Abstract
Calanthe arcuata is an endemic terrestrial orchid in China with high value of ornament and breeding. Here, we reported the first complete chloroplast genome of this plant in this research. The circular genome is 158,735 bp in length and includes large single-copy (LSC) region of 87,348 bp, small single-copy (SSC) region of 18,489 bp, and a pair of invert repeats (IR) regions of 26,449 bp. It contains 136 genes, including 88 protein-coding (PCG), 38 transfer RNA (tRNA), and eight ribosomal RNA (rRNA) gene. The phylogenetic analysis indicated that C. arcuata is the sister to C. davidii and C. triplicata.
Keywords: Calanthe arcuata, chloroplast genome, Orchidaceae, Illumina sequencing, phylogenetic
Calanthe arcuata is an endemic terrestrial orchid widely distributed in China (Gansu, Hubei, Shaanxi, Guizhou, Sichuan, Yunnan, Tibet, and Taiwan) (Chen et al. 2010). Calanthe arcuata usually grows in soil or rocks along streams in humus-rich valleys, under mixed forests at altitudes between 2700 and 3450 m (Clayton and Cribb 2013). The distribution of this species has the characteristics of continental and island discontinuous distribution. Due to climate change, human destruction, and habitat loss or fragmentation, wild populations of C. arcuata sharply decreased in recent years. As a result, this species has been listed as a vulnerable species in the Red List (IUCN 2018). In order to better protection of this orchid and understand their genetic information, we assembled complete chloroplast genome data, which would be helpful for the population genetic and phylogenetic research of C. arcuata.
We assembled the complete chloroplast genome of C. arcuata using a silicon dried sample, which was collected from Taibai Mountain, Shaanxi, China (N 34°05′, E 107°75′). DNA was stored at Fujian Agriculture and Forestry University (specimen voucher S69). The total genomic DNA was extracted using a modified CTAB method (Doyle and Doyle 1987) and sequenced based on the Illumina pair-end technology. The clean reads were firstly aligned to C. triplicata (GenBank accession No. KF753635) (Yang et al. 2014). Filtered reads were then assembled into contigs in CLC Genomics Workbench version 8.0 (CLC Bio, Aarhus, Denmark). The genome was automatically annotated using DOGMA (Wyman et al. 2004), then adjusted using Geneious version 11.1.15 (Kearse et al. 2012) and submitted to GenBank with accession number MK934523. The complete chloroplast genome of C. arcuata is 158,735 base pairs (bp) in length, containing a large single-copy (LSC) region of 87,348 bp, a small single-copy (SSC) region of 18,489 bp, and two inverted repeat (IR) regions of 26,449 bp. A total of 136 genes were annotated, including 88 protein-coding (PCG), 38 transfer RNA (tRNA), and eight ribosomal RNA (rRNA) gene. The complete genome GC content was 36.60%, whereas the corresponding values of the LSC, SSC, and IR regions are 34.2%, 29.7%, and 43%, respectively.
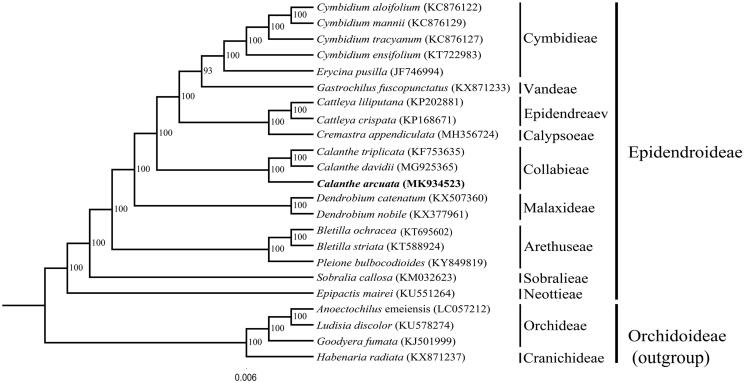
In order to study the phylogenetic position of C. arcuata, a phylogenetic analysis was carried out with 22 complete chloroplast genomes of Orchidaceae species, which included 18 Epidendroideae species (Bletilla ochracea, B. striata, Calanthe davidii, C. triplicata, Cattleya crispata, C. liliputana, Cremastra appendiculata, Cymbidium aloifolium, C. ensifolium, C. mannii, C. tracyanum, Dendrobium catenatum, D. nobile, Epipactis mairei, Erycina pusilla, Gastrochilus fuscopunctatus, Pleione bulbocodioides, and Sobralia callosa) and four Orchidoideae species (Goodyera fumata, Anoectochilus emeiensis, Ludisia discolor and Habenaria radiata) as outgroup. All of the data was downloaded from NCBI GenBank. The sequences were aligned using HomBlocks pipeline (Bi et al. 2017). RAxML-HPC2 on XSEDE version 8.2.10 (Stamatakis 2014) was used to construct a maximum likelihood tree, the branch support was computed with 1,000 bootstrap replicates. The ML tree analysis indicated that C. arcuata is sister to C. davidii (MG925365) and C. triplicata (KF753635) with 100% bootstrap support (Figure 1).
Figure 1.
The maximum-likelihood (ML) tree based on 18 species of Epidendroideae, based on whole chloroplast genome sequences, with Orchidoideae: Goodyera fumata, Anoectochilus emeiensis, Ludisia discolor, and Habenaria radiata as outgroup. The bootstrap value based on 1000 replicates is shown on each node, and the position of Calanthe arcuata is shown in bold.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Bi G, Mao Y, Xing Q, Cao M. 2017. HomBlocks: a multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 110:18–22. [DOI] [PubMed] [Google Scholar]
- Chen S, Liu Z, Zhu G. 2010. Orchidaceae flora of China. Vol. 25. Beijing: Science Press, Missouri Botanical Garden Press. [Google Scholar]
- Clayton D, Cribb P. 2013. The genus Calanthe. Borneo: Natural History Publications. [Google Scholar]
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15. [Google Scholar]
- IUCN. 2018. The IUCN Red List of Threatened Species. Version. 2018-2; [accessed 2018 Mar 20]. www.iucnredlist.org.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255. [DOI] [PubMed] [Google Scholar]
- Yang JB, Li DZ, Li HT. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Resour. 14:1024–1031. [DOI] [PubMed] [Google Scholar]