Abstract
The complete mitochondrial genome of sand-hopper Trinorchestia longiramus was analyzed in this study, which is the first for the genus within the family Talitridae. The mitogenome sequence is 15,401 bp in length containing two ribosomal RNA genes, 22 transfer RNA genes, 13 protein-coding genes, and a control region as found in most amphipods. The gene order showed that T. longiramus has a unique control region location compared to other amphipods. Phylogenetic analysis using the maximum likelihood method positioned T. longiramus within the monophyletic clades of the family Talitridae.
Keywords: Sand-hopper, Talitridae, mitochondria, phylogeny, Trinorchestia longiramus
The sand-hopper Trinorchestia longiramus is mostly found around South Korea (Woo et al. 2016) and also found from the northern Hokkaido to southern Tohoku in Japan (Sasago 2011). This talitrid species mostly inhabits in sandy beaches and is considered as an important trophic link between primary producers and higher consumers, helps in the energy flow within these ecosystems (Jeong et al. 2009). Mitochondrial genome (mitogenome) based phylogenetic analysis would improve our understanding of the evolutionary relationship with talitrids. However, only two talitrid mitogenomes i.e. Platorchestia parapacifica and Platorchestia japonica are available. In this study, we sequenced and analyzed the complete mitochondrial DNA sequence of T. longiramus. This is the first complete mitogenome for the genus Trinorchestia within the family Talitridae.
Trinorchestia longiramus were captured by hand from sandy beach (37°41′29″N, 129°2′2.7″E) of South Korea. The specimen is available with the storage number CR00246482 at the National Marine Biodiversity Institute of Korea. DNA was isolated from the specimens by conventional phenol-chloroform method (Sambrook et al. 1989). Approximately 32 Gb of reads were generated by paired-end (PE) sequencing with 251 bp read length performed in an Illumina HiSeq 2500 sequencer. Mitogenome was assembled from the PE reads using NOVOPlasty (Dierckxsens et al. 2017), where P. parapacifica (GenBank accession MG010371) used as the seed sequence. The mitogenome was annotated using MITOS webserver (Bernt et al. 2013); some genes were annotated manually. The mitogenome was submitted to NCBI GenBank and is available with accession number MH542431. Trinorchestia longiramus mitogenome is 15,401 bp in length containing 13 protein-coding genes (PCGs), two ribosomal RNAs (rRNA), 22 transfer RNAs (tRNAs), and a control region (CR), common in all amphipods.
A unique gene arrangement pattern observed in T. longiramus as well as in the family Talitridae, in which trnL2 is found between Nad2 and Nad3, different from amphipods (Romanova et al. 2016). Generally, the CR is located between small rRNA and Nad2 in pancrustacea mt genomes (Kilpert and Podsiadlowski 2006). However, in T. longiramus, CR is found between 16S and 12S rRNA genes, which is unique among all amphipods. Among PCGs, four (CytB, Cox3, Nad3, and Nad4) started with ATG, four (Cox2, Atp6, Atp8, and Nad6) started with ATA, two (Nad2 and Nad4L) started with ATT, one (Cox1) with ATC, one (Nad5) with GTG, and one (Nad1) with TTG, which have been found as start codons in crustacean mitogenomes (Lavrov et al. 2000). Ten genes ended with TAA as stop codon, two (Nad1 and Nad5) ended with TAG, and one (Cox2) ended with T(AA). In Cox2, TAA stop codon is completed by the addition of A residues to the mRNA. The overall base composition is 37.0% for A, 34.2% for T, 10.4% for G, and 18.4% for C.
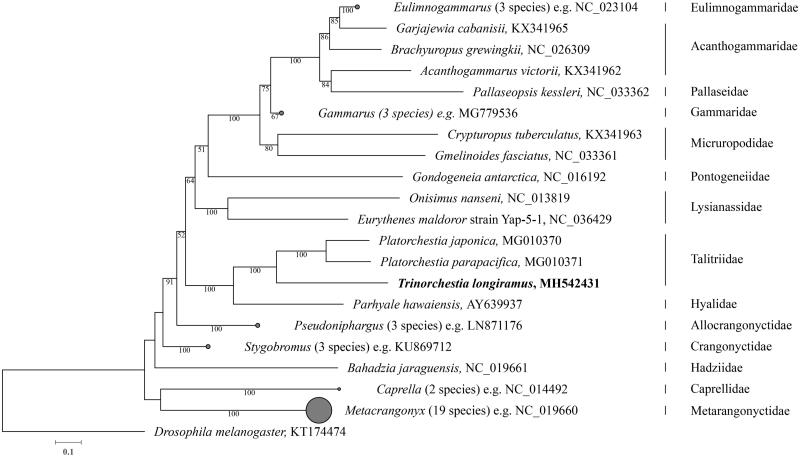
A maximum likelihood phylogenetic analysis with 45 amphipod mitogenomes placed T. longiramus among talitrids with bootstrap value = 100% (Figure 1). Trinorchestia longiramus formed monophyletic clade with other Platorchestia species. Further study with diverse taxonomic sampling from the talitrids will help in understanding phylogenetic status of the family.
Figure 1.
Maximum-likelihood (ML) phylogeny of 45 amphipods analyzed with the aligned and concatenated nucleotide sequences 13 protein-coding genes (11,984 bases). Trinorchestia longiramus is shown in bold. Sequences were aligned separately for each gene by MUSCLE v3.8.425 (Edgar 2004). ML tree was generated with RAxML v8.2.11 (Stamatakis 2014) using GTR + Γ +I substitution model supported by 1000 bootstrap replicates.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319. [DOI] [PubMed] [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong SJ, Yu OH, Suh H-L. 2009. Reproductive patterns and secondary production of Gammaropsis japonicus (Crustacea, Amphipoda) on the seagrass Zostera marina of Korea. Hydrobiologia. 623:63–76. [Google Scholar]
- Kilpert F, Podsiadlowski L. 2006. The complete mitochondrial genome of the common sea slater, Ligia oceanica (Crustacea, Isopoda) bears a novel gene order and unusual control region features. BMC Genomics. 7:241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavrov DV, Boore JL, Brown WM. 2000. The complete mitochondrial DNA sequence of the horseshoe crab Limulus polyphemus. Mol Biol Evol. 17:813–824. [DOI] [PubMed] [Google Scholar]
- Romanova EV, Aleoshin VV, Kamaltynov RM, Mikhailov KV, Logacheva MD, Sirotinina EA, Gornov AY, Anikin AS, Sherbakov DY. 2016. Evolution of mitochondrial genomes in Baikalian amphipods. BMC Genomics. 17:1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch EF., Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold spring harbor laboratory press. [Google Scholar]
- Sasago Y. 2011. Study for distribution and molecular phylogenetic analysis of the talitrid amphipods in Japan [M.Sc. thesis]. Mie University, Tsu. [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo J, An H, Lim B-J, Song HY, Kim M-S, Jung TW, Jeong S, Cho I-Y, Oh S, Han D, Yoon M. 2016. Demographic history of Trinorchestia longiramus (Amphipoda, Talitridae) in South Korea inferred from mitochondrial DNA sequence variation. Crustaceana. 89:1559–1573. [Google Scholar]