Summary
The spread of protein aggregates during disease progression is a common theme underlying many neurodegenerative diseases. The microtubule-associated protein tau (MAPT) plays a central role in the pathogenesis of several forms of dementia known as tauopathies, including Alzheimer’s disease (AD), frontotemporal dementia (FTD) and chronic traumatic encephalopathy (CTE)1. Progression of these diseases is characterized by the sequential spread and deposition of protein aggregates in a predictable pattern that correlates with clinical severity2. This observation and complementary experimental studies3,4 have suggested that tau can spread in a prion-like manner by passing to naïve cells where it templates misfolding and aggregation. However, while tau propagation has been extensively studied, the underlying cellular mechanisms remain poorly understood. Here we show that the low-density lipoprotein (LDL) receptor-related protein 1 (LRP1) controls tau endocytosis and subsequent spread. Knockdown of LRP1 significantly reduced tau uptake in H4 neuroglioma cells and iPS-derived neurons. The interaction between tau and LRP1 is mediated by lysine residues in the microtubule binding repeat region of tau. Furthermore, we find that downregulation of LRP1 in an in vivo mouse model of tau spread effectively reduced tau propagation between neurons. Our results identify LRP1 as a key regulator of tau spread in the brain and, thus, as a novel target for diseases of tau spread and aggregation.
Results
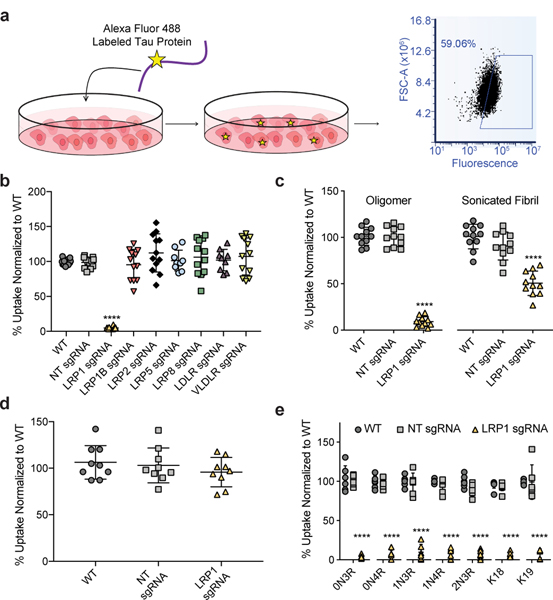
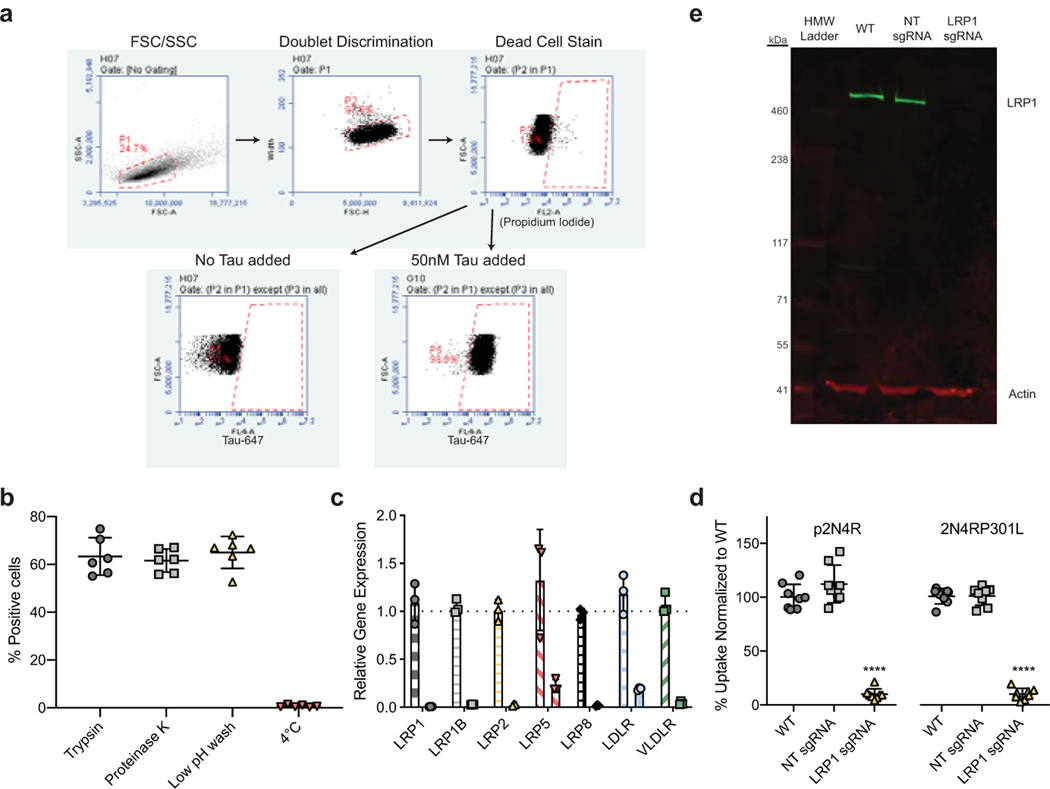
Based on recent work highlighting the importance of heparan sulfate proteoglycans (HSPGs) in tau uptake5,6, and the known role of LDL receptors to work in conjunction with HSPGs7 we set out to test if any of the LDL receptor family members could influence tau internalization. Using CRISPRi technology, we efficiently repressed gene expression of various LDLR family members (LRP1, LRP1B, LRP2, LRP5, LRP8, LDLR, VLDLR - Extended Data Fig. 1c) in H4 neuroglioma cells and tested the ability of these cells to endocytose monomeric tau5 (Fig. 1a,b). Genetic silencing of LRP1 almost completely blocked the uptake of full-length soluble monomeric tau (2N4R isoform), whereas no other LDLR family member showed a significant effect (Fig. 1b). Different uptake mechanisms have been proposed for soluble and aggregated tau8. LRP1 knockdown, however, was also sufficient to inhibit the uptake of tau oligomers, and reduced but did not completely inhibit the uptake of sonicated tau fibrils (Fig. 1c). Uptake of a disease-relevant mutant of tau and of phosphorylated tau were also affected by LRP1 knockdown (Extended Data Fig. 1d). To show that LRP1 knockdown was specific for tau endocytosis, we analyzed transferrin uptake in H4 cells and demonstrated that LRP1 knockdown had no effect on the endocytosis of transferrin (Fig. 1d). Knockdown of LRP1 also prevented the uptake of the smaller isoforms of tau (0N3R, 0N4R, 1N3R, 1N4R, 2N3R) as well as fragments of tau that contain only the microtubule binding repeat region (K18; 4 repeats, K19; 3 repeats, Fig. 1e), highlighting the microtubule binding region as the potential interaction site.
Figure 1: Tau uptake is mediated by LRP1.
a, Diagram of tau uptake assay. Fluorescently labeled tau is added to the cellular medium where it can be endocytosed by cells. The uptake of tau can be measured via fluorescence using flow cytometry of single cells. See methods for more details. b, Flow cytometry measurement of tau uptake (50nM, 1hr) in H4 CRISPRi neuroglioma cells (H4i) with various LDLR family proteins knocked-down, NT sgRNA=Non-targeting sgRNA (n=12, LRP5/LDLR n=9). c, Tau oligomer and sonicated fibril endocytosis in H4i cells (50nM, 100nM respectively, 1hr, n=11). d, Transferrin uptake (500nM, 1hr, n=9) in H4i cells (ANOVA; p-value = 0.4464). e, Tau isoform endocytosis (50nM, 1hr, n=6, K18 n=9) in H4i cells. All experiments in b-e were performed in biological replicates over three independent experiments and normalized to WT uptake (100%). All data expressed as mean ± s.d. One-way ANOVA with Tukey’s method, two-sided was performed to determine significance. Displayed are the comparisons against WT, ****=p-value < 0.0001. Comparison to NT sgRNA resulted in the same level of statistical significance.
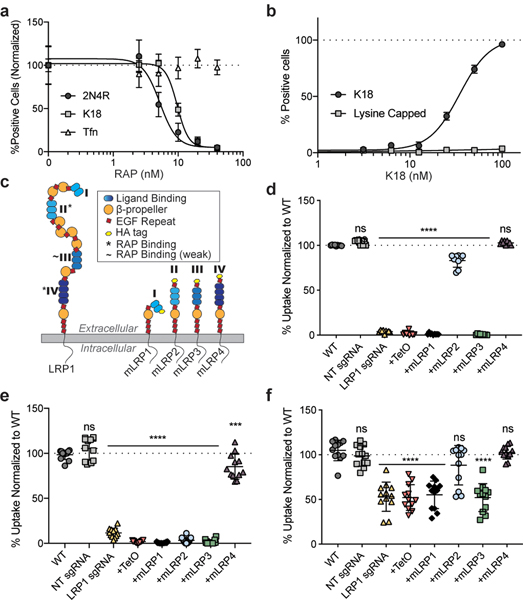
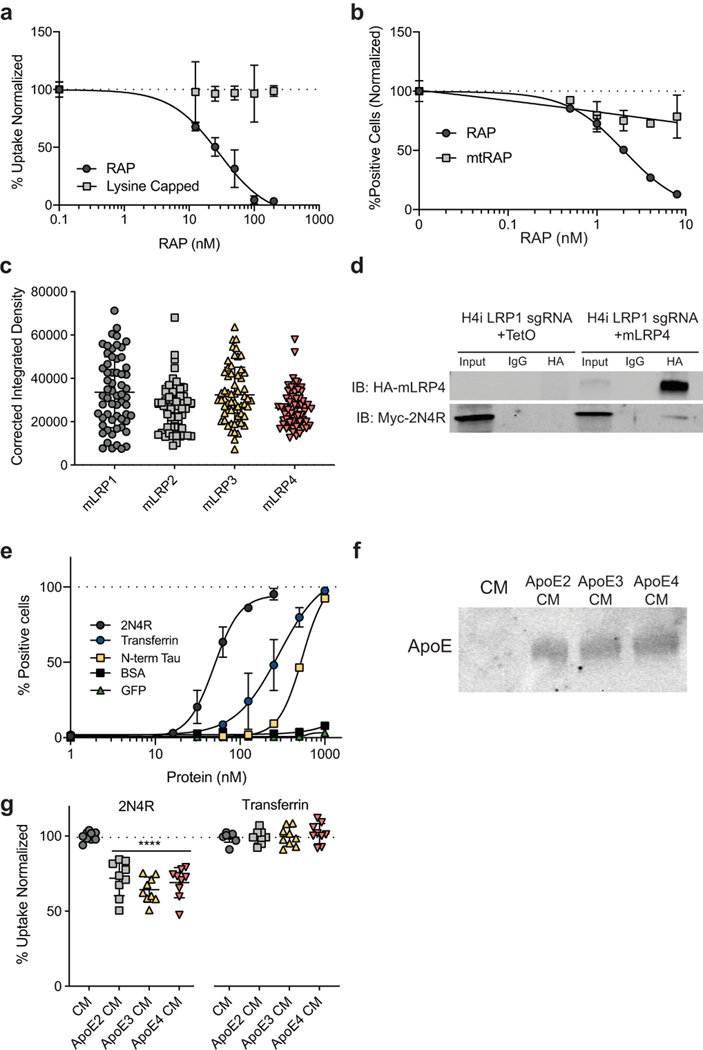
To further support the results from the LRP1 knockdown cell lines, a well-known LRP1 binding protein, receptor-associated protein (RAP), was used as a competitor for tau uptake in wild-type (WT) H4 cells. RAP is a small 39kDa chaperone for LRP1 that is known to bind tightly to LRP1 (KD=9nM)9. Increasing concentrations of RAP in the culture medium, concurrent with tau addition, were highly effective at inhibiting the uptake of both full-length and K18 tau (IC50=4.9nM and 9.6nM, respectively), but did not influence the internalization of transferrin (Fig. 2a). Point mutations of key residues in RAP needed for its interaction with LRP110 (K256A, K270A – “mtRAP”) were sufficient to reduce this competitive effect (Extended Data Fig. 2a,b).
Figure 2: Tau interacts with LRP1 similar to known ligands.
a, Uptake of K18, 2N4R tau, and transferrin (Tfn) in the presence of increasing RAP concentrations in the cell media. b, Uptake of K18 and lysine capped K18 in WT H4 cells c, Schematic of LRP1 ectodomains. d, Uptake of 2N4R tau with mLRP ectodomain expression (n=9, WT vs. NT; p-value = 0.2232, WT vs. mLRP4; p-value = 0.5450). e, Uptake of K18 with mLRP ectodomain expression (n=12, WT vs. NT; p-value = 0.2648, WT vs. mLRP4; p-value = 0.0006). f, Uptake of N-terminus of tau with mLRP ectodomain expression (n=12, WT vs NT; p-value = 0.9713, WT vs. mLRP2; p-value = 0.1289, WT vs. mLRP4; p-value = >0.9999). Experiments in a-b were performed in biological duplicates over three independent experiments (n=6) and representative examples are shown. Experiments in d-f were performed in biological triplicates over at least three independent experiments and normalized to WT uptake (100%, indicated with line). All data expressed as mean ± s.d. One-way ANOVA with Tukey’s method, two-sided was performed to determine significance. Displayed is the multiple comparison against WT, ns=not-significant, ***=p-value ≤0.001, ****=p-value < 0.0001. Comparison to NT sgRNA resulted in the same level of statistical significance.
In an attempt to understand how tau may interact with LRP1, we examined known crystal structures of the LDLR family bound to its ligands11,12. LRP1 (and other LDLR family members) contain cysteine-rich complement-type repeats (CRs) to bind and internalize their ligands. Each CR is composed of approximately 40 amino acids, six cysteines and an acidic residue cluster (normally aspartic acid) that coordinates Ca+2 and interacts with lysine residues on ligands through salt bridges13. Tau has a high lysine content, 44 lysines in 441 amino acids (10% content), and 20 of those are located within the microtubule binding region (K18: ~15% content). Furthermore, cryo-EM structures of tau fibrillar aggregates from AD and CTE brains14,15 show 10 or 11 resolved lysines with all but one exposed to the exterior and thus available for interactions with other proteins. Therefore, to assess if lysine salt bridges with LRP1 were necessary for tau uptake we capped all lysine residues on K18 using a sulfo-NHS acetate and tested endocytosis. Capping of lysine residues on K18 prevented uptake of tau in WT H4 cells (Fig. 2b), indicating that, similar to other LRP1 ligands, lysine residues are critical for this interaction.
LRP1 is a large (600kDa) member of the LDLR family, and it contains 31 CR repeats divided into four different ligand binding domains16. To ask which of these ligand binding domains influenced tau uptake, we designed ectodomain constructs, called mini-LRPs (mLRPs) to assess the ability of individual subdomains (Fig. 2c) to rescue tau internalization in our CRISPRi LRP1 knockdown cells. By expressing these mLRPs in the absence of full length LRP1 (Extended Data Fig. 2c), we found that full-length tau uptake was completely rescued with subdomain 4 (mLRP4) and partially rescued with mLRP2 (Fig. 2d). Further, mLRP4 was able to co-immunoprecipitate tau confirming an interaction between the two (Extended Data Fig. 2d). Only mLRP4 improved K18 uptake (Fig. 2e), indicating that two motifs in tau could possess the ability to bind LRP1, one within the microtubule binding region (amino acids 244–372) that interacts with mLRP4 and one in the N-terminal half (amino acids 1–243) or C-terminal end (amino acids 373–441) that interacts with mLRP2. To test this further, we examined the uptake of the N-terminus of tau (amino acids 1–243) and found that both mLRP4 and to a lesser extent, mLRP2 could rescue N-terminal uptake (Fig. 2f). This suggests a model where a primary interaction site on mLRP4 mediates tau uptake, but that the N-terminus of tau can also mediate interactions with a secondary site on mLRP2. We also noted that while LRP1 knockdown was sufficient to reduce uptake of the N-terminus, we did not observe the striking inhibition observed with constructs containing the microtubule binding repeat region. This may be attributed to the higher concentrations of N-terminus needed under our experimental assay conditions (Extended Data Fig. 2e), and could suggest a secondary route of tau endocytosis mediated by a slower LRP1-independent pathway. To determine if other known ligands of mLRP2 and mLRP4 are able to compete for uptake we overexpressed various ApoE isoforms in HEK293T cells and harvested the conditioned medium containing ApoE (Extended Data Fig. 2f). Results showed that incubation of H4 cells in ApoE conditioned medium significantly reduced uptake of full-length tau, yet failed to reduce transferrin uptake (Extended Data Fig. 2g).
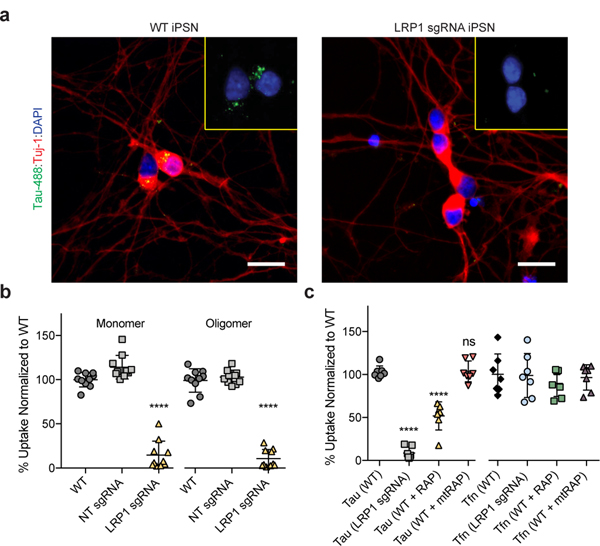
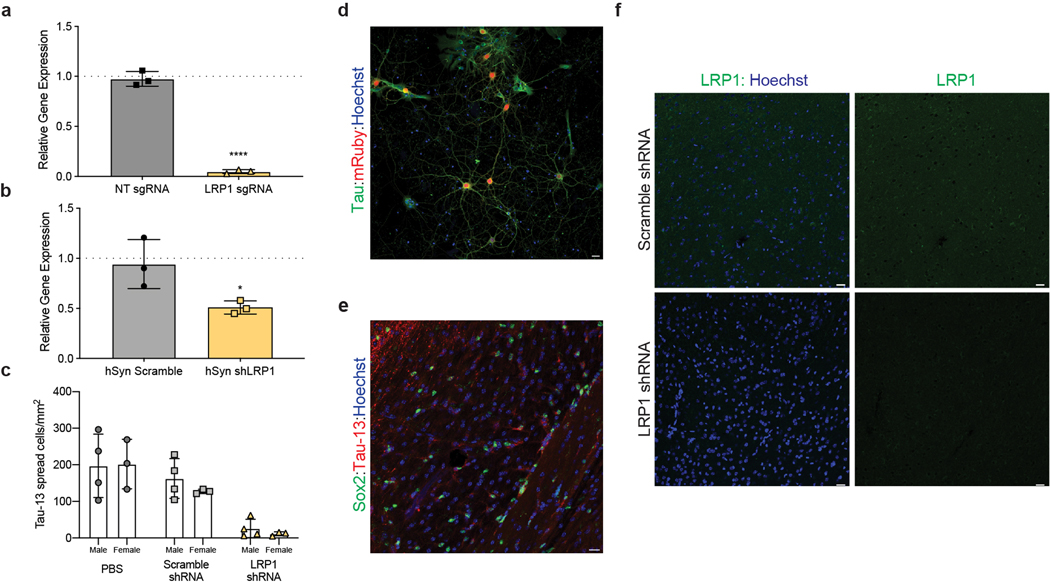
Native LRP1 is highly expressed in neurons at the post-synaptic density17,18, and previous work has highlighted that spread of tau in vivo is likely mediated trans-synaptically. Therefore, we asked if uptake of tau in neurons was also regulated by LRP1. We reduced the expression of LRP1 in human iPS-derived neurons (iPSNs) using CRISPRi (Extended Data Fig. 3a). Microscopy of adherent iPSNs showed that LRP1 knockdown efficiently reduced the amount of internalized tau (Tau-488; green; Fig. 3a). Quantification of tau endocytosis by flow cytometry confirmed the reduction of tau uptake upon LRP1 knockdown or upon addition of RAP, but not mtRAP into the culture medium (Fig. 3b,c). Similarly, to H4 cells, LRP1 knockdown or the addition of RAP had no effect on transferrin uptake in iPSNs (Fig. 3c).
Figure 3: LRP1 mediates tau uptake in neurons.
a, Representative images of WT or LRP1 sgRNA iPSN after tau uptake (green, Tau-488; red, neuronal marker Tuj-1; blue, DAPI; scale bar 20μm). b, Uptake of 2N4R monomers and oligomers in CRISPRi iPSN (n=10). c, Uptake of 2N4R tau (WT vs. mtRAP; p-value = >0.9999) or Tfn (ANOVA; p-value = 0.7815) in the presence of RAP or mtRAP (n=7). All experiments in a-c were performed over three independent experiments and normalized to WT uptake (100%). Data expressed as mean ± s.d. One-way ANOVA with Tukey’s method, two-sided was performed to determine significance. Displayed is the multiple comparison against WT, ns=not-significant, ****=p-value < 0.0001. Comparison to NT sgRNA resulted in the same level of statistical significance.
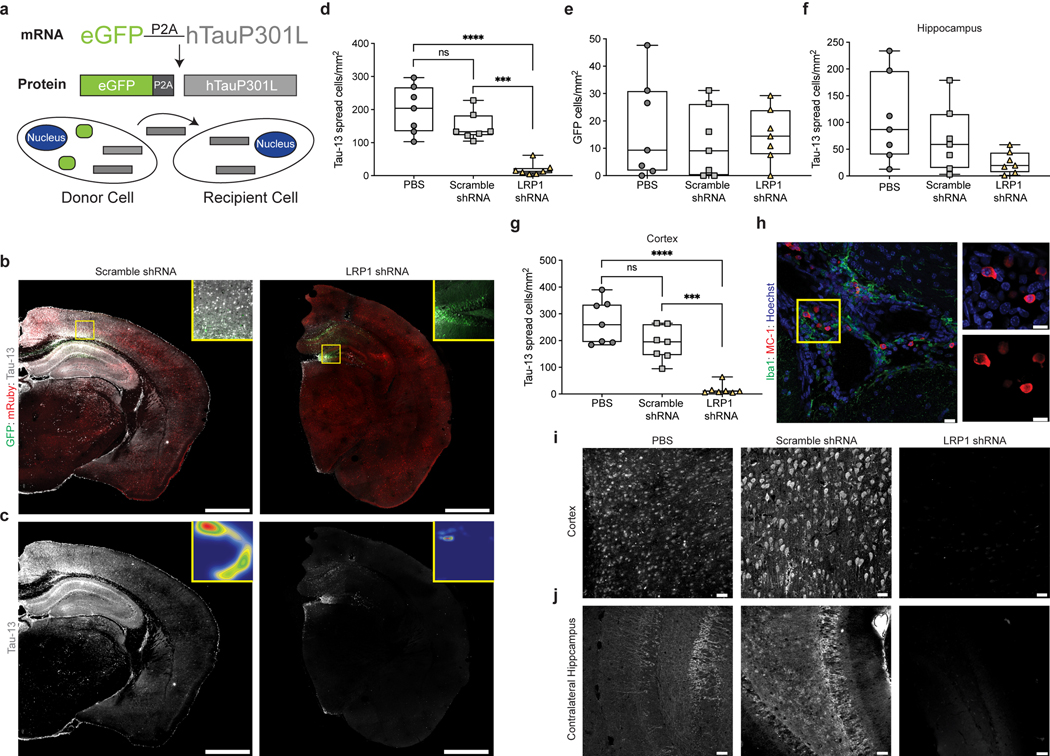
Based on these in vitro results, we sought to determine if LRP1 was also critical for tau spread in the brain. A newly developed model of tau spread has been described recently that utilizes an adeno-associated virus (AAV) and allows reliable discrimination of neurons that have been transduced to express human tau (hTau) versus neurons that receive hTau protein through spread19. The AAV encodes for one mRNA, GFP-P2A-hTau under control of a CMV promoter, but produces two proteins, GFP and hTau (Fig. 4a). The P2A peptide self-cleaves during translation20 and, thus, transduced neurons can be identified by the presence of GFP and hTau whereas cells that have taken up tau protein via spread mechanisms can be identified by the absence of GFP but the presence of hTau (Fig. 4a). To regulate LRP1 expression in parallel, an AAV (PHP.eB serotype) carrying an shRNA for LRP1 under the human synapsin (hSyn) promoter was used to knockdown LRP1 in neurons (Extended Data Fig. 3b,d). We retro-orbitally injected AAVs coding either the LRP1 shRNA or a scramble control shRNA into six-week old wildtype mice, and two weeks later performed stereotactic injections of the AAV GFP-2A-hTau virus into the hippocampus. After three weeks of incubation, the mice were sacrificed and spread was determined by immunofluorescence (Fig. 4b,c). Scramble and LRP1 shRNA AAVs contained the fluorescent protein reporter mRuby which allowed us to visualize their expression throughout the brain (Fig. 4b). Animals expressing LRP1 shRNA showed reduced expression for LRP1 as determined by qPCR and IHC (Extended Data Fig. 3b,f). To quantify tau spread, the number of hTau+/GFP- cells were counted after immunostaining for human tau (Fig. 4c,d). We observed a substantial amount of tau spread in PBS and scramble injected animals (mean ± s.d. = 199 ± 73 and 147 ± 43 hTau+/GFP- cells/mm2, respectively), whereas spread was greatly diminished in LRP1 knockdown animals (19 ± 19 hTau+/GFP- cells/mm2) (Fig. 4d). This was not due to differences in the number of transduced cells, as we observed equivalent amount of GFP+ cells across all animals (Fig. 4e). When analyzing tau spread by brain region, we found that while LRP1 shRNA animals had similar numbers of spread cells in the ipsilateral hippocampus (Fig. 4f), spread was significantly diminished in the cortex (Fig. 4g,i). We observed no effect of sex on the spread phenotype (Extended Data Fig. 3c). LRP1 is highly expressed in neurons, but it has also been shown that LRP1 is present in other cell types including astrocytes and microglia21. While not a focus of this work, we did observe instances of hTau+ astrocytes (Extended Data Fig. 3e) that will be a topic of future studies. Consistent with previous reports19, we found examples of MC-1 (antibody recognizing an altered conformation of tau) reactivity in PBS and scramble injected animals (Fig. 4h). Tau spread was also observed in the contralateral hippocampus in PBS and scramble injected mice, but was absent in our LRP1 shRNA animals (Fig. 4j).
Figure 4: LRP1 knockdown reduces tau spread in vivo.
a, Diagram of AAV used to study tau spread in vivo. b, Representative mosaic of a scramble shRNA injected mouse and LRP1 shRNA injected mouse (green, GFP; red, shRNA; white, hTau, scale bar 1mm) c, hTau spread in either scramble or LRP1 shRNA mice, insets represent false color densitometric maps of hTau staining d, Quantification of total number of hTau+/GFP- cells/mm2 of PBS, scramble shRNA or LRP1 shRNA mice (PBS vs. scramble shRNA; p-value = 0.1582). e, Quantification of total number of GFP+ cells/mm2 (ANOVA; p-value = 0.8007) f, Quantification of ipsilateral hippocampal hTau+/GFP- cells/mm2 in PBS, scramble shRNA or LRP1 shRNA mice (ANOVA; p-value = 0.0692) g, Quantification of ipsilateral cortical hTau+/GFP- cells/mm2 in PBS, scramble shRNA or LRP1 shRNA mice (PBS vs. scramble shRNA; p-value = 0.0548). h, Image of MC-1 positive tau in PBS injected animal (green, Iba1; red, MC-1; blue, Hoechst; scale bar 20μm) i, Representative images of ipsilateral cortex from PBS, scramble shRNA or LRP1 shRNA mice (white, hTau; scale bar 20μm). j, Representative images of contralateral hippocampus from PBS, scramble shRNA or LRP1 shRNA mice (white, hTau; scale bar 20μm). Experiments in b-j n=7 animals for each group (PBS, scramble shRNA, LRP1 shRNA). Data is shown as box-plots with the median displayed as a line and the whiskers representing min/max. One-way ANOVA with Tukey’s method, two-sided was performed to determine significance, ns=not-significant, ***=p-value ≤0.001, ****=p-value < 0.0001.
Taken together, our study identifies LRP1 as a master regulator of tau protein endocytosis in neurons, with an important role for tau spread in the brain. Targeting neuronal LRP1 led to a significant reduction of tau spread in vivo and could point to a novel therapeutic approach for tau-related neurodegenerative diseases. The importance of LRP1 in AD has previously been appreciated for its regulatory role in Aβ degradation/production and ApoE internalization22. ApoE4 has been shown to exacerbate Aβ pathology dependent on LRP1 expression23 and recently ApoE4 has also been shown to worsen tau pathology24. ApoE4 is known to have the lowest levels of circulating protein in the brain25, suggesting it would be the least effective in inhibiting tau-LRP1 interactions and, thus, suppressing tau spread. Altogether, our results point to LRP1 as a common factor that can help to explain the co-occurrence of amyloid and tau lesions. Recently, tau uptake studies have focused on mechanisms dependent on heparan sulfate proteoglycans (HSPGs)5,6. However, HSPGs are known to act both independently and in concert with co-receptors to mediate endocytosis26. Therefore, this work improves our understanding of tau spread, it highlights LRP1 as a critical determinant for tau propagation, and it suggests that interventions along a single pathway or at a single nidus, in which tau, ApoE and HSPGs operate dynamically in conjunction with LRP1, may remediate both plaque and tangle pathologies.
Methods:
Cloning
sgRNA (single guide RNA, used for dCas9-KRAB gene repression) constructs for individual genes were cloned into the pLG15 vector, according to previously published methods27. mLRP constructs were cloned into a pIRES-EGFP-puro (Addgene# 45567) from human cDNA. All mLRP constructs contained the N-terminal signaling peptide, residues 1–5, hemagglutinin (HA) tag, and the C-terminal transmembrane domain/tail (residues 3765–4525)28. Individual mLRP constructs were cloned to contain their respective subdomains in between the HA tag and C-terminal domain (mLRP1 residues 6–94; mLRP2 residues 787–1164; mLRP3 residues 2462–2923; mLRP4 residues 3264–3764). RAP was cloned into the pMCSG7 vector from human cDNA. Mutagenesis was performed on various constructs under standard protocols. Correct DNA sequences were confirmed (Genewiz).
Protein Purification and Labeling
Tau proteins were purified as described previously5. RAP, mtRAP, and the N-terminus of tau were cloned into the pMCSG7 and purified on Nickel-nitrilotriacetic acid (Ni-NTA) Agarose (Qiagen). The his-tag was removed by Tobacco Etch Virus (TEV) protease cleavage and subsequent clean-up on Ni-NTA agarose. Transferrin was purchased from Millipore and labeled in house. Protein was labeled with Alexa Fluor® 488 or 647 5-SDP ester (Life Technologies) according to the supplier’s instructions. After labeling, 100mM glycine was added to quench the reaction and the proteins were subjected to Zeba desalting columns (Thermo Scientific) to remove any unreacted label. Average label incorporation was between 1 and 1.5 moles/mole of protein, as determined by measuring fluorescence and protein concentration (Amax x MW of protein / [protein] x εdye). Lysine capped proteins were prepared with Sulfo-NHS-Acetate (Thermo) according to manufacturer’s instructions. Phosphorylation of tau was achieved by incubation with mouse brain extract in accordance with published work29 and confirmed via western blotting. To prepare tau fibrils and oligomers, 10 uM protein, in PBS 1 mM DTT pH 7.4, was mixed with heparin (0.05 mg/ml) and incubated with shaking at 37°C. The formation of oligomers was observed after 4h of shaking, whereas fibril formation occurred after 5 days as previously described30. To make sonicated fibril samples, fibrillized protein was sonicated using a Model 120 Sonicator (Fisher) at 30% amplitude for 30 × 1 s pulses.
Cell Culture
Cell lines (H4, HEK293T) were obtained from the ATCC and were maintained in DMEM supplemented with 10% FBS, 100ug/ml penicillin/streptomycin. H4 cells constitutively expressing CRISPRi machinery (H4i)5 were cultured in DMEM supplemented with 10% FBS, 100μg/ml penicillin/streptomycin. Cultures were maintained in a humidified atmosphere of 5% CO2 at 37°C. H4i cells were infected with indicated sgRNA constructs and selected with puromycin (1ug/ml). Knockdown was confirmed with qPCR and western blot analysis using anti-LRP1 (Sigma L2295; 1:1000) and anti-actin (Sigma A5441; 1:10000) as a loading control (Extended Data Fig. 1b,d). LRP1 sgRNA H4i cells were infected with mLRP constructs and sorted for GFP expression using a SH800 cell sorter (Sony Biotechnology). Expression of mLRP constructs was confirmed via western blot analysis using anti-HA primary antibody (Sigma H3663; 1:1000) and anti-GAPDH (Abcam 181602; 1:10000) as a loading control and quantification of surface expression was analyzed via immunocytochemistry (Extended Data Fig. 2c). HEK293T cells were transfected with various ApoE pCMV4 constructs (Addgene #87085–7) using Lipofectamine 2000 (Thermo Fisher) and conditioned media (CM) was harvested two days later. ApoE presence in media was confirmed via western blot (anti-ApoE; Abcam 52607; 1:1000; Extended Data Fig. 2f). Cells routinely tested negative for mycoplasma contamination.
iPS Culturing and Differentiation
CRISPRi iPSc with a constitutive dCas9-BFP-KRAB and doxycycline-inducible Neurogenin2 (Ngn2)31 were maintained in 6-well Matrigel (Corning) coated plates with E8 media (STEMCELL Technologies) and split with ReLeSR (STEMCELL Technologies) at a 1:20 ratio every 4–5 days. To induce differentiation, cells were plated in 6-well Matrigel coated plates with N2 predifferentiation media (Knockout DMEM/F12, NEAA, N2, 10ng/ml NT-3, 10ng/ml BDNF, 1μg/ml mouse laminin) in the presence of doxycycline (2μg/ml). Three days later, cells were lifted again and replated on PLL coated 24-well plates in BrainPhys Neuronal medium (STEMCELL Technologies). iPS neurons were assayed between days 14–18 of maturity as described below.
Tau Uptake Assay
H4 cells were plated at 50,000 cells/well, iPSN were plated at 100,000 cells/well in a 24-well plate. The next day media was replaced, and cells were treated with Alexa Fluor 488 or Alexa Fluor 647-labeled tau protein (50nM) or Alexa Fluor 488-labeled transferrin (500nM) for 1hr at 37°C (unless otherwise indicated in the figure legend). Cells were then washed twice with PBS and trypsinized to lift cells from the plate. Lifted cells were analyzed using an Accuri-C6 Flow Cytometer (BD Biosciences) and propidium iodide was used to exclude dead cells from the analysis (Extended Data Fig. 1a). Experiments were run in biological duplicates or triplicates and data was normalized to WT uptake to combine data sets from multiple experiments. Internalization controls are provided in Extended Data Fig. 1b. For RAP competition, recombinant RAP protein was added into the media at indicated concentrations at the same time as labeled tau. For ApoE competition, ApoE CM was used as the replacement media before tau addition.
AAV Production and Purification
For shRNA, PHP.eB AAVs were generated and harvested as previously described32. This engineered capsid allows efficient transduction of the central nervous system via the vasculature33. Sequences for LRP1 shRNA (CCGGGCTGAACACATTCTTTGGTAACTCGAGTTACCAAAGAATGTGTTCAGCTTTTTG), and scramble shRNA (CCGGCAACAAGATGAAGAGCACCAACTCGAGTTGGTGCTCTTCATCTTGTTGTTTTTG) were obtained from Sigma and were tested prior to AAV cloning in mouse primary culture. Purified viruses were concentrated, washed in PBS, sterile filtered and titered using qPCR. EGFP-2A-hTauP301L constructs were packaged into AAV2/8 (Massachusetts Eye and Ear Institute vector core).
Animals
All procedures were done with approval from the Institutional Animal Care and Use Committee and in compliance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals, the Animal Welfare Act, and guidelines from the University of California, Santa Barbara. Animals were maintained in ventilated cages, on a standard rodent diet of chow and water ad libitum and housed under a 12-hour light/dark cycle.
Experiments were performed starting at 6w of age on a sex mixed (4 males and 3 females per condition) cohort of in-house bred mice (FVB/B6). Animals were injected retro-orbitally with either PBS or an AAV PHP.eB of mRuby-shRNA with either a scramble or LRP1 sequence. After two weeks, animals were intracranially injected with AAVs encoding EGFP-2A-hTauP301L into the hippocampus (1.5μL unilaterally). Injections were performed under standard aseptic surgery conditions. Animals were anesthetized with isoflurane (2%) and injections were made at the coordinates (from bregma A/P −2mm, M/L −1.5mm, D/V −1.5mm). A Hamilton needle was used to inject the AAV solution at 0.5μL/min. Afterward, the skin over the injection site was sutured and animals were allowed to recover on a warming pad. Mice received meloxicam for two days after surgery. No calculations were performed to predetermine sample size, sample sizes were based on previously published work and chosen to support meaningful conclusions and no specific method of randomization was used.
Immunohistochemistry
Mice were perfused using 4% paraformaldehyde in 0.1M sodium cacodylate pH 7.4 for 15 mins. at room temperature. Brains were then immersion fixed for 48 hrs at 4°C. Immunocytochemistry was performed as described elsewhere34, briefly, samples were rinsed 3 × 5 mins., 1 × 1hr in cold phosphate buffered saline (PBS; pH: 7.4) and then sectioned at 100μm using a vibratome (Leica). Sections were immersed in normal donkey serum (Jackson ImmunoResearch Laboratories) 1:20 in PBS containing 0.5% BSA, 0.1% Triton-X 100, and 0.1% sodium azide (PBTA) at 4°C on a rotator overnight agitation. Next, sections were immersed in primary antibodies, anti-Tau13 (Biolegend 835201; 1:200), anti-Iba1 (Wako laboratory chemicals 019–19741; 1:200), MC-1 (Peter Davies; 1:200), and anti-Sox2 (Abcam 97959; 1:200) diluted in PBTA. Subsequently, sections were rinsed in PBTA 5 × 5 mins, 1 × 1hr in PBTA and then placed in corresponding secondary antibodies (Jackson ImmunoResearch Laboratories; 1:200). Lastly, sections were rinsed and mounted using Vectashield (Vector Laboratories Inc.) on a glass slide and sealed under an #0 coverslip (Thomas Scientific) using nail polish.
Large-scale mosaic acquisition and image registration
Samples were imaged using an Olympus FV1000 confocal microscope equipped with a precision motorized stage (Applied Scientific Instrumentation, Inc.). Mosaics were captured using an UPlanSApo 20x oil immersion lens N.A. 0.85 at 1μm intervals in the z-dimension and a pixel display of 800 × 800 along the x-y axes. Image stacks were collected sequentially using the Olympus Fluoview software version 4.2 with 5% overlap between individual tiles. Image registration of individual z-stacks was performed in a semi-automated fashion using the bio-image software Imago 1.5 (Mayachitra Inc.). False color densitometric maps were generated using x,y coordinates from annotated images using the visualization open source package ggplot2 version 3.2.1 as part of RStudio desktop version 1.2.5001 and subsequently rendered using R version 3.6.1. Animal mosaics were quantified under blinding conditions, file names were withheld until after image analysis was completed.
Statistical Analysis
All statistical analysis was performed using Prism 8 software. To determine statistical significance, Shapiro-Wilk tests were first used to evaluate the assumption of normality of the data. Given a p>0.05 then normality assumptions were not rejected and ANOVA tests were used. In certain cases, outliers were removed to meet the normality assumption and then an ANOVA test was performed, however all data is displayed in full. Post-hoc analysis of ANOVA was performed using Tukey’s method in order to compare experimental samples to both WT and NT sgRNA (or Scramble shRNA) controls. For qPCR data unpaired t-tests were used to determine significance.
Data availability
Source data of graphs plotted in Figs 1–4 and Extended Data Figs 1-3 are available as source data files. Fully scanned western blot gels can be found in the Supplementary Fig. 1. All other data are available from the corresponding author upon reasonable request
Supplementary Material
Extended Data
Extended Data Figure 1. Tau uptake is regulated by LRP1.
a, Gating strategy for tau uptake assay. First, cells were gated on FSC/SSC (Mean FSC-A: ~ 8,000,000/Mean SSC-A: ~800,000). Cells were then gated on FSC-H vs. Width to discriminate doublets. Dead cells were removed from analysis using propidium iodide as a stain, and positive cells were determined by gating on a negative (no tau added) population. b, Internalization controls for tau uptake assay (n=6). c, qPCR analysis of various genes with sgRNA in H4i cells. First column represents NT sgRNA control for each target (n=3). d, Uptake of phosphorylated (p2N4R) or mutated (2N4RP301L) full length tau in H4i cells (n=8). e, Western blot analysis of LRP1 in WT, NT sgRNA or LRP1 sgRNA H4i cells. All results in b-e were performed in three independent experiments and normalized to WT uptake (100%). Data is expressed as mean ± s.d. One-way ANOVA with Tukey’s method, two-sided was performed to determine significance, ****=p-value < 0.0001.
Extended Data Figure 2. LRP1 ligands influence tau uptake.
a, Uptake of 2N4R tau with RAP or lysine capped RAP competition. b, Uptake of 2N4R tau with RAP or mtRAP competition. c, Corrected Integrated Density of surface HA staining for different ectodomain cell lines (n=60). d, Co-immunoprecipitation of HA-mLRP4 with myc-2N4R (n=3). e, Uptake of various ligands at indicated concentrations (1hr). f, Western blot analysis of conditioned media from HEK293T cells mock treated or overexpressing ApoE (n=3). g, Uptake of full-length tau and transferrin in conditioned media from f (n=9). All results in a-b,e were performed in biological duplicates over three independent experiments (n=6) with representative experiments shown. Data is expressed as mean ± s.d. One-way ANOVA with Tukey’s method, two-sided was performed to determine significance, ****=p-value < 0.0001.
Extended Data Figure 3. LRP1 expression influences tau spread.
a, qPCR analysis of LRP1 expression in iPSN (n=3; p-value < 0.0001). b, qPCR analysis of LRP1 expression from mouse cortex transduced with hSyn Scramble shRNA or LRP1 shRNA (n=3; p-value = 0.0412). c, Quantification of tau spread in animals broken down by sex (Males n=4, Females n=3; Two-way ANOVA, sex = ns, p-value = 0.5335). d, Immunofluorescence of mouse primary culture transduced with AAVmRuby-hSyn-shLRP1 (green, Tau; red, mRuby; blue, Hoechst; scale bar 20μm, n=3). d, Immunohistochemistry of hTau+ astrocytes in PBS injected animal (green, Sox2; red, hTau; blue, Hoechst; scale bar 20μm, n=3). e, Immunohistochemistry of scramble shRNA and LRP1 shRNA animals (green, LRP1; blue; Hoechst, scale bar 20μm, n=3). Unpaired t-test, two-tailed was performed to determine statistical significance for qPCR in a,b, *=p-value ≤0.05, ****=p-value < 0.0001.
Acknowledgements
This study was funded by the National Institutes of Health (NIH), K99 AG064116 (J.N.R.), DP2 GM119139 (M.K.), R01 AG062359 (M.K.), R56 AG057528 (M.K.), U54 NS 100717 (M.K., K.S.K.), Tau Consortium (M.K., K.S.K.), German Center for Neurodegenerative Diseases (S.W.), Ben Barres Early Career Acceleration Award from the Chan Zuckerberg Initiative (M.K.), Tri-counties Blood Bank (J.N.R.), Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (K.S.K.), Larry L. Hillblom Foundation (K.S.K.), the Edward N. & Della L. Thome Memorial Foundation (K.S.K.). We thank the Laboratory for Stem Cell Biology and Engineering at UCSB for use of their flow cytometer and the Neuroscience Research Institute Microscopy Facility for the use of their microscopes. We thank Jason Dong for his help in the early stages of this project. We thank Peter Davies for providing the MC-1 Antibody.
References:
- 1.Goedert M, Eisenberg DS & Crowther RA Propagation of Tau Aggregates and Neurodegeneration. Annu Rev Neurosci 40, 189–210, doi: 10.1146/annurev-neuro-072116-031153 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Braak H & Braak E Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol 82, 239–259, doi: 10.1007/bf00308809 (1991). [DOI] [PubMed] [Google Scholar]
- 3.de Calignon A et al. Propagation of tau pathology in a model of early Alzheimer’s disease. Neuron 73, 685–697, doi: 10.1016/j.neuron.2011.11.033 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo JL et al. Unique pathological tau conformers from Alzheimer’s brains transmit tau pathology in nontransgenic mice. J Exp Med 213, 2635–2654, doi: 10.1084/jem.20160833 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rauch JN et al. Tau Internalization is Regulated by 6-O Sulfation on Heparan Sulfate Proteoglycans (HSPGs). Sci Rep 8, 6382, doi: 10.1038/s41598-018-24904-z (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holmes BB et al. Heparan sulfate proteoglycans mediate internalization and propagation of specific proteopathic seeds. Proc Natl Acad Sci U S A 110, E3138–3147, doi: 10.1073/pnas.1301440110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanekiyo T et al. Heparan sulphate proteoglycan and the low-density lipoprotein receptor-related protein 1 constitute major pathways for neuronal amyloid-beta uptake. J Neurosci 31, 1644–1651, doi: 10.1523/JNEUROSCI.5491-10.2011 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans LD et al. Extracellular Monomeric and Aggregated Tau Efficiently Enter Human Neurons through Overlapping but Distinct Pathways. Cell Rep 22, 3612–3624, doi: 10.1016/j.celrep.2018.03.021 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Nardis C et al. Recombinant Expression of the Full-length Ectodomain of LDL Receptor-related Protein 1 (LRP1) Unravels pH-dependent Conformational Changes and the Stoichiometry of Binding with Receptor-associated Protein (RAP). J Biol Chem 292, 912–924, doi: 10.1074/jbc.M116.758862 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dolmer K, Campos A & Gettins PG Quantitative dissection of the binding contributions of ligand lysines of the receptor-associated protein (RAP) to the low density lipoprotein receptor-related protein (LRP1). J Biol Chem 288, 24081–24090, doi: 10.1074/jbc.M113.473728 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nikolic J et al. Structural basis for the recognition of LDL-receptor family members by VSV glycoprotein. Nat Commun 9, 1029, doi: 10.1038/s41467-018-03432-4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fisher C, Beglova N & Blacklow SC Structure of an LDLR-RAP complex reveals a general mode for ligand recognition by lipoprotein receptors. Mol Cell 22, 277–283, doi: 10.1016/j.molcel.2006.02.021 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Lillis AP, Van Duyn LB, Murphy-Ullrich JE & Strickland DK LDL receptor-related protein 1: unique tissue-specific functions revealed by selective gene knockout studies. Physiol Rev 88, 887–918, doi: 10.1152/physrev.00033.2007 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fitzpatrick AWP et al. Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature 547, 185–190, doi: 10.1038/nature23002 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falcon B et al. Novel tau filament fold in chronic traumatic encephalopathy encloses hydrophobic molecules. Nature 568, 420–423, doi: 10.1038/s41586-019-1026-5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xing P et al. Roles of low-density lipoprotein receptor-related protein 1 in tumors. Chin J Cancer 35, 6, doi: 10.1186/s40880-015-0064-0 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andersen OM & Willnow TE Lipoprotein receptors in Alzheimer’s disease. Trends Neurosci 29, 687–694, doi: 10.1016/j.tins.2006.09.002 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Nakajima C et al. Low density lipoprotein receptor-related protein 1 (LRP1) modulates N-methyl-D-aspartate (NMDA) receptor-dependent intracellular signaling and NMDA-induced regulation of postsynaptic protein complexes. J Biol Chem 288, 21909–21923, doi: 10.1074/jbc.M112.444364 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wegmann S et al. Experimental evidence for the age dependence of tau protein spread in the brain. Sci Adv 5, eaaw6404, doi: 10.1126/sciadv.aaw6404 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szymczak AL et al. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat Biotechnol 22, 589–594, doi: 10.1038/nbt957 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Auderset L, Cullen CL & Young KM Low Density Lipoprotein-Receptor Related Protein 1 Is Differentially Expressed by Neuronal and Glial Populations in the Developing and Mature Mouse Central Nervous System. PLoS One 11, e0155878, doi: 10.1371/journal.pone.0155878 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shinohara M, Tachibana M, Kanekiyo T & Bu G Role of LRP1 in the pathogenesis of Alzheimer’s disease: evidence from clinical and preclinical studies. J Lipid Res 58, 1267–1281, doi: 10.1194/jlr.R075796 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tachibana M et al. APOE4-mediated amyloid-beta pathology depends on its neuronal receptor LRP1. J Clin Invest 129, 1272–1277, doi: 10.1172/JCI124853 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi Y et al. ApoE4 markedly exacerbates tau-mediated neurodegeneration in a mouse model of tauopathy. Nature 549, 523–527, doi: 10.1038/nature24016 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahley RW Central Nervous System Lipoproteins: ApoE and Regulation of Cholesterol Metabolism. Arterioscler Thromb Vasc Biol 36, 1305–1315, doi: 10.1161/ATVBAHA.116.307023 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christianson HC & Belting M Heparan sulfate proteoglycan as a cell-surface endocytosis receptor. Matrix Biol 35, 51–55, doi: 10.1016/j.matbio.2013.10.004 (2014). [DOI] [PubMed] [Google Scholar]
Additional References
- 27.Horlbeck MA et al. Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. Elife 5, doi: 10.7554/eLife.19760 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Obermoeller-McCormick LM et al. Dissection of receptor folding and ligand-binding property with functional minireceptors of LDL receptor-related protein. J Cell Sci 114, 899–908 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Despres C et al. Identification of the Tau phosphorylation pattern that drives its aggregation. Proc Natl Acad Sci U S A 114, 9080–9085, doi: 10.1073/pnas.1708448114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Usenovic M et al. Internalized Tau Oligomers Cause Neurodegeneration by Inducing Accumulation of Pathogenic Tau in Human Neurons Derived from Induced Pluripotent Stem Cells. The Journal of neuroscience : the official journal of the Society for Neuroscience 35, 14234–14250, doi: 10.1523/JNEUROSCI.1523-15.2015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tian R et al. CRISPR Interference-Based Platform for Multimodal Genetic Screens in Human iPSC-Derived Neurons. Neuron, doi: 10.1016/j.neuron.2019.07.014 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Challis RC et al. Systemic AAV vectors for widespread and targeted gene delivery in rodents. Nat Protoc 14, 379–414, doi: 10.1038/s41596-018-0097-3 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Chan KY et al. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat Neurosci 20, 1172–1179, doi: 10.1038/nn.4593 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luna G et al. The effects of transient retinal detachment on cavity size and glial and neural remodeling in a mouse model of X-linked retinoschisis. Invest Ophthalmol Vis Sci 50, 3977–3984, doi: 10.1167/iovs.08-2910 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data of graphs plotted in Figs 1–4 and Extended Data Figs 1-3 are available as source data files. Fully scanned western blot gels can be found in the Supplementary Fig. 1. All other data are available from the corresponding author upon reasonable request