Abstract
Complete chloroplast genome of candidate new species from Rosa rugosa, named as Rosa angusta, is 156,989 bp long and has four subregions: 86,227 bp of large single copy (LSC) and 18,816 bp of small single copy (SSC) regions are separated by 25,793 bp of inverted repeat (IR) regions including 130 genes (85 protein-coding genes, eight rRNAs, and 37 tRNAs). The overall GC content of this chloroplast genome is 37.2% and in the LSC, SSC, and IR regions are 35.2%, 31.1%, and 42.8%, respectively. Phylogenetic trees show that R. angusta is close to R. rugosa with enough number of sequence variations.
Keywords: Rosa rugosa, Rosa angusta, chloroplast genome, Rosaceae
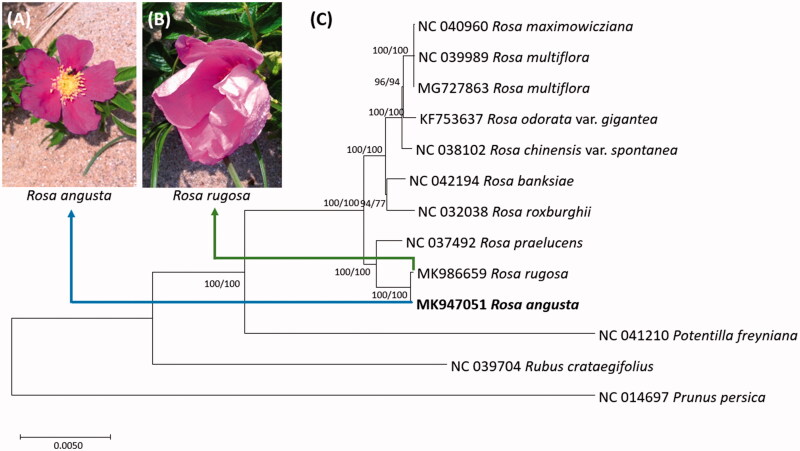
Genus Rosa belonging to family Rosaceae consists of around 200 species distributed in the temperate and subtropical regions of the Northern hemisphere (Rehder 1949) and its taxonomic treatment is complicated due to highly diverged characteristics (Wissemann and Ritz 2005). In 2013, Mr. Suhwan Nam, one of the authors, found a small population of Rosa rugosa in Hakampo beach of Taean-gun in Chungcheongnam-do, presenting that leaflets are elliptic and petals are not overlapped; while those of R. rugosa are widely elliptic and petals are overlapped (Figure 1A,B). We suspected that this population is a candidate new species, named as Rosa angusta. To decipher its genetic background, its chloroplast genome was completed.
Figure 1.
(A) Picture of Rosa augsta flower, (B) Picture of Rosa rugosa flower, (C) Neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) phylogenetic trees of ten Rosa chloroplast genomes and three outgroup species: Rosa angusta (MK947051 in this study), Rosa rugosa (MK986659), Rosa praelucens (NC_037492), Rosa roxburghii (NC_032038), Rosa banksiae (NC_042194), Rosa chinensis var. spontanea (NC_038102), Rosa odorata var. gigantea (KF753637), Rosa multiflora (NC_039989 and MG727863), Rosa maximowicziana (NC_040960), and three outgroup species: Potentilla freyniana (NC_041210), Rubus crataegifolius (NC_039704), and Prunus persica (NC_014697). Phylogenetic tree was drawn based on neighbor joining tree. The numbers above branches indicate bootstrap support values of maximum likelihood and neighbor joining phylogenetic tree, respectively.
Its total DNA isolated from Hagampo coast, Wonbuk-myeon, Taean-gun, Chungcheongnam-do, Republic of Korea, was extracted from fresh leaves using a DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Voucher was deposited in InfoBoss Cyber Herbarium (IN; IB-90006). Genome was sequenced using HiSeqX at Macrogen Inc., Korea, and de novo assembly and confirmation were performed by Velvet 1.2.10 (Zerbino and Birney 2008), SOAPGapCloser 1.12 (Zhao et al. 2011), BWA 0.7.17 (Li 2013), and SAMtools 1.9 (Li et al. 2009). Geneious R11 11.0.5 (Biomatters Ltd., Auckland, New Zealand) was used for annotation based on Rosa praelucens chloroplast (NC_037492; Jian et al. 2018).
Chloroplast genome of R. angusta (Genbank accession is MK947051) is 156,989 bp long (GC ratio is 37.2%) and has four subregions: 86,227 bp of large single copy (35.2%) and 18,816 bp of small single copy (SSC; 31.1%) regions are separated by 25,793 bp of inverted repeat (IR; 42.8%). It contains 130 genes (85 protein-coding genes, eight rRNAs, and 37 tRNAs); 17 genes (seven protein-coding gene, four rRNAs, and six tRNAs) are duplicated in IR regions.
Based on raw reads of R. rugosa (SRR1660458), chloroplast genome (Genbank accession is MK986659) was reconstructed. Pair-wise alignment between R. rugosa and R. angusta chloroplast genomes was conducted under the Plant Chloroplast Database (PCD; Park et al., in preparation), resulting 40 single nucleotide polymorphisms and 224 insertions and deletions. They present enough differences between two neighbor species supported by various researches showing less number of intraspecies variations on chloroplast genomes (Kim et al. 2019; Min et al. 2019; Park and Kim 2019; Park, Kim, Kwon, et al. 2019; Park, Kim, Lee 2019; Park, Kim, Xi 2019; Park, Kim, Xi, Heo 2019; Park, Kim, Xi, Nho, et al. 2019; Park et al. 2019b, 2019a; Park, Xi, et al. 2019).
Ten Rosa chloroplast genomes including that of R. angusta, and three outgroup chloroplast genomes were used for constructing neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) phylogenic trees using MEGA X (Kumar et al. 2018) after aligning whole chloroplast genomes by MAFFT 7.388 (Katoh and Standley 2013) with fixing SSC directions of Rosa maximowicziana and Rosa multiflora (Jeon and Kim 2019). Phylogenetic trees show that R. augusta is similar to R. rugosa with reasonable number of sequence varaitions (Figure 1C). Chloroplast genome of candidate new species (Heo et al. 2019; Kim et al. 2019; Oh et al. 2019) can provide additional evidence to clarify its taxonomical position which is similar to the cases (Heo et al. 2019; Oh et al. 2019).
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Heo K-I, Park J, Kim Y. 2019. The complete chloroplast genome of new variety candidate in Korea, Potentilla freyniana var. chejuensis (Rosoideae). Mitochondrial DNA B. 4:1354–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J-H, Kim S-C. 2019. Comparative analysis of the complete chloroplast genome sequences of three closely related East-Asian Wild Roses (Rosa sect. Synstylae; Rosaceae). Genes. 10:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian H-y, Zhang S-d, Zhang T, Qiu X-q, Yan H-j, Li S-b, Wang Q-g, Tang K-x. 2018. Characterization of the complete chloroplast genome of a critically endangered decaploid rose species, Rosa praelucens (Rosaceae). Conserv Genet Resour. 10:851–854. [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Heo K-I, Park J. 2019. The second complete chloroplast genome sequence of Pseudostellaria palibiniana (Takeda) Ohwi (Caryophyllaceae): intraspecies variations based on geographical distribution. Mitochondrial DNA B. 4:1310–1311. [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv Preprint arXiv:13033997. [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J, Park J, Kim Y, Kwon W. 2019. The complete chloroplast genome of Artemisia fukudo Makino (Asteraceae): providing insight of intraspecies variations. Mitochondrial DNA B. 4:1510–1512. [Google Scholar]
- Oh S-H, Suh HJ, Park J, Kim Y, Kim S. 2019. The complete chloroplast genome sequence of a morphotype of Goodyera schlechtendaliana (Orchidaceae) with the column appendages. Mitochondrial DNA B. 4:626–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Kim Y. 2019. The second complete chloroplast genome of Dysphania pumilio (R. Br.) mosyakin & clemants (Amranthaceae): intraspecies variation of invasive weeds. Mitochondrial DNA B. 4:1428–1429. [Google Scholar]
- Park J, Kim Y, Kwon W, Nam S, Song MJ. 2019. The second complete chloroplast genome sequence of Nymphaea alba L. (Nymphaeaceae) to investigate inner-species variations. Mitochondrial DNA B. 4:1014–1015. [Google Scholar]
- Park J, Kim Y, Lee K. 2019. The complete chloroplast genome of Korean mock strawberry, Duchesnea chrysantha (Zoll. & Moritzi) Miq. (Rosoideae). Mitochondrial DNA B. 4:864–865. [Google Scholar]
- Park J, Kim Y, Xi H. 2019. The complete chloroplast genome of aniseed tree, Illicium anisatum L. (Schisandraceae). Mitochondrial DNA B. 4:1023–1024. [Google Scholar]
- Park J, Kim Y, Xi H, Heo K-I. 2019. The complete chloroplast genome of ornamental coffee tree, Coffea arabica L. (Rubiaceae). Mitochondrial DNA B. 4:1059–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Kim Y, Xi H, Nho M, Woo J, Seo Y. 2019. The complete chloroplast genome of high production individual tree of Coffea arabica L. (Rubiaceae). Mitochondrial DNA B. 4:1541–1542. [Google Scholar]
- Park J, Kim Y, Xi H, Oh YJ, Hahm KM, Ko J. 2019a. The complete chloroplast genome of common camellia tree in Jeju island, Korea, Camellia japonica L. (Theaceae): intraspecies variations on common camellia chloroplast genomes. Mitochondrial DNA Part B. 4:1292–1293. [Google Scholar]
- Park J, Kim Y, Xi H, Oh YJ, Hahm KM, Ko J. 2019b. The complete chloroplast genome of common camellia tree, Camellia japonica L. (Theaceae), adapted to cold environment in Korea. Mitochondrial DNA B. 4:1038–1040. [Google Scholar]
- Park J, Xi H, Kim Y, Heo K-I, Nho M, Woo J, Seo Y, Yang JH. 2019. The complete chloroplast genome of cold hardiness individual of Coffea arabica L. (Rubiaceae). Mitochondrial DNA B. 4:1083–1084. [Google Scholar]
- Rehder A. 1949. Bibliography of cultivated trees and shrubs hardy in the cooler temperate regions of the northern hemisphere. Jamaica Plain (MA): Arnold Arboretum of Harvard University. [Google Scholar]
- Wissemann V, Ritz CM. 2005. The genus Rosa (Rosoideae, Rosaceae) revisited: molecular analysis of nrITS-1 and atp B-rbc L intergenic spacer (IGS) versus conventional taxonomy. Bot J Linn Soc. 147:275–290. [Google Scholar]
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12:S2. [DOI] [PMC free article] [PubMed] [Google Scholar]