Abstract
Hibiscus taiwanensis S. Y. Hu is an ornamental plant of Hibiscus, native to Taiwan. Here, we reported the complete chloroplast genome of H. taiwanensis. The chloroplast genome of H. taiwanensis was 161,056 bp in length, containing a couple of inverted repeat (IR) regions of 26,300 bp, a large single-copy (LSC) region of 89,538 bp and a small single-copy (SSC) region of 18,918 bp. The complete chloroplast genome annotation revealed a total of 131 genes, including 85 protein-coding genes, 7 rRNA genes, and 37 tRNA genes. The entire GC content was 36.9%. Phylogenetic tree analyses indicated that H. taiwanensis was closely clustered with H. rosa-sinensis and H. syriacus.
Keywords: Hibiscus taiwanensis, chloroplast, genome
Hibiscus taiwanensis S. Y. Hu, a deciduous tree or shrub, which belongs to the genus Hibiscus of the family Malvaceae. This plant, 3–8 m high, is not stellate, but densely strigose and scabrous (Tang et al. 2007). The H. taiwanensis grows everywhere below the altitude of 1200 m all over Taiwan and has been distributed elsewhere in Southeast Asia (Lim 2014). It is introduced as an ornamental plant by the world's major botanical gardens. Its blossom is not only seductive but also esculent. The stem and root of H. taiwanensis have been used as antifungal, analgesic, antipyretic, anti-inflammatory, and anthelmintic agents in traditional Chinese medicine (Gan 1965). Previous studies on H. taiwanensis have focused on its chemical composition and medicinal value (Lim 2014). However, there are a few reports on the taxonomy and phylogeny of H. taiwanensis. Here, we first report the complete chloroplast genome sequence of H. taiwanensis to provide genomic sources for further researching phylogenetic relationships of Hibiscus.
The fresh and healthy leaves were collected from the Chengdu Botanical Garden (30°45′52″N, 104°8′11″E), Sichuan Province, China. Voucher specimens were deposited in the Herbarium of Sichuan University (SZ, XXR20181221). The total genomic DNA was extracted from the above leaves with the modified CTAB method (Doyle and Doyle 1987). The total genomic DNA was sequenced using the Illumina Hiseq Platform (Illumina, San Diego, CA, USA). We used the raw data to assemble the complete chloroplast genome by NOVOPlasty (Dierckxsens et al. 2017), with the complete chloroplast genome of H. rosa-sinensis as the reference (GenBank accession no. MK382984). We annotated the assembled chloroplast genome via Geneious 11.0.4 with the sequence of H. rosa-sinensis as the reference and corrected the annotation result manually (Kearse et al. 2012). Finally, the complete chloroplast sequences of 13 species which belong to Malvales were aligned by MUFFT (Katoh et al. 2002). The sequences which have been aligned were applied to build the maximum-likelihood (ML) tree with 1000 bootstrap replicates by RaxML (Stamatakis 2006).
The circular chloroplast genome of H. taiwanensis was 161,056 bp in length (GenBank accession no.MK937807), divided into four regions. A couple of inverted repeat (IR) regions of 26,300 bp separated by the large single-copy (LSC) region of 89,538 bp and small single-copy (SSC) region of 18,918 bp. The chloroplast genome detected a total of 131 genes, including 85 protein-coding genes, 7 rRNA genes, and 37 tRNA genes. The entire GC content of H. taiwanensis cp genome was 36.9% with the corresponding values of LSC, SSC, and IR regions being 34.7%, 31.5%, and 42.6%, respectively.
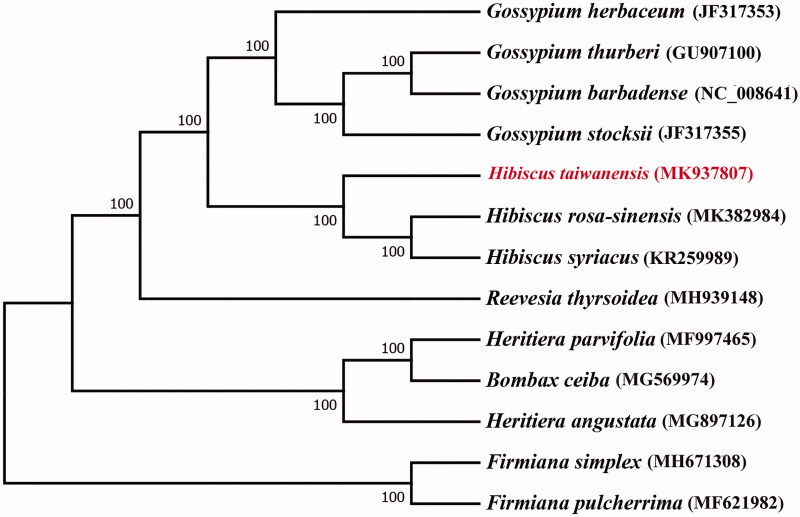
As a few complete chloroplast sequences of Malvaceae have been released, we choose the complete chloroplast sequences of 13 species which belong to Malvales to construct the phylogenetic tree. The result showed that H. taiwanensis is closely clustered with H. rosa-sinensis and H. syriacus (Figure 1), which was in accordance with early studies (Tang et al. 2014). This complete chloroplast genome can be further used for studying the value of H. taiwanensis and the phylogenetic relationships among the genus Hibiscus L.
Figure 1.
ML phylogenetic tree of H. taiwanensis with 12 species of Malvales was constructed by chloroplast genome sequences. Numbers on the nodes are bootstrap values from 1000 replicates. Two species of Firmiana were selected as outgroup.
Acknowledgements
The authors appreciate the opened raw cp genome data in NCBI. The authors thank the materials and help which were provided by Botanical staff. The authors thank Dengfeng Xie, Danmei Su, Xin Yang for their help of sequence analysis.
Disclosure statement
The authors declare no conflicts of interest and are responsible for the content.
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15. [Google Scholar]
- Gan WS. 1965. Manual of medicinal plants in Taiwan. Vol. 3 Taiwan: National Research Institute of Chinese Medicine; p. 516. [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim TK. 2014. Hibiscus taiwanensis Vol. 8 In: Edible medicinal and non-medicinal plants. Netherlands: Springer; 381–384. [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690. [DOI] [PubMed] [Google Scholar]
- Tang LD, Yuan MM, Li W, et al. 2014. Phylogenetic classification of Hibiscus based on morphological traits. J Henan Agric Sci. 43:105–111. [Google Scholar]
- Tang Y, Gilbert MG, Dorr LJ. 2007. Hibiscus linnaeus Vol.12 In: Flora of China.; 290–291. [Google Scholar]