrote the manuscript with assistance from all co‐authors.
Supporting information
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Phylogenetic tree based on nodD gene sequences of the 210 pea and faba bean Rlv isolates.
Fig. S2 Quantitative estimates of nodD sequence abundance in soils.
Fig. S3 Genomic diversity of Rhizobium leguminosarum sv. viciae isolates using dDDH analysis.
Fig. S4 Phylogenetic tree based on concatenated nodABCDEFIJLMN gene sequences.
Fig. S5 Monoinoculation experiments with uncompetitive strains (CFN = 0).
Methods S1 Procedure to estimate a chromosome‐wide P‐value threshold in the association analysis of pea/faba bean CFN phenotype with SNP data; procedure to test for the random spatial clustering of significant SNPs within a given genomic region.
Table S1 Composition of soil pools used for trapping experiments.
Table S2 Agronomic history of plots used for sampling in the five European sites.
Table S3 Pea and faba bean cultivars.
Table S4 Primers and DNA spike used in this study.
Table S5 nodD sequences, nodD groups and allele numbers of isolates and references strains used in this work.
Table S6 Distribution and enrichment of nodD alleles and groups.
Table S7 Pea and faba bean symbiotic efficiency of selected isolates as the function of their phylogenic classes and their geographic origins.
Table S8 Genomic data of Rhizobium bacteria sequenced in this study.
Table S9 Presence/absence of candidate genes correlated with the CFN with P. sativum or V. faba.
Table S10 Presence/absence of candidate genes correlated with the CFN with P. sativum or V. faba.
Acknowledgements
This paper is dedicated to our colleague Gisèle Laguerre who opened the way for this work. This work was supported by the FP7 European Community's Seventh Framework Programme ‘LEGumes for the Agriculture of TOmorrow’ under the grant agreement no. FP7‐613551 and by the GrasP grant of the Agence Nationale de la Recherche.
The copyright line for this article was changed on 23 October 2020 after original online publication.
References
- Amarger N. 1981. Competition for nodule formation between effective and ineffective strains of Rhizobium meliloti . Soil Biology and Biochemistry 13: 475–480. [Google Scholar]
- Andrews M, De Meyer S, James EK, Stępkowski T, Hodge S, Simon MF, Young JPW. 2018. Horizontal transfer of symbiosis genes within and between rhizobial genera: occurrence and importance. Genes 9: 321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baev N, Schultze M, Barlier I, Ha DC, Virelizier H, Kondorosi E, Kondorosi A. 1992. Rhizobium nodM and nodN genes are common nod genes: nodM encodes functions for efficiency of nod signal production and bacteroid maturation. Journal of Bacteriology 174: 7555–7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloemberg GV, Kamst E, Harteveld M, van der Drift KM, Haverkamp J, Thomas‐Oates JE, Lugtenberg BJ, Spaink HP. 1995. A central domain of Rhizobium NodE protein mediates host specificity by determining the hydrophobicity of fatty acyl moieties of nodulation factors. Molecular Microbiology 16: 1123–1136. [DOI] [PubMed] [Google Scholar]
- Bourion V, Heulin‐Gotty K, Aubert V, Tisseyre P, Chabert‐Martinello M, Pervent M, Delaitre C, Vile D, Siol M, Duc G et al. 2018. Co‐inoculation of a pea core‐collection with diverse rhizobial strains shows competitiveness for nodulation and efficiency of nitrogen fixation are distinct traits in the interaction. Frontiers in Plant Science 8: 2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer E, Rennie DA, Rennie RJ. 1988. Dinitrogen fixation of lentil, field pea and fababean under dryland conditions. Canadian Journal of Soil Science 68: 553–562. [Google Scholar]
- Broughton WJ, Jabbouri S, Perret X. 2000. Keys to symbiotic harmony. Journal of Bacteriology 182: 5641–5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavassim MIA, Moeskjaer S, Moslemi C, Fields B, Bachmann A, Vilhjalmsson B, Schierup MH, Young JPW, Andersen SU. 2019. The genomic architecture of introgression among sibling species of bacteria. bioRxiv: 526707. doi: 10.1101/526707. [DOI] [Google Scholar]
- Deakin WJ, Broughton WJ. 2009. Symbiotic use of pathogenic strategies: rhizobial protein secretion systems. Nature Reviews. Microbiology 7: 312–320. [DOI] [PubMed] [Google Scholar]
- Debellé F, Maillet F, Vasse J, Rosenberg C, de Billy F, Truchet G, Dénarié J, Ausubel FM. 1988. Interference between Rhizobium meliloti and Rhizobium trifolii nodulation genes: genetic basis of R. meliloti dominance. Journal of Bacteriology 170: 5718–5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demont N, Debellé F, Aurelle H, Dénarié J, Promé JC. 1993. Role of the Rhizobium meliloti nodF and nodE genes in the biosynthesis of lipo‐oligosaccharidic nodulation factors. Journal of Biological Chemistry 268: 20134–20142. [PubMed] [Google Scholar]
- Dénarié J, Debellé F, Rosenberg C. 1992. Signaling and host range variation in nodulation. Annual Review of Microbiology 46: 497–531. [DOI] [PubMed] [Google Scholar]
- Devine TE, Kuykendall LD, Breithaupt BH. 1980. Nodulation of soybeans carrying the nodulation‐restrictive gene, rj1, by an incompatible Rhizobium japonicum strain upon mixed inoculation with a compatible strain. Canadian Journal of Microbiology 26: 179–182. [DOI] [PubMed] [Google Scholar]
- Downie JA, Surin BP. 1990. Either of two nod gene loci can complement the nodulation defect of a nod deletion mutant of Rhizobium leguminosarum bv viciae . Molecular & general genetics: MGG 222: 81–86. [DOI] [PubMed] [Google Scholar]
- Economou A, Hamilton WD, Johnston AW, Downie JA. 1990. The Rhizobium nodulation gene nodO encodes a Ca2(+)‐binding protein that is exported without N‐terminal cleavage and is homologous to haemolysin and related proteins. EMBO Journal 9: 349–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fesenko AN, Provorov NA, Orlova IF, Orlov VP, Simarov BV. 1995. Selection of Rhizobium leguminosarum bv. viceae strains for inoculation of Pisum sativum L. cultivars: analysis of symbiotic efficiency and nodulation competitiveness. Plant and Soil 172: 189–198. [Google Scholar]
- Fields B, Moeskjær S, Friman V‐P, Andersen SU, Young JPW. 2019. MAUI‐seq: multiplexed, high‐throughput amplicon diversity profiling using unique molecular identifiers. bioRxiv: 538587. doi: 10.1101/538587. [DOI] [Google Scholar]
- Firmin JL, Wilson KE, Carlson RW, Davies AE, Downie JA. 1993. Resistance to nodulation of cv. Afghanistan peas is overcome by nodX, which mediates an O‐acetylation of the Rhizobium leguminosarum lipo‐oligosaccharide nodulation factor. Molecular Microbiology 10: 351–360. [DOI] [PubMed] [Google Scholar]
- González V, Santamaría RI, Bustos P, Pérez‐Carrascal OM, Vinuesa P, Juárez S, Martínez‐Flores I, Cevallos MÁ, Brom S, Martínez‐Romero E et al. 2019. Phylogenomic rhizobium species are structured by a continuum of diversity and genomic clusters. Frontiers in Microbiology 10: 910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg B, Davies AE, Wilson KE, Bisseling T, Downie JA. 2002. Competitive nodulation blocking of cv. Afghanistan pea is related to high levels of nodulation factors made by some strains of Rhizobium leguminosarum bv. viciae . Molecular Plant–Microbe Interactions 15: 60–68. [DOI] [PubMed] [Google Scholar]
- Jain C, Rodriguez‐R LM, Phillippy AM, Konstantinidis KT, Aluru S. 2018. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nature Communications 9: 5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorrin B, Imperial J. 2015. Population genomics analysis of legume host preference for specific rhizobial genotypes in the Rhizobium leguminosarum bv. viciae symbioses. Molecular Plant–Microbe Interactions 28: 310–318. [DOI] [PubMed] [Google Scholar]
- Kang HM, Sul JH, Service SK, Zaitlen NA, Kong S‐Y, Freimer NB, Sabatti C, Eskin E. 2010. Variance component model to account for sample structure in genome‐wide association studies. Nature Genetics 42: 348–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaharada Y, Kelly S, Nielsen MW, Hjuler CT, Gysel K, Muszyński A, Carlson RW, Thygesen MB, Sandal N, Asmussen MH et al. 2015. Receptor‐mediated exopolysaccharide perception controls bacterial infection. Nature 523: 308–312. [DOI] [PubMed] [Google Scholar]
- Kiers ET, Rousseau RA, West SA, Denison RF. 2003. Host sanctions and the legume–rhizobium mutualism. Nature 425: 78–81. [DOI] [PubMed] [Google Scholar]
- Kumar N, Lad G, Giuntini E, Kaye ME, Udomwong P, Shamsani NJ, Young JPW, Bailly X. 2015. Bacterial genospecies that are not ecologically coherent: population genomics of Rhizobium leguminosarum . Open Biology 5: 140133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguerre G, Depret G, Bourion V, Duc G. 2007. Rhizobium leguminosarum bv. viciae genotypes interact with pea plants in developmental responses of nodules, roots and shoots. New Phytologist 176: 680–690. [DOI] [PubMed] [Google Scholar]
- Laguerre G, Heulin‐Gotty K, Brunel B, Klonowska A, Le Quéré A, Tillard P, Prin Y, Cleyet‐Marel J‐C, Lepetit M. 2012. Local and systemic N signaling are involved in Medicago truncatula preference for the most efficient Sinorhizobium symbiotic partners. New Phytologist 195: 437–449. [DOI] [PubMed] [Google Scholar]
- Laguerre G, Louvrier P, Allard M‐R, Amarger N. 2003. Compatibility of rhizobial genotypes within natural populations of Rhizobium leguminosarum biovar viciae for nodulation of host legumes. Applied and Environmental Microbiology 69: 2276–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis‐Henderson WR, Djordjevic MA. 1991a. nodT, a positively‐acting cultivar specificity determinant controlling nodulation of Trifolium subterraneum by Rhizobium leguminosarum biovar trifolii . Plant Molecular Biology 16: 515–526. [DOI] [PubMed] [Google Scholar]
- Lewis‐Henderson WR, Djordjevic MA. 1991b. A cultivar‐specific interaction between Rhizobium leguminosarum bv. trifolii and subterranean clover is controlled by nodM, other bacterial cultivar specificity genes, and a single recessive host gene. Journal of Bacteriology 173: 2791–2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linhartová I, Bumba L, Mašín J, Basler M, Osička R, Kamanová J, Procházková K, Adkins I, Hejnová‐Holubová J, Sadílková L et al. 2010. RTX proteins: a highly diverse family secreted by a common mechanism. Federation of European Microbiology Societies 34: 1076–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipuma J, Cinege G, Bodogai M, Oláh B, Kiers A, Endre G, Dupont L, Dusha I. 2014. A vapBC‐type toxin‐antitoxin module of Sinorhizobium meliloti influences symbiotic efficiency and nodule senescence of Medicago sativa . Environmental Microbiology 16: 3714–3729. [DOI] [PubMed] [Google Scholar]
- McAnulla C, Edwards A, Sanchez-Contreras M, Sawers RG, Downie JA. 2007. Quorum-sensingregulated transcriptional initiation of plasmid transfer and replication genes in Rhizobium leguminosarum biovar viciae . Microbiology 153: 2074–2082. [DOI] [PubMed] [Google Scholar]
- McKenzie RH, Middleton AB, Solberg ED, DeMulder J, Flore N, Clayton GW, Bremer E. 2001. Response of pea to rhizobia inoculation and starter nitrogen in Alberta. Canadian Journal of Plant Science 81: 637–643. [Google Scholar]
- Meade J, Higgins P, O'Gara F. 1985. Studies on the inoculation and competitiveness of a Rhizobium leguminosarum strain in soils containing indigenous rhizobia. Applied and Environmental Microbiology 49: 899–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier‐Kolthoff JP, Auch AF, Klenk H‐P, Göker M. 2013. Genome sequence‐based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergaert P, Montagu MV, Holsters M. 1997. Molecular mechanisms of Nod factor diversity. Molecular Microbiology 25: 811–817. [DOI] [PubMed] [Google Scholar]
- Moawad HA, Ellis WR, Schmidt EL. 1984. Rhizosphere response as a factor in competition among three serogroups of indigenous Rhizobium japonicum for nodulation of field‐grown soybeans. Applied and Environmental Microbiology 47: 607–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutch LA, Young JPW. 2004. Diversity and specificity of Rhizobium leguminosarum biovar viciae on wild and cultivated legumes. Molecular Ecology 13: 2435–2444. [DOI] [PubMed] [Google Scholar]
- Naamala J, Jaiswal SK, Dakora FD. 2016. Antibiotics resistance in Rhizobium: type, process, mechanism and benefit for agriculture. Current Microbiology 72: 804–816. [DOI] [PubMed] [Google Scholar]
- Ozer EA, Allen JP, Hauser AR. 2014. Characterization of the core and accessory genomes of Pseudomonas aeruginosa using bioinformatic tools Spine and AGEnt. BMC genomics 15: 737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter SS, Chang PL, Conow CA, Dunham JP, Friesen ML. 2017. Association mapping reveals novel serpentine adaptation gene clusters in a population of symbiotic Mesorhizobium . The ISME Journal 11: 248–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Jurkiewicz A, Fukai E, Quistgaard EM, Albrektsen AS, James EK, Thirup S, Stougaard J. 2007. LysM domains mediate lipochitin–oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO Journal 26: 3923–3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivilla R, Sutton JM, Downie JA. 1995. Rhizobium leguminosarum NodT is related to a family of outer‐membrane transport proteins that includes TolC, PrtF, CyaE and AprF. Gene 161: 27–31. [DOI] [PubMed] [Google Scholar]
- Robleto EA, Kmiecik K, Oplinger ES, Nienhuis J, Triplett EW. 1998. Trifolitoxin production increases nodulation competitiveness of Rhizobium etli CE3 under agricultural conditions. Applied and Environmental Microbiology 64: 2630–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saïdi S, Ramírez‐Bahena M‐H, Santillana N, Zúñiga D, Álvarez‐Martínez E, Peix A, Mhamdi R, Velázquez E. 2014. Rhizobium laguerreae sp. nov. nodulates Vicia faba on several continents. International Journal of Systematic and Evolutionary Microbiology 64: 242–247. [DOI] [PubMed] [Google Scholar]
- Sallet E, Gouzy J, Schiex T. 2014. EuGene‐PP: a next‐generation automated annotation pipeline for prokaryotic genomes. Bioinformatics 30: 2659–2661. [DOI] [PubMed] [Google Scholar]
- Spaink HP, Sheeley DM, van Brussel AA, Glushka J, York WS, Tak T, Geiger O, Kennedy EP, Reinhold VN, Lugtenberg BJ. 1991. A novel highly unsaturated fatty acid moiety of lipo‐oligosaccharide signals determines host specificity of Rhizobium. Nature 354: 125–130. [DOI] [PubMed] [Google Scholar]
- Sulima AS, Zhukov VA, Afonin AA, Zhernakov AI, Tikhonovich IA, Lutova LA. 2017. Selection signatures in the first exon of paralogous receptor kinase genes from the Sym2 region of the Pisum sativum L. Genome. Frontiers in Plant Science 8: 1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surin BP, Downie JA. 1988. Characterization of the Rhizobium leguminosarum genes nodLMN involved in efficient host‐specific nodulation. Molecular Microbiology 2: 173–183. [DOI] [PubMed] [Google Scholar]
- Surin BP, Watson JM, Hamilton WDO, Economou A, Downie JA. 1990. Molecular characterization of the nodulation gene, nodT, from two biovars of Rhizobium leguminosarum . Molecular Microbiology 4: 245–252. [DOI] [PubMed] [Google Scholar]
- Sutton JM, Lea EJ, Downie JA. 1994. The nodulation‐signaling protein NodO from Rhizobium leguminosarum biovar viciae forms ion channels in membranes. Proceedings of the National Academy of Sciences, USA 91: 9990–9994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian CF, Young JPW, Wang ET, Tamimi SM, Chen WX. 2010. Population mixing of Rhizobium leguminosarum bv. viciae nodulating Vicia faba: the role of recombination and lateral gene transfer. FEMS Microbiology Ecology 73: 563–576. [DOI] [PubMed] [Google Scholar]
- Triplett EW, Sadowsky MJ. 1992. Genetics of competition for nodulation of legumes. Annual Review of Microbiology 46: 399–428. [DOI] [PubMed] [Google Scholar]
- Wakelin S, Tillard G, van Ham R, Ballard R, Farquharson E, Gerard E, Geurts R, Brown M, Ridgway H, O'Callaghan M. 2018. High spatial variation in population size and symbiotic performance of Rhizobium leguminosarum bv. trifolii with white clover in New Zealand pasture soils. PLoS ONE 13: e0192607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Tang F, Gao M, Krishnan HB, Zhu H. 2010. R gene‐controlled host specificity in the legume‐rhizobia symbiosis. Proceedings of the National Academy of Sciences, USA 107: 18735–18740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yost CK, Del Bel KL, Quandt J, Hynes MF. 2004. Rhizobium leguminosarum methyl‐accepting chemotaxis protein genes are down‐regulated in the pea nodule. Archives of Microbiology 182: 505–513. [DOI] [PubMed] [Google Scholar]
- Yost CK, Rochepeau P, Hynes MF. 1998. Rhizobium leguminosarum contains a group of genes that appear to code for methyl‐accepting chemotaxis proteins. Microbiology 144: 1945–1956. [DOI] [PubMed] [Google Scholar]
- Young JPW. 2016. Bacteria are smartphones and mobile genes are apps. Trends in Microbiology 24: 931–932. [DOI] [PubMed] [Google Scholar]
- Zézé A, Mutch LA, Young JP. 2001. Direct amplification of nodD from community DNA reveals the genetic diversity of Rhizobium leguminosarum in soil. Environmental Microbiology 3: 363–370. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Phylogenetic tree based on nodD gene sequences of the 210 pea and faba bean Rlv isolates.
Fig. S2 Quantitative estimates of nodD sequence abundance in soils.
Fig. S3 Genomic diversity of Rhizobium leguminosarum sv. viciae isolates using dDDH analysis.
Fig. S4 Phylogenetic tree based on concatenated nodABCDEFIJLMN gene sequences.
Fig. S5 Monoinoculation experiments with uncompetitive strains (CFN = 0).
Methods S1 Procedure to estimate a chromosome‐wide P‐value threshold in the association analysis of pea/faba bean CFN phenotype with SNP data; procedure to test for the random spatial clustering of significant SNPs within a given genomic region.
Table S1 Composition of soil pools used for trapping experiments.
Table S2 Agronomic history of plots used for sampling in the five European sites.
Table S3 Pea and faba bean cultivars.
Table S4 Primers and DNA spike used in this study.
Table S5 nodD sequences, nodD groups and allele numbers of isolates and references strains used in this work.
Table S6 Distribution and enrichment of nodD alleles and groups.
Table S7 Pea and faba bean symbiotic efficiency of selected isolates as the function of their phylogenic classes and their geographic origins.
Table S8 Genomic data of Rhizobium bacteria sequenced in this study.
Table S9 Presence/absence of candidate genes correlated with the CFN with P. sativum or V. faba.
Table S10 Presence/absence of candidate genes correlated with the CFN with P. sativum or V. faba.


