Abstract
In this study, the complete mitochondrial genome of Phyllotreta striolata (Coleoptera, Chrysomeloidea, Chrysomelidae) was first determined. The complete genome is 15,689 bp in length. It contains 13 protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes, and a control region (A + T-rich region). The gene organization, nucleotide composition, and codon usage are similar to other Chrysomelidae mitogenomes. The overall nucleotide composition was 39.90% A, 35.94% T, 15.27% C, and 8.89% G, respectively. Phylogenetic analysis both highly supported that P. striolata showed a close relationship with P. undulata. The measure of complete mitogenome sequence of P. striolata will provide fundamental data for the phylogenetic and biogeographic studies of the Chrysomeloidea and Coleoptera.
Keywords: Chrysomeloidea, complete mitochondrial genome, Phyllotreta striolata
The striped flea beetle (SFB), Phyllotreta striolata (Fabricius) (Coleoptera, Chrysomelidae, Galerucinae), is a notorious vegetable pest worldwide. It is harmful to many plants, especially cruciferous vegetables (Feeny et al. 1970). In the south of China, the cultivation area of cruciferous vegetables is expanding year by year, which results in the continuous occurrence of the striped flea beetles. In addition, the long-term use of a single chemical control method in production has led to rapid and widespread improvement of pesticide resistance in the population of P. striolata, and its occurrence is becoming more and more serious (Liu et al. 2018). So, it is important to sequence and annotate the mitochondrial genome (mito-genome) of P. striolata. Here, we report the complete mito-genome sequence of P. striolata by next-generation sequencing (NGS). The annotated mitochondrial DNA (mtDNA) sequence has been deposited in GenBank under accession number MK256930.
In this study, the specimens were collected from Wenzhou, Zhejiang province (28°5′18.11″N, 120°30′55.57″E) of China in September 2018 and were deposited in the insect collection of Wenzhou Vocational College of Science and Technology (Wenzhou, China).
The specimens were initially preserved in 100% ethanol and then stored at 4 °C prior to DNA extraction. Total genomic DNA was extracted from the individual adult specimen using a DNeasy tissue kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. One library was constructed with VAHTS™ Universal DNA Library Prep Kit for Illumina® and sequenced with Illumina HiSeq X Ten sequencer (150 bp pared-end) by Novogene (Beijing, China).
The complete mitochondrial genome of P. striolata was a circular molecule with 15,689 bp in length (GenBank Accession number MK256930). The mitogenome of P. striolata contained two rRNA genes, 13 protein-coding genes (PCGs), 22 tRNA genes, and a control region. The overall nucleotide composition was 39.90% A, 35.94% T, 15.27% C, and 8.89% G, with a slight AT bias of 75.84%. Five PCGs initiation codons were ATT, four PCGs initiation codons were ATG, two PCGs initiation codons were ATA, two PCGs began with ATC, different from P. undulata (cox2, nad4l, and nad6 with ATA, nad4 with ATG, cob with ATT) and P. parallela (cox2 and nad5 with ATA, nad4 with ATG, cob with ATC) (Gomez-Rodriguez et al. 2015). Correspondingly, six PCGs stopped with the complete termination codon TAA, two PCGs stopped with the complete termination codon TAG, cox2 stopped with an incomplete termination codon TA while the rest of PCGs ended with an incomplete termination codon T–– (atp8, cox 3, nad4, and nad5), which was different from P. undulata and P. parallela (nad2, nad3, and cox1 with T, atp8 and cox3 with TAA) (Gomez-Rodriguez et al. 2015). Moreover, the 22 tRNA genes ranged in size from 59 bp (tRNASer(UCU)) to 73 bp (tRNALys). The two rRNA genes of P. striolata were determined by sequence alignment with other species. The lrRNA is located between the tRNALeu(UAG) and tRNAVal with a 1262bp in length while the srRNA is located between the tRNAVal and the control region with a 746 bp in length.
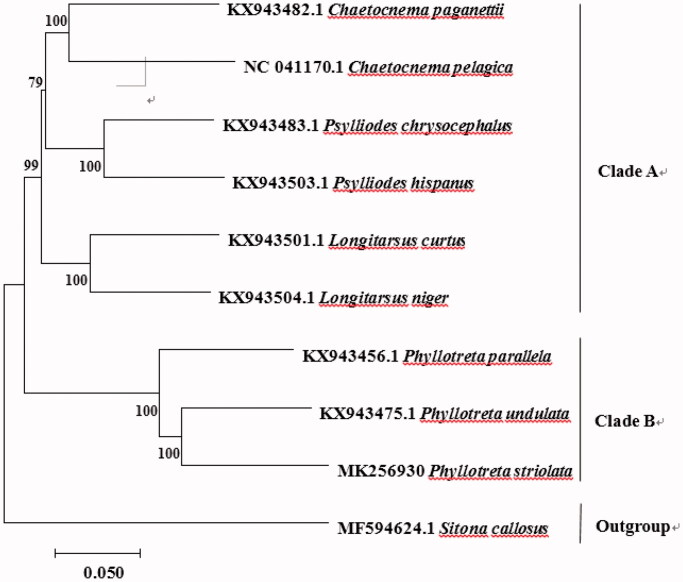
To confirm the phylogenetic relationships between P. striolata and other Chrysomelidae, phylogenetic analysis was performed on the concatenated dataset of 13 PCGs at neighbour-joining (NJ) method that produced NJ tree (Gomez-Rodriguez et al., 2015; Nie et al., 2018; Zhang et al., 2017) (Figure 1). Sitona callosus (Curculionidae, Entiminae) was defined as an outgroup. The other nine species were divided into two clades. Phyllotreta parallela, P. undulata and P. striolata were grouped in one clade, suggested the close relationship of these species, and further confirmed that P. striolata belongs to the subfamily Galerucinae. Phyllotreta striolata was evolutionarily closest to P. undulata, and P. parallela was a sister of P. striolata (Figure 1).
Figure 1.
Neighbour-joining (NJ) phylogenetic tree of Phyllotreta striolata and nine other species using Sitona callosus as an outgroup. The number above each node indicates the NJ bootstrap support values.
Acknowledgements
We are grateful to Dr Song Shengnan from Oil Crops Research Institute, Chinese Academy of Agricultural Science (Wuhan), Dr Wang Sheng from Wenzhou Vocational College of Science and Technology, Dr Tang Pu and Zheng Boying from Zhejiang University for their help in this study.
Disclosure statement
The authors declare no conflict of interest. The authors alone are responsible for the content and writing of the paper.
References
- Feeny P, Paauwk KL, Demony NJ. 1970. Flea beetles and mustard oils: Host plant specificity of Phyllotreta cruciferae and P. striotata adults (Coleoptera: Chrysomelidae). Ann Entomol Soc Am. 63:841–847. [Google Scholar]
- Gomez-Rodriguez C, Crampton-Platt A, Timmermans M, Baselga A, Vogler AP. 2015. Validating the power of mitochondrial metagenomics for community ecology and phylogenetics of complex assemblages. Methods Ecol Evol. 6:883–894. [Google Scholar]
- Liu H, Xiang YL, Zhao XF, Chen JH, Chen YP, Luo JF, Zhang MX, Li ZQ. 2018. Research and demonstration of Phyllotreta striolata Fabricius integrated prevention and control technology. J Environ Entomol. 40:461–467. (In Chinese with English Abstract.) [Google Scholar]
- Nie RE, Breeschoten T, Timmermans M, Nadein K, Xue HJ, Bai M, Huang Y, Yang XK, Vogler AP. 2018. The phylogeny of Galerucinae (Coleoptera: Chrysomelidae) and the performance of mitochondrial genomes in phylogenetic inference compared to nuclear rRNA genes. Cladistics. 34:113–130. [DOI] [PubMed] [Google Scholar]
- Zhang L, Wang J, Yang XZ, Li XP, Feng RQ, Yuan ML. 2017. Mitochondrial genome of Sitona callosus (Coleoptera: Curculionidae) and phylogenetic analysis within Entiminae. Mitochondrial DNA B Resour. 2:538–539. [DOI] [PMC free article] [PubMed] [Google Scholar]