Abstract
The complete chloroplast (cp) genome sequence of Actinidia eriantha was sequenced and assembled using Illumina paired-end data. The cp genome from A. eriantha is 156,964 bp in length, composed of a pair of 23,892 bp inverted repeat regions (IR) separated by a large single copy region (LSC) of 88,639 bp and a small single copy region (SSC) of 20,541 bp. The cp genome contained 113 unique genes, including 79 protein-coding genes, 30 tRNA genes, and four ribosomal RNA genes. The phylogenetic position of A. eriantha based the cp genome data was sister to the group A. rufa, A. deliciosa, and A. chinensis.
Keywords: Actinidia eriantha, chloroplast genome, Actinidiaceae, phylogenetic analysis
Actinidia Lindl. belong to family Actinidiaceae and consists of 54 species and 21 varieties (Kumar et al. 2016) found mainly in East and South Asia. Actinidia eriantha is endemic to China and has been listed in the second category of key protected wild plants (Guo et al. 2017). The fruit traits of A. eriantha have several advanatges, such as high vitamin C, easiness of pelling the pericarp and good storage (Zhong et al. 2012). It is recognized as a valuable species for commercial kiwifruit improvement as well as having been used in traditional Chinese medicine (Zhong et al. 2012; Sun et al. 2015). However, little is known the genetic diversity and genetic differentiation among the populations of A. eriantha. Such information is essential for developing optimum conservation and management strategies for A. eriantha. In this study, we report the first complete plastome of A. eriantha, and assessed its phylogenetic position within Actinidaceae.
One individual of A. eriantha was sampled from National Kiwifruit Germplasm Genebank of China, Wuhan Botanical Garden, the Chinese Academy of Sciences (30°33′48.6″ N, 114°25′1.2″ E). Total DNA was isolated using the DNeasy plant Mini Kit (Quiagen, Carlsbad, CA) and stored in a DNA bank in the Wuhan Botanical Garden, the Chinese Academy of Sciences. The strategy for sequencing, assembly and annotation the chloroplast genome was adapted from Yao et al. (2015). The sequence of chloroplast genomes was deposited in GenBank (accession numbers KY100978).
The complete cp genome sequence of A. eriantha is 156,964 bp in length. The complete cp genome exhibit the typical quadripartite structure, including a pair of IRs (23,892 bp) separated by the LSC (88,639 bp) and SSC (20,541 bp) regions. The cp genomes encode an identical set of 113 predicted functional genes when duplicated genes are calculated only once, including 79 protein-coding genes, 30 transfer RNA (tRNA) genes, and four rRNA genes. Five protein-coding (i.e. rps7, ndhB, ycf15, ycf2, and rps12), eight tRNA (i.e. trnH-GUG, trnI-CAU, trnLCAA, trnI-GAU, trnV-GAC, trnA-UGC, trnR-ACG, and trnN-GUU) and all four rRNA genes are duplicated in the IR regions, and one tRNA gene [trnfM (CAU)] is duplicated in the LSC regions. The LSC region contains 62 protein-coding and 21 tRNA genes, and the SSC region contains 12 protein-coding and one tRNA gene. The GC content of A. eriantha cp genomes is 37.2%, which is similar to the other reported Actinidia cp genomes (Yao et al. 2015; Lan et al. 2018). The GC content of the IR regions (43.1%) is higher than that of the LSC and SSC regions (35.4% and 31.1%, respectively).
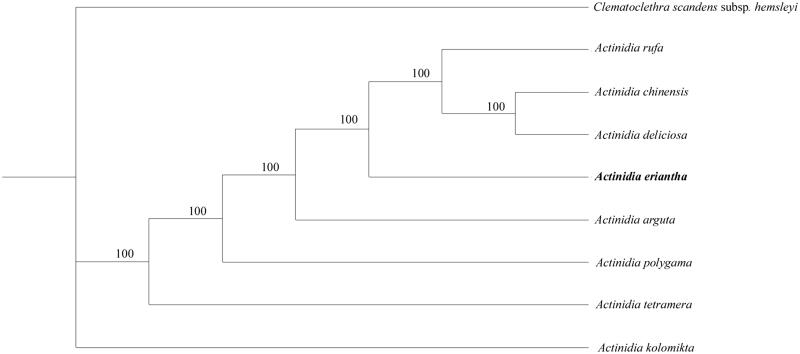
To confirm the phylogenetic position of A. eriantha, nine complete chloroplast genome sequences of Actinidiaceae were aligned using MAFFT v.7 (Katoh and Standley 2013) and maximum parsimony (MP) analysis was conducted using with PAUP software (version 4.0b10) with 1000 bootstrap replicates (Swofford 2003). The MP tree showed that A. eriantha was sister to the group including the A. rufa, A. deliciosa, and A. chinensis (Figure 1).
Figure 1.
Phylogenetic position of Actinidia eriantha in Actinidia as inferred by MP analyses of chloroplast genome sequences. Numbers above the lines indicate the maximum likelihood bootstrap value >50% for each clade. Accession Numbers: Actinidia chinensis (NC026690), A. deliciosa (NC026691), A. rufa (NC039973), A. tetramera (NC031187), A. arguta (NC034913), A. polygama (NC031186), A. kolomikta (NC034915), and Clematoclethra scandens subsp. hemsleyi (KX345299).
Disclosure statement
The authors report no declaration of interest. The authors alone are responsible for the content and writing of this article.
References
- Guo R, Landis JB, Moore MJ, Meng A, Jian S, Yao X, Wang H. 2017. Development and application of transcriptome-derived microsatellites in Actinidia eriantha (Actinidiaceae). Front Plant Sci. 8:1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y, Cheng L, Huang W, Cao Q, Zhou Z, Luo A, Hu G. 2018. The complete chloroplast genome sequence of Actinidia kolomikta from North China. Conserv Genet Resour. 10:475–477. [Google Scholar]
- Sun HX, Zhang J, Chen FY, Chen XF, Zhou ZH, Wang H. 2015. Activation of RAW264.7 macrophages by the polysaccharide from the roots of Actinidia eriantha and its molecular mechanisms. Carbohydr Polym. 121:388–402. [DOI] [PubMed] [Google Scholar]
- Swofford DL. 2003. PAUP*. Phylogenetic analysis using parsimony (* and other methods), Version 4b 10. Sunderland (MA): Sinauer Associates. [Google Scholar]
- Yao X, Tang P, Li Z, Li D, Liu Y, Huang H. 2015. The first complete chloroplast genome sequences in Actinidiaceae: genome structure and comparative analysis. PloS One. 10:e0129347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong C, Wang M, Jiang Z, Huang H. 2012. ‘Jinyan’, an interspecific hybrid kiwifruit with brilliant yellow flesh and good storage quality. HortScience. 47:1187–1190. [Google Scholar]