Abstract
Corydalis trisecta Franch. is an endemic plant found in China. In this study, we presented the first complete chloroplast genome of C. trisecta, which was assembled and characterized based on Illumina pair-end sequencing data. The complete chloroplast genome was 161,410 bp in length, with a GC content of 41.4% in total. Its structure contained a large single copy (LSC) region of 89,127 bp and a small single copy (SSC) region of 16,993 bp, which were separated by a pair of extremely inverted repeats (IRs) of 27,645 bp each, with GC content 39.8, 87.2, and 45.2%, respectively. The phylogenetic analysis indicated that C. triscta was sister to Lamprocapons spectabilis in Papaveraceae.
Keywords: Cordalis trisecta Franch., phylogenetic relationship, chloroplast genome
Corydalis trisecta Franch. is a perennial herbaceous plant belonging to the family Papaveraceae included in order Ranunculales. This species is endemic to Qinling Mountain with an altitude of 2500–3300 m in China (Franch, 1894). Recently, it is investigated to be a key species based on field investigation (Wu et al. 2015). However, in spite of its ecological importance, till date, genomic studies have been hindered due to lack of information about the complete chloroplast (cp) genome of C. trisecta. In this study, we assembled the cp genome of C. trisecta Franch. based on Illumina paired-end sequencing to improve an appreciation of its genomics.
Leaves were collected from a single individual of C. trisecta Franch. at the 34°07′24″N, 107°53′31″E, and were dried using silica gel. The genomic DNA was extracted using a modified CTAB protocol (Yang et al. 2014), and genome sequencing was performed by the Illumina Hiseq 2000 Platform (Illumina, San Diego, CA). DNA sample and voucher specimen (No. PHLZH2017105) of C. trisecta Franch. were deposited in the Northwest University Museum (NUM). The program NGSQCToolkit_version 2.3.3 was used to trimming all raw reads (Patel and Jain 2012). Then the clean reads were assembled by MIRA version 4.0.2 after dislodging the low quality reads (Chevreux et al. 2004). In total, 1,683,089 bp raw reads were obtained, and cp genome was assembled by MITObim v1.8 (Hahn et al. 2013) with the published sequences of Coreanomecon hylomeconid cp genome and Lamprocapons spectabilis cp genome as the initial references. The cp genome of C. trisecta was annotated using software Geneious v 9.0.2 (Biomatters Ltd., Auckland, NewZealand) by comparison with the cp genome of C. hylomeconid. We deposited the annotated cp genome of C. trisecta to Genebank with the accession number MK713939.
The whole cp genome was 161,410 bp in length, containing a pair of inverted repeats (IRs) of 27,645 bp each, a large single copy region (LSC, 89,127 bp), and a small single copy region (SSC, 16,993 bp). The cp genome encoded 134 genes, including 85 proteins-coding genes, 37 tRNA genes, and 8 rRNA genes. The nucleotide composition was (28.9% A, 21.0% C, 20.4% G, and 29.7% T) with overall GC content of 41.4%. The GC content in whole cp genome, LSC region, SSC region, and IR region were 38.5, 36.5, 32.6, and 38.5%, respectively.
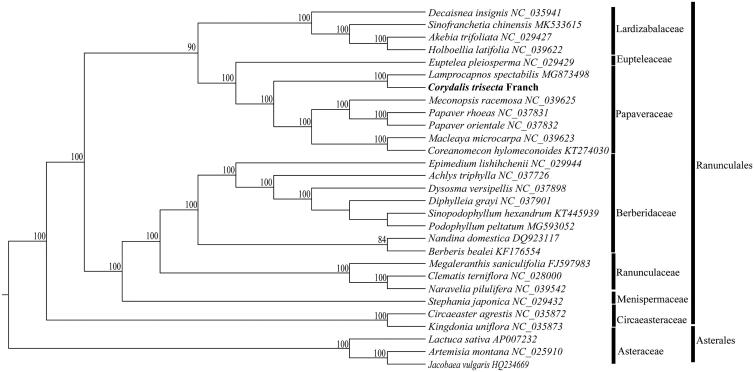
In order to investigate the phylogenetic status of C. trisecta, the available complete cp genomes of 29 species were aligned using MAFFT (Katoh and Standley 2013) with the default parameters. A maximum likelihood (ML) analysis was reconstructed from all of the 29 complete cp genome sequences by RAxML version 7.2.8 (Stamatakis 2006) with 500 bootstrap replicates. The result of a phylogenetic analysis indicated that C. trisecta was sister to L. spectabilis (Figure 1). Furthermore, the complete cp genome of C. trisecta will provide useful genomic information for detailed population genetic studies in the future.
Figure 1.
Maximum-likelihood phylogenetic tree based on 29 complete cp genome sequences. The bootstrap values are indicated next to the branches.
Disclosure statement
The authors declare that they have no competing interests.
References
- Chevreux B, Pfisterer T, Drescher B, Driesel AJ, Muller WE, Wetter T, Suhai S. 2004. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 14:1147–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franch 1894. Morot. Journ the Bot. 8:284. [Google Scholar]
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach . Nucl Acids Res. 41:e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Boil Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 7:e30619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690. [DOI] [PubMed] [Google Scholar]
- Wu Z, Yi H, Sun J, Zhao L, Liu J, Chang Z. 2015. A revision of Corydalis in Qinling Mountains. Acta Bot Boreal-Occident Sin. 35:2343–2348. [Google Scholar]
- Yang J-B, Li D-Z, Li H-T. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Resour. 14:1024–1031. [DOI] [PubMed] [Google Scholar]