Abstract
Exorista japonica (Townsend, 1909), a dipteran tachinid fly, is an endoparasitoid of lepidopteran larvae as a potential biological agent. We have determined a 17,663 bp mitogenome of E. japonica which includes 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNAs, and a single large non-coding region of 2,773 bp. The base composition was AT-biased (81.4%). Phylogenetic trees present monophyly of Tachinidae and Exoristinae, where two Exorista species are clustered in single clade, Exoristini. The E. japonica mitogenome will be a good resource for understanding phylogenetic relationship among species of Tachinidae presenting morphological and ecological complexities.
Keywords: Exorista japonica, mitochondrial genome, Exorista, phylogenetic relationship, Tachinidae
Exorista japonica (Townsend, 1909), a dipteran endoparasitoid, belongs to Exoristini and Exoristinae in Tachinidae where there are more than 8,500 species and 1,520 genera (O’Hara 2012). Tachinidae presents diverse host ranges (Hemiptera, Lepidoptera, Coleoptera, and Hymenoptera) and ovipositional strategies (Stireman et al. 2006; O’Hara 2013). Exorista japonica is distributed in Inida, Nepal, Vietnam, China, Taiwan, Japan, and Korea (Crosskey 1976; Park et al. 2016). Its female adult directly attaches milky eggs to the larval integument of 18 lepidopteran families (mainly Lymantriidae, Lasiocampidae, Noctuidae, and Arctiidae), e.g. Mythimna separata, Spodoptera litura, Lymantria dispar, and Mamestra brassicae. Hatched E. japonica larva penetrates host body resulting in death of host finally (Nakamura 1994; Dindo and Nakamura 2018).
Recently, molecular phylogenetic analyses combining quantitative morphologies or ecological traits have been conducted in Tachinidae at tribal or subfamily level utilizing several loci (Stireman 2002; Tachi and Shima 2010; Cerretti et al. 2014; Stireman et al. 2018). Several unresolved relationships within Dexiinae, Tachininae, and Exoristinae (Stireman et al. 2018) require more molecular marker sequences; however, only six Tachinidae mitogenomes are available till now (Shao et al. 2012).
To compensate this short-coming, we completed mitogenomes of E. japonica collected in Gimje-si, South Korea in 2018 (35°46′11″N, 126°49′20″E; Voucher was deposited in InfoBoss Cyber Herbarium (IN; Seo BY, INH-00020; South Korea)). Genomic DNA was extracted using CTAB-based DNA extraction method (iNtRON biotechnology, Inc., Korea). HiSeqX was used for sequencing (Macrogen Inc., Korea). Filtering, de novo assembly, and gap filing processes were done by Velvet 1.2.10 (Zerbino and Birney 2008), Trimmomatic 0.33 (Bolger et al. 2014), SOAPGapCloser 1.12 (Zhao et al. 2011), BWA 0.7.17 (Li 2013), and SAMtools 1.9 (Li et al. 2009). Geneious R11 11.1.5 (Biomatters Ltd, Auckland, New Zealand) and ARWEN (Laslett and Canbäck 2008) were used to annotate E. japonica mitogenome based on both Nemorilla maculosa (NC_039823) and E. sorbillans mitogenomes (NC_014704; Shao et al. 2012).
Exorista japonica mitogenome (GenBank accession is MK903727) is 17,663 bp long and GC ratio is 18.6%. It contains 13 protein-coding genes, 2 rRNAs, and 22 tRNAs (63bp to 72 bp in size). Gene order is same to those of other Tachinids. Interestingly, its control region (2,773 bp), single long non-coding AT-rich region (AT ratio is 84.3%), is the longest among seven mitogenomes of Tachinidae while E. sorbillans is the shortest (105 bp). This extreme difference within same genus suggests several evolutionary events in AT-rich region inside this genus.
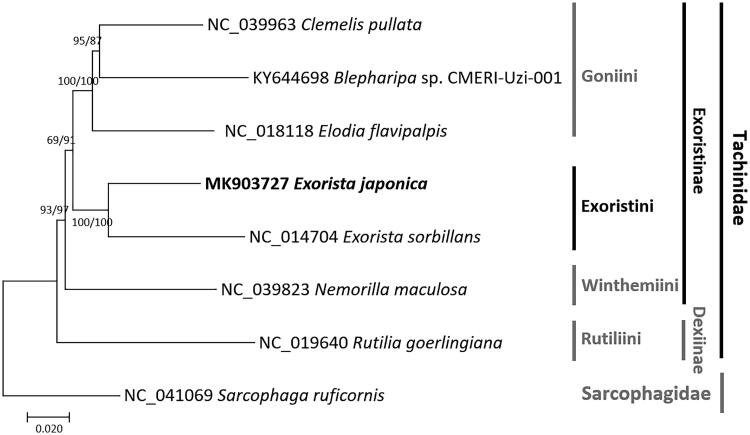
We inferred the phylogenetic relationship of seven Tachinids including E. japonica and one outgroup species, Sarcophaga ruficornis (Sarcophagidae). Multiple sequence alignment was conducted by MAFFT 7.388 (Katoh and Standley 2013) using these complete mitogenomes. Bootstrapped neighbor-joining and maximum likelihood trees were constructed using MEGA X (Kumar et al. 2018). E. japonica closely clustered with E. sorbillans with high bootstrap value (Figure 1). Additional mitochondrial genomes of Exorista are strongly required for understanding inter-species distribution of extreme difference in AT-rich region. In addition, these mitochondrial genomes are expected to give an insight into phylogenetic relationship and evolutionary history of Exorista species in morphologically and ecologically complex Tachinidae.
Figure 1.
Neighbor-joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1000) phylogenetic tree of seven Tachinidae species and one Oestroidea species: Exorista japonica (MK903727 in this study), E. sorbillans (NC_014704), Elodia flavipalpis (NC_018118), Clemelis pullata (NC_039963), Blepharipa sp. CMERI-Uzi-001 (KY644698), Nemorilla maculosa (NC_039823), Rutilia goerlingiana (NC_019640), and Sarcophaga ruficornis (NC_041069). Phylogenetic tree was drawn based on neighbor-joining phylogenetic tree. Grey bars in right side indicate tribes, subfamilies, and families, respectively. The numbers above branches indicate bootstrap support values of maximum likelihood and neighbor-joining phylogenetic trees, respectively.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerretti P, Giudice GL, O’hara JE. 2014. A new Loewia Egger (Diptera: Tachinidae) from Turkey, with taxonomic and nomenclatural remarks on congeners. Zootaxa. 3754:450–460. [DOI] [PubMed] [Google Scholar]
- Crosskey RW. 1976. A taxonomic conspectus of the Tachinidae (Diptera) of the Oriental Region.Bull Brit Meuseum. 26:3–357. [Google Scholar]
- Dindo ML, Nakamura S. 2018. Oviposition strategies of tachinid parasitoids: two Exorista species as case studies. Int J Insect Sci. 10:1179543318757491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability . Mol Biol Evol.. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175. [DOI] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 13033997. [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S. 1994. Parasitization and life history parameters of Exorista japonica (Diptera: Tachinidae) using the common armyworm, Pseudaletia separata (Lepidoptera: Noctuidae) as a host. Appl Entomol Zool. 29:133–140. [Google Scholar]
- O’Hara JE. 2012. World genera of the Tachinidae (Diptera) and their regional occurrence. http://www.nadsdiptera.org/Tach/WorldTachs/Genera/Gentach_ver8.pdf
- O’Hara JE. 2013. History of tachinid classification (Diptera, Tachinidae). ZooKeys. 316:1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C, Seo B, Choi B. 2016. The temperature-dependent development of the parasitoid fly, Exorista Japonica (Townsend)(Diptera: Tachinidae). Korean J Appl Entomol. 138:175–183. [Google Scholar]
- Shao Y-J, Hu X-Q, Peng G-D, Wang R-X, Gao R-N, Lin C, Shen W-D, Li R, Li B. 2012. Structure and evolution of the mitochondrial genome of Exorista sorbillans: the Tachinidae (Diptera: Calyptratae) perspective. Mol Biol Rep. 39:11023–11030. [DOI] [PubMed] [Google Scholar]
- Stireman JO. 2002. Learning in the generalist tachinid parasitoid Exorista mella Walker (Diptera: Tachinidae). J Insect Behav. 15:689–706. [Google Scholar]
- Stireman III JO, Cerretti P, O'Hara JE, Blaschke JD, Moulton JK. 2018. Molecular phylogeny and evolution of world Tachinidae (Diptera). Mol Phylogenet Evol. pii:S1055-7903(18)30604-3. [DOI] [PubMed] [Google Scholar]
- Stireman II J, O'Hara JE, Wood DM. 2006. Tachinidae: evolution, behavior, and ecology. Annu Rev Entomol. 51:525–555. [DOI] [PubMed] [Google Scholar]
- Tachi T, Shima H. 2010. Molecular phylogeny of the subfamily Exoristinae (Diptera, Tachinidae), with discussions on the evolutionary history of female oviposition strategy. Syst Entomol. 35:148–163. [Google Scholar]
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q-Y, Wang Y, Kong Y-M, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12:S2. [DOI] [PMC free article] [PubMed] [Google Scholar]