Abstract
Hylomecon japonica, a widespread species in East Asia, is a valuable horticultural and medicinal plant. Here, we obtained the first complete sequence of the H. japonica chloroplast genome. The complete cp genome was 160,011 bp long, with a large single-copy region (LSC, 88,165 bp) and a small single copy region (SSC, 18,378 bp) separated by a pair of inverted repeats (IRs, 26,734 bp). The cp genome contained 114 unique genes, including 80 protein-coding genes, 30 tRNA genes, and four rRNA genes. The phylogenetic analysis indicated that H. japonica is close related with Coreanomecon hylomeconoides.
Keywords: Coreanomecon hylomeconoides, spring ephemeral plant, plastome, phylogenomics
Hylomecon japonica (Thunb.) Prantl et Kündig, the sole species of its genus, is a perennial spring ephemeral plant distributed in China, Japan, Korea and the Russian Far East (Zhang and Christopher 2008; Xu and Wang 2017). It is characterized by bright yellow flowers and various leaf morphology, which has developed as an ornamental plant (Xiao et al. 2013). Meanwhile, as a Chinese folk medicine, it is used for the treatment of arthritis, neuralgia, and eczema (Kim et al. 2003), which contains various active compounds, such as flavonol glycosides and saponins (Lee et al. 2012; Wang 2017). Despite the importance of the species, only one nDNA (NADPH gene) and two cpDNA markers (rpoB–trnC and trnG intron regions) have been used for phylogenetic analysis at intraspecific taxonomic level (Xu and Wang 2017), less is known about the chloroplast genome in the genus. In this study, we reported and characterized the complete chloroplast genome of H. japonica based on the Illumina paired-end sequencing data. Moreover, the phylogeny of Ranunculales was reconstructed by utilizing the published related species’ chloroplast genome sequences.
Total genomic DNA was extracted from silica-dried leaves collected from Mt. Tianmushan in Zhejiang province, China using a modified CTAB method (Doyle and Doyle 1987). The voucher specimen (Pan Li, LP185533-1) was collected and deposited in the Herbarium of Zhejiang University (HZU). DNA libraries preparation and pair-end 125 bp read length sequencing were performed on the Illumina HiSeq 2500 platform. About 6.8 Gb of raw data were trimmed and assembled into contigs using CLC Genomics Workbench 8. Then, all the contigs were mapped to the reference cp genome of Coreanomecon hylomeconoides Nakai (KT274030; Kim & Kim 2016) using BLAST (NCBI BLAST v2.2.31) search and the draft cp genome of H. japonica was constructed by connecting overlapping terminal sequences in Geneious R11 software (Biomatters Ltd., Auckland, New Zealand). Gene annotation was performed via the online program Dual Organellar Genome Annotator (DOGMA; Wyman et al. 2004).
The complete cp genome of H. japonica (GenBank accession MK251463) was 160,011 bp long consisting of a pair of inverted repeat regions (IRs with 26,734 bp) divided by two single-copy regions (LSC with 88,165 bp; SSC with 18,378 bp). The overall GC content of the total length, LSC, SSC, and IR regions were 38.8%, 37.4%, 33.2%, and 43.2%, respectively. The cp genome encoded a total of 132 genes, of which 114 were unique and 18 were duplicated in the IR regions. The 114 unique genes contained 80 protein-coding genes, 30 tRNA genes, and 4 rRNA genes.
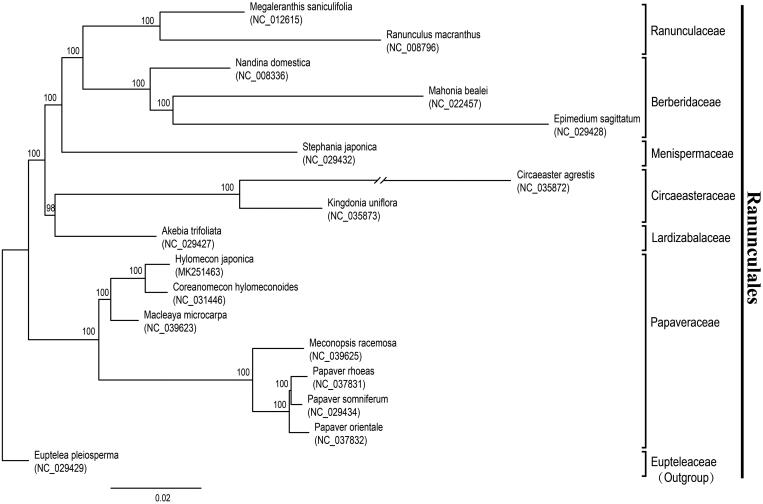
Maximum likelihood (ML) analyses were performed on a data set that included 80 protein-coding genes for 17 taxa in Ranunculales using RAxML v8.2.10 on CIPRES (http://www.phylo.org) under the GTR + G model. The phylogenetic result (Figure 1) is consistent with the prior phylogenetic study on Ranunculales (Wang et al. 2009; Sun et al. 2017). Hylomecon japonica exhibited the closest relationship with Coreanomecon hylomeconoides.
Figure 1.
Phylogenetic tree reconstruction of 17 taxa of Ranunculales using ML method. Relative branch lengths are indicated. Numbers near the nodes represent ML bootstrap value.
Disclosure statement
The authors are really grateful to the opened raw genome data from public database. The authors report no conflicts of interest and are responsible for the content and writing of the paper.
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15. [Google Scholar]
- Kim SW, In DS, Kim TJ, Liu JR. 2003. High frequency somatic embryogenesis and plant regeneration in petiole and leaf explant cultures and petiole-derived embryogenic cell suspension cultures of Hylomecon vernalis. Plant Cell Tissue Organ Cult. 74:163–167. [Google Scholar]
- Kim HW, Kim KJ. 2016. Complete plastid genome sequences of Coreanomecon hylomeconoides Nakai (Papaveraceae), a Korea endemic genus. Mitochondr DNA Part B. 1:601–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SY, Kim KH, Lee IK, Lee KH, Choi SU, Lee KR. 2012. A new flavonol glycoside from Hylomecon vernalis. Arch Pharm Res. 35:415–421. [DOI] [PubMed] [Google Scholar]
- Sun Y, Moore MJ, Lin N, Adelalu KF, Meng A, Jian S, Yang L, Li J, Wang H. 2017. Complete plastome sequencing of both living species of Circaeasteraceae (Ranunculales) reveals unusual rearrangements and the loss of the ndh gene family. BMC Genom. 18:592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Lu AM, Ren Y, Endress ME, Chen ZD. 2009. Phylogeny and classification of Ranunculales: Evidence from four molecular loci and morphological data. Perspect Plant Ecol Evol Syst. 11:81–110. [Google Scholar]
- Wang J. 2017. Studies on the saponins from Hylomecon japonica (Thunb.) Prantl et Kündig (I) (In Chinese). Jilin University. China. [Google Scholar]
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255. [DOI] [PubMed] [Google Scholar]
- Xiao Z, Zhu JY, Zhou Y, Liang Y. 2013. Application of herbaceous ornamental plant of Changbai Mountain in garden landscape (In Chinese). J Tonghua Normal Univ. 34:41–43. [Google Scholar]
- Xu XD, Wang D. 2017. Are there two varieties in Hylomecon japonica (Papaveraceae)? Morphological and molecular evidence. Annales Botanici Fennici. 54:391–399. [Google Scholar]
- Zhang ML, Christopher GW. 2008. Papaveraceae – Hylomecon. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China, Vol. 7 Beijing (China): Science Press; p. 285–286. [Google Scholar]