Abstract
Potentilla freyniana Bornm. has radical leaves with petioles and trifoliolate distinguished from Potentilla fragarioides and Duchesnea indica. In this study, we presented first complete chloroplast genome of P. freyniana to understand its phylogenetic position. Its length is 156,381 bp long and has four subregions: 85,724 bp of large single copy (LSC) and 18,617 bp of small single copy (SSC) regions are separated by 26,020 bp of inverted repeat (IR) regions including 129 genes (84 protein-coding genes, 8 rRNAs, and 37 tRNAs). The overall GC content of the chloroplast genome is 36.9% and those in the LSC, SSC, and IR regions are 34.8%, 30.7%, and 42.7%, respectively. Phylogenetic trees show that phylogenetic position of P. freyniana disagrees with three phylogenetic studies, showing that more Potentilla chloroplast genomes are required for clarifying Potentilla phylogeny.
Keywords: Potentilla freyniana, chloroplast genome, Rosaceae, Potentilla, phylogeny
Genus Potentilla, belonging to Rosaceae, covers around 300 species and is distributed in field of mountain from temperate and arctic regions (Rydberg 1898). Potentilla freyniana Bornm. has radical leaves with petioles and trifoliolate (Chen and Craven 2007), which is an important key to distinguish from Potentilla fragarioides and Duchesnea indica.
P. freyniana was clustered with Potentilla fragaiodes using chloroplast markers (Heo et al. under review), considering that it belongs to Fragarioides/C/P6 clade (Töpel et al. 2011; Feng et al. 2017; Heo et al., under review; Figure 1). Since three phylogenetic studies utilized limited number of molecular markers, complete chloroplast genome of P. freyniana will be utilized to understand its phylogenetic position together with available Potentilla (Ferrarini et al. 2013; Zhang et al. 2017; Park, Heo, Kim, and Kwon under review) and Duchesnea chloroplast genomes (Heo et al. under review; Park, Heo et al., under review). Here, we reported completed chloroplast genome of P. freyniana.
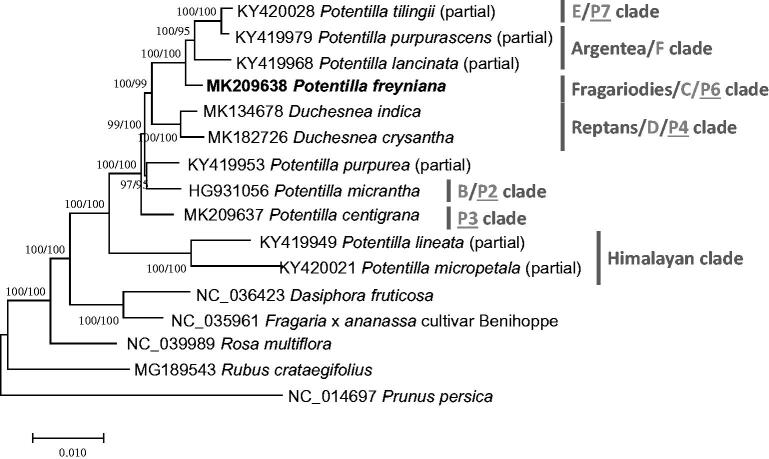
Figure 1.
Neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) phylogenetic tree of sixteen Rosaceae partial or complete chloroplast genomes: Potentilla freyniana (MK209638; in this study), Potentilla centigrana (MK209637), Duchesnea chrysantha (MK182726), Duchesnea indica (MK134678), Potentilla tilingii (KY420028; partial genome), Potentilla lancinata (KY419968; partial genome), Potentilla purpurea (KY419953; partial genome), Potentila purpurascens (KY419979; partial genome), Potentilla micropetala (KY420021; partial genome), Potentilla micrantha (HG931056), Potentilla lineata (KY419949; partial genome), Dasiphora fruticosa (NC 036423), Fragaria x ananassa cultivar Benihoppe (NC_035961), Rubus crataegifolius (MG189543), and Rosa multiflora (NC_039989), and Prunus persica (NC_014697). The numbers above branches indicate bootstrap support values of maximum likelihood and neighbor joining phylogenetic tree, respectively. Clade names were presented as dark grey, light grey, and underlined characters, defined by Töpel et al. (2011), Feng et al. (2017), Heo et al. (under review), respectively.
Total DNA of P. freyniana collected in Korea [Voucher in InfoBoss Cyber Herbarium (IN); K-I. Heo, IB-00573] was extracted from fresh leaves by using a DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Genome sequencing was performed using HiSeq2000 at Macrogen Inc., Seoul, Korea, and de novo assembly was done by Velvet 1.2.10 (Zerbino and Birney 2008), gaps were filled by SOAPGapCloser 1.12 (Zhao et al. 2011). Assembled sequences were confirmed by BWA 0.7.17 (Li 2013) and SAMtools 1.9 (Li et al. 2009). Geneious R11 11.0.5 (Biomatters Ltd., Auckland, New Zealand) was used for chloroplast genome annotation from that of Potentilla centigrana chloroplast genome (MK209637).
The chloroplast genome of P. freyniana (Genbank accession is MK209638) is 156,381 bp long and has four subregions: 85,724 bp of large single copy (LSC) and 18,617 bp of small single copy (SSC) regions are separated by 26,020 bp of inverted repeat (IR). It contained 129 genes (84 protein-coding genes, 8 rRNAs, and 37 tRNAs); 17 genes (6 protein-coding genes, 4 rRNAs, and 7 tRNAs) are duplicated in IR regions. The overall GC content of P. freyniana chloroplast genome is 36.9% and those in the LSC, SSC, and IR regions are 34.8%, 30.7%, and 42.7%, respectively.
Nine partial or complete Potentilla (Ferrarini et al. 2013; Zhang et al. 2017), two Duchesnea (Heo et al., under review; Park et al., under review), and five Rosaceae chloroplast genomes were used for constructing phylogenic trees. Whole chloroplast genome sequences were aligned by MAFFT 7.388 (Katoh and Standley 2013) for constructing neighborjoining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) trees using MEGA X (Kumar et al. 2018). Phylogenetic trees show that P. freyniana shows independent lineage (Figure 1), supported by three phylogenic studies (Töpel et al. 2011; Feng et al. 2017; Heo et al., under review). In addition, trees support that Fragaiodies/C/P6 clade is clustered with Argentea/F and E/P7 clades; while three phylogenetic studies indicated that Reptans/D/P4 clades were clustered with Argentea/F and E/P7 clades (Töpel et al. 2011; Feng et al. 2017; Heo et al. under review). It reflects that phylogenetic relationship of Potentilla species should be analyzed with more samples with complete chloroplast genome for better resolution.
Disclosure statement
The authors declare that they have no competing interests.
References
- Chen J, Craven L. 2007. Flora of China. Flora of China. 13:321–328. [Google Scholar]
- Feng T, Moore MJ, Yan MH, Sun YX, Zhang HJ, Meng AP, Li XD, Jian SG, Li JQ, Wang HC. 2017. Phylogenetic study of the tribe Potentilleae (Rosaceae), with further insight into the disintegration of Sibbaldia. J Syst Evol. 55:177–191. [Google Scholar]
- Ferrarini M, Moretto M, Ward JA, Šurbanovski N, Stevanović V, Giongo L, Viola R, Cavalieri D, Velasco R, Cestaro A, Sargent DJ. 2013. An evaluation of the PacBio RS platform for sequencing and de novo assembly of a chloroplast genome. BMC Genomics. 14:670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo K-I, Kim Y, Maki M, Park J. Under review. The Complete Chloroplast Genome of Mock Strawberry, Duchesnea indica (Andrews) Th.Wolf (Rosoideae). doi: 10.1080/23802359.2018.1553527 [DOI] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability . Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol. 35:1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:1303.3997. [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Heo K-I, Kim Y, Kwon W. Under review. The complete chloroplast genome of Potentilla centigrana Maxim. Rosaceae. doi: 10.1080/23802359.2019.1573113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Heo K-I, Kim Y, Maki M, Lee S. Under review. The Complete Chloroplast Genome, Duchesnea chrysantha (Zoll. & Moritzi) Miq. Rosoideae. doi: 10.1080/23802359.2019.1565964 [DOI] [Google Scholar]
- Rydberg PA. 1898. Monograph of the North American Potentilleae. Lancaster, USA: New Era Printing Co. [Google Scholar]
- Töpel M, Lundberg M, Eriksson T, Eriksen B. 2011. Molecular data and ploidal levels indicate several putative allopolyploidization events in the genus Potentilla (Rosaceae). PLoS Curr. 3:RRN1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SD, Jin JJ, Chen SY, Chase MW, Soltis DE, Li HT, Yang JB, Li DZ, Yi TS. 2017. Diversification of Rosaceae since the Late Cretaceous based on plastid phylogenomics. New Phytologist. 214:1355–1367. [DOI] [PubMed] [Google Scholar]
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12:S2. [DOI] [PMC free article] [PubMed] [Google Scholar]