Abstract
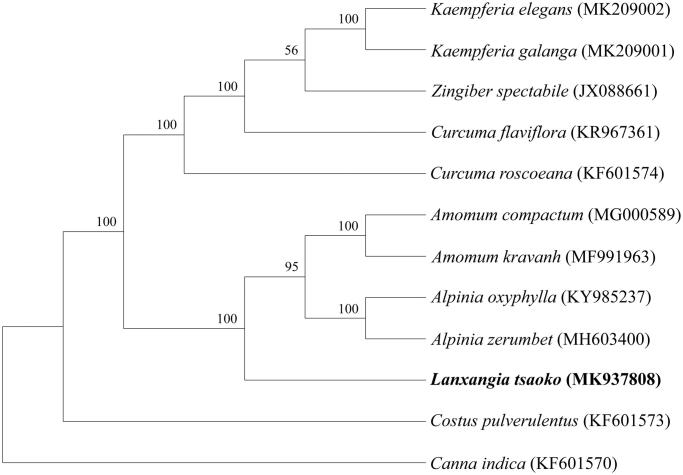
Lanxangia tsaoko is a famous Chinese traditional medicine with long history. In this study, the complete chloroplast (cp) genome of L. tsaoko was characterized and assembled using a high-throughput sequencing method. The complete cp genome is 164,008 bp in length, including a large single copy (LSC) region of 89,134 bp and a small single copy (SSC) region of 15,372 bp separated by two inverted repeat (IR) regions of 29,751 bp. A total of 137 genes were identified, including 88 protein-coding genes (PCG), 38 transfer RNA genes, 8 ribosomal RNA genes, and 3 pseudogenes. The phylogenetic analysis revealed that L. tsaoko is located in Zingiberaceace, closely related to Amomum and Alpinia with a 100% bootstrap support.
Keywords: Lanxangia tsaoko, complete chloroplast genome, phylogenetic analysis, traditional Chinese medicine
Lanxangia tsaoko (Crevost & Lemarié) M. F. Newman & Škorničk. (Zingiberaceae) is a perennial herb of Lanxangia M. F. Newman & Škorničk., which is geographically restricted to southern China (Yunnan), northern Laos, and Vietnam (Boer et al. 2018). It was once one of the members of Amomum Roxb. according to morphological characters in the past, but has long been challenged. Combined with the molecular data and morphological characters, new genera Lanxangia was described and separated from Amomum (Boer et al. 2018). Lanxangia tsaoko has been used for centuries as food, spice, and perfume in China, Japan, and Korea. Its fruit has been used in traditional Chinese medicine to treat inflammatory and infectious diseases, such as throat infections, malaria, abdominal pain, and diarrhea. Phytochemicals have been reported, such as bicyclononane, tsaokoin, isotsaokoin, diarylheptanoids, and so no (Song et al. 2001; Moon et al. 2004; Starkenmann et al. 2007; Lee et al. 2008). To better understand genetics information for further development and utilization strategy, we reported and characterized the complete cp genome of L. tsaoko (GenBank Accession Number: MK937808).
Fresh leaves of L. tsaoko were collected from Puer (Yunnan, China; 28°49′53″ N, 100°58′23″ E). Voucher specimen was deposited in the Herbarium of KUN. Total genomic DNA was extracted using modified CTAB method (Doyle and Doyle 1987). Reads of the complete cp genome were assembled using CLC Genomic Workbench v10 (CLC Bio., Aarhus, Denmark). All the contigs were checked against the reference genome of Amomum compactum (MG000589) using BLAST (https://blast.ncbi.nlm.nih.gov/) and aligned contigs were oriented according to the reference genome. The complete cp genomes were then constructed using Geneious v4.8.5 (Biomatters Ltd., Auckland, New Zealand) and was automatically annotated using DOGMA (http://dogma.ccbb.utexas.edu/). To identify the phylogenetic position of L. tsaoko, the maximum likelihood (ML) tree was conducted by MEGA v7.0 (Kumar et al. 2016) with 1000 bootstrap replicates based on the alignments created by the online program MAFFT (https://mafft.cbrc.jp/alignment/server/index/index.html) using already published complete cp genomes.
The complete cp genome of L. tsaoko is 164,008 bp in length, comprising of a large single copy (LSC) region of 89,134 bp and a small single copy (SSC) region of 15,372 bp, separated by two inverted repeat regions (IRs) of 29,751 bp. The overall GC content is 36.0%. The genome contained total of 137 genes, including 88 protein coding genes, 38 tRNA genes, 8 rRNA genes, and 3 pseudogenes.
To investigate the phylogenetic position of L. tsaoko, 9 other published complete cp genomes of Zingiberaceae were used to construct a phylogenetic tree, using Costus pulverulentus (KF601573) in Costaceae and Canna indica (KF601570) in Cannaceae as outgroups. The phylogenetic analysis revealed that L. tsaoko is located in Zingiberaceace, closely related to Amomum and Alpinia with a 100% bootstrap support, which was in accordance with the previously result (Figure 1) (Boer et al. 2018). This complete cp genome can be subsequently used for phylogenetic and genetic engineering studies of L. tsaoko, and would be fundamental to formulate potential development and management strategies for this important medicine herb.
Figure 1.
Phylogenetic analysis of 12 species based on maximum likelihood (ML) of the complete chloroplast genome sequences. Bootstrap support values (%) are indicated on each node.
Acknowledgements
We appreciate the Laboratory of Molecular Biology in the Germplasm Bank of Wild Species in Southwest China, Kunming Institute of Botany, Chinese Academy of Sciences, for providing experimental platform.
Disclosure statement
The authors are really grateful to the opened raw genome data from public GenBank. No potential conflict of interest was reported by the authors.
References
- Boer H, Newman M, Poulsen AD. 2018. Convergent morphology in Alpinieae (Zingiberaceae): recircumscribing Amomum as a monophyletic genus. Taxon. 67:6–36. [Google Scholar]
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15. [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Kim S, Sung S, Kim Y. 2008. Inhibitory constituents of lipopolysaccharide-induced nitric oxide production in BV2 microglia isolated from Amomum tsaoko. Planta Medica. 74:867–869. [DOI] [PubMed] [Google Scholar]
- Moon SS, Lee JY, Cho SC. 2004. Isotsaokoin, an antifungal agent from Amomum tsao-ko. J Nat Prod. 67:889–891. [DOI] [PubMed] [Google Scholar]
- Song QS, Teng RW, Liu XK. 2001. Tsaokoin, a new bicyclic nonane from Amomum tsaoko. Chin Chem Lett. 12:227–230. [Google Scholar]
- Starkenmann C, Mayenzet F, Brauchli R, Wunsche L, Vial C. 2007. Structure elucidation of a pungent compound in black cardamom: Amomum tsaoko crevost et lemarié (Zingiberaceae). J Agri Food Chem. 55:10902–10907. [DOI] [PubMed] [Google Scholar]