Abstract
The complete mitochondrial genome was sequenced in two individuals of Taranetz charr Salvelinus taranetzi. The genome sequences are 16,654 bp in size, and the gene arrangement, composition, and size are very similar to the charr genomes published previously. The difference between the two genomes studied is low, 0.05%. The present study confirms the independent taxonomic status of S. taranetzi within the genus Salvelinus. The level of sequence divergence between S. taranetzi and Salvelinus alpinus inferred from the complete mitochondrial genomes is relatively low (1.1%), indicating recent divergence of the species.
Keywords: charr genus Salvelinus, Taranetz charr Salvelinus taranetzi, Arctic charr Salvelinus alpinus, mtDNA, phylogeny
The Taranetz charr Salvelinus taranetzi was described by Kaganovsky (1955) from Lake Achchen in the Chukotka Peninsula, Russia. Originally described as a separate species, it was subsequently synonymized with Arctic charr Salvelinus alpinus taranetzi (Behnke 1984). However, Glubokovsky and Chereshnev (Glubokovsky and Chereshnev 1981; Chereshnev 1982) pointed out that S. taranetzi is different from S. alpinus and may be regarded as a separate species. However, there is still a lack of molecular data to prove this opinion (Oleinik et al. 2017). Most of the previous studies of S. taranetzi along with other charr are restricted to an analysis of only short fragments of few mitochondrial and nuclear genes (e.g. Osinov et al. 2017, and references therein).
We sequenced and described two complete mitochondrial genomes of S. taranetzi for the first time in this study, for further study and more precise phylogenetic analysis. Specimens of Taranetz charr were collected from Lake Achchen, Chukchi Peninsula (64°50′ N, 174°36′ W), and Lake Pekulineiskoe, Chukchi Peninsula (62°33′ N, 177°17′ E); one specimen was collected from the type locality. The fish specimens are stored in the collection of the Genetics Laboratory, National Scientific Center of Marine Biology FEB RAS, Vladivostok, Russia (www.imb.dvo.ru). Totally 5 pairs of primers were used (sequences are available upon request), which were designed based on public sequences available in GenBank for salmonid fishes. The sequenced fragments were de novo assembled into complete mitochondrial genome and annotated by comparing with published genome sequences of charr using Geneious R11 (http://www.geneious.com/).
The complete mitochondrial genome of S. taranetzi was 16,654 bp in length (GenBank accession numbers MK695630 and MK695631). The genome organization was identical to that of typical salmon mitochondrial genomes. Similar to charr mitochondrial genomes (Balakirev et al. 2016), the overall base composition was 28.0% of A, 26.4% of T, 28.6% of C, and 17.0% of G with a slight A + T bias (54.5%). We detected 18 single-nucleotide and no length differences between the sequences MK695630 and MK695631; two substitutions were detected in the control region and 12S rRNA, other single-nucleotide substitutions were found in overall protein-coding sequences. The proportion of variable sites was highest for the NADH dehydrogenase subunit genes (55.6%). Total sequence divergence (Dxy) was 0.0011 ± 0.0002.
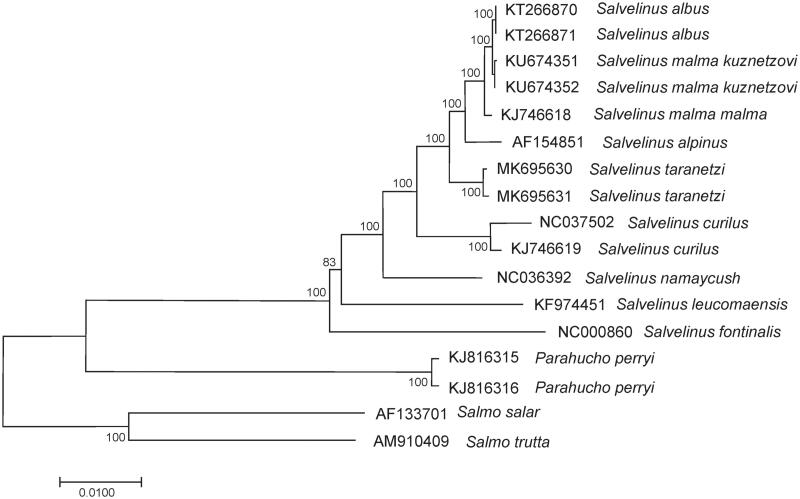
The comparison of mitochondrial genomes now obtained with mitochondrial genomes of related groups available in GenBank including genera Salvelinus (AF154851, KF974451, KJ746618, KJ746619, KT266870, KT266871, KU674351, KU674352, NC000860, NC036392, and NC037502), Salmo (AF133701, and AM910409), and Parahucho (KJ816315, and KJ816316) point to the independent taxonomic status of S. taranetzi within the genus Salvelinus (Figure 1). The level of divergence (Dxy) between S. taranetzi and taxa within the phylogenetic group was in the range from 0.0095 ± 0.0006 to 0.0442 ± 0.0019. Salvelinus taranetzi specimens showed similar sequence divergence (0.0103 ± 0.0007 on average) from S. malma malma, S. malma kuznetzovi, S. albus, and S. alpinus. These values corresponded to the level of intraspecific variability in the genus, and all charr taxa showed similar phylogenetic relationships to those found by Oleinik et al. (2015).
Figure 1.
Maximum likelihood (ML) tree constructed based on the comparison of complete mitochondrial genome sequences of Salvelinus taranetzi and other GenBank representatives of the family Salmonidae. The tree is based on the TrN93 plus gamma model of nucleotide substitution. Genbank accession numbers for all sequences are listed in the figure. Numbers at the nodes indicate bootstrap probabilities from 1000 replications (values below 80% are omitted).
Disclosure statement
No potential conflict of interest was reported by the authors. The research on mitochondrial genome sequencing was conducted at the School of Natural Sciences, Far Eastern Federal University, Vladivostok, Russia. The data analysis and manuscript preparation were conducted at the National Scientific Center of Marine Biology, Vladivostok, Russia.
References
- Balakirev ES, Parensky VA, Kovalev MYu, Ayala FJ. 2016. Complete mitochondrial genome of the stone char Salvelinus kuznetzovi (Salmoniformes, Salmonidae). Mitochondrial DNA (Part B). 1:287–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke RJ. 1984. Organizing the diversity of the Arctic charr complex In: Johnson L, Burns B, editors. Biology of the Arctic charr. Winnipeg: University of Manitoba Press; p. 3–21. [Google Scholar]
- Chereshnev IA. 1982. To the taxonomic status of sympatric anadromous chars of the genus Salvelinus (Salmonidae) from eastern Chukotka. Vopr Ikhtiol. 22:922–936. [Google Scholar]
- Glubokovsky MK, Chereshnev IA. 1981. Controversial issues of the phylogeny in charrs of the genus Salvelinus from the Holarctic. I. The data on anadromous charr from the East Siberian Sea basin. Vopr Ichthyol. 21:771–786. [Google Scholar]
- Kaganovsky AG. 1955. Charr from the Bering Sea basin. Vopr Ikhtiol. 3:54–56. [Google Scholar]
- Oleinik AG, Skurikhina LA, Brykov VA. 2015. Phylogeny of charrs of the genus Salvelinus based on mitochondrial DNA data. Russ J Genet. 51:55–68. [PubMed] [Google Scholar]
- Oleinik AG, Skurikhina LA, Kukhlevsky AD. 2017. Secondary contact between two divergent lineages of charrs of the genus Salvelinus in the Northwest Pacific. Russ J Genet. 53:1221–1233. [Google Scholar]
- Osinov AG, Volkov AA, Alekseyev SS, Sergeev AA, Oficerov MV, Kirillov AF. 2017. On the origin and phylogenetic position of Arctic charr (Salvelinus alpinus complex, Salmonidae) from Lake Cherechen’ (middle Kolyma River basin): controversial genetic data. Polar Biol. 40:777–786. [Google Scholar]