Abstract
Suaeda salsa, an annual succulent halophytic herb, is one of the major halophyte widely distributed in both saline inland and the intertidal zone. In this study, we report the complete chloroplast genome (plastome) of S. salsa. The plastome was 151,642 bp in length and comprises a large single-copy region (83,502 bp), a small single-copy region (17,780 bp), and a pair of inverted repeats (25,180 bp). It encodes 113 unique genes, including 79 protein-coding genes (PCGs), 30 tRNAs, and four rRNAs. The overall GC content of this plastome was 36.4%. Phylogenomic analysis based on 20 plastomes revealed that S. salsa was closely related to S. malacosperma.
Keywords: Suaeda salsa, plastome, phylogenomics
Suaeda salsa (Amaranthaceae/Chenopodiaceae) is an annual leaf-succulent halophytic herb with tolerance to salt (Chen et al. 2010; Song and Wang 2015). This species is distributed in Europe and Asia, and it is one of the major halophyte widely distributed in both saline inland and the intertidal zone of northern China (Sui et al. 2010; Song et al. 2016). Suaeda salsa grows better in littoral saline soils than in saline inland soils of arid zones (Li et al. 2012; Liu et al. 2018), and it has high salt tolerance during seed germination and seedling stage (Zhou et al. 2016; Song et al. 2017). With no salt glands or bladders (Yang et al. 2010), S. salsa is adapted to saline soils through its ability to hyper-accumulate Na+ and Cl− in its succulent leaves (Guo et al. 2015) and is capable of removing salts and heavy metals from saline soils (Wang et al. 2015). Furthermore, S. salsa roots of the intertidal population could accumulate more Na+ and Cl− in both the cortex and the stele than that of the inland population (Song et al. 2011). In addition, salinity can improve chilling resistance and reproductive capacity of S. salsa (Cheng et al. 2014; Guo et al. 2018). Suaeda salsa is valuable and has been applied as a model halophyte for understanding salt tolerance (Chen et al. 2016). In this study, we reported the plastome of S. salsa, which would provide fundamental genetic resource for studying this important species.
Fresh leaves of S. salsa were collected from Kenli District (Shandong, China; 37°42′N, 118°58′E). Voucher specimen (hgwz-1) was deposited at College of Life Sciences, Shandong Normal University. Total genomic DNA was extracted by the modified CTAB method described in Wang et al. (2013). Due to limited fresh sample, the plastid DNA was not directly extracted (Liu et al. 2017). The total genomic DNA was used for library preparation and paired-end (PE) sequencing by the Illumina MiSeq instrument at Novogene (Beijing, China). The plastome was assembled using Organelle Genome Assembler (OGA, https://github.com/quxiaojian/OGA) described in Qu (2019). Annotation was performed with Plastid Genome Annotator (PGA, https://github.com/quxiaojian/PGA) (Qu et al., 2019), coupled with manual correction using Geneious v9.1.4 (Kearse et al. 2012). To determine the phylogenetic placement of S. salsa, a maximum-likelihood (ML) tree was reconstructed using RAxML v8.2.10 (Stamatakis 2014), including tree robustness assessment using 1000 rapid bootstrap replicates with the GTRGAMMA substitution model, based on the alignment of 79 shared PCGs using MAFFT v7.313 (Katoh and Standley 2013).
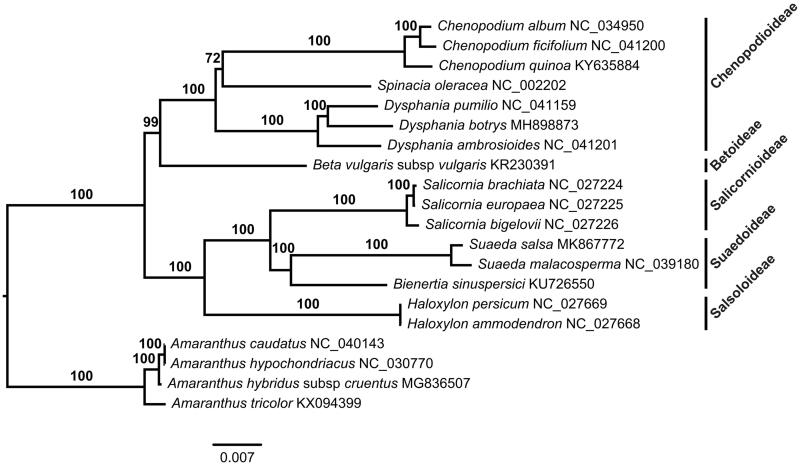
The complete plastome of S. salsa (GenBank accession number: MK867772) was 151,642 bp in length and comprises a large single-copy region (LSC: 83,502 bp), a small single-copy region (SSC: 17,780 bp), and a pair of inverted repeats (IR: 25,180 bp). This plastome encodes 113 unique genes, including 79 protein-coding genes (PCGs), 30 tRNAs, and four rRNAs. The overall GC content was 36.4%. A total of 113 unique genes were annotated in this plastome, including 79 protein-coding genes (PCGs), 30 tRNAs, and four rRNAs. Among them, eleven PCGs and six tRNAs contained introns, in which nine PCGs and six tRNAs contained one intron and two PCGs contained two introns. There were 18 duplicated genes in the IR. The ML phylogenetic tree showed that S. salsa was closely related to S. malacosperma (Figure 1).
Figure 1.
A maximum likelihood (ML) tree inferred from 79 plastome genes is shown. Four Amaranthus species from Amaranthaceae are used as outgroup. The numbers on branches are bootstrap support values.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Chen M, Song J, Wang BS. 2010. NaCl increases the activity of the plasma membrane H+-ATPase in C3 halophyte Suaeda salsa callus. Acta Physiol Plant. 32:27–36. [Google Scholar]
- Chen TS, Yuan F, Song J, Wang BS. 2016. Nitric oxide participates in waterlogging tolerance through enhanced adventitious root formation in the euhalophyte Suaeda salsa. Funct Plant Biol. 43:244–253. [DOI] [PubMed] [Google Scholar]
- Cheng S, Yang Z, Wang MJ, Song J, Sui N, Fan H. 2014. Salinity improves chilling resistance in Suaeda salsa. Acta Physiol Plant. 36:1823–1830. [Google Scholar]
- Guo JR, Li YD, Han GL, Song J, Wang BS. 2018. NaCl markedly improved the reproductive capacity of the euhalophyte Suaeda salsa. Funct Plant Biol. 45:350–361. [DOI] [PubMed] [Google Scholar]
- Guo JR, Suo SS, Wang BS. 2015. Sodium chloride improves seed vigour of the euhalophyte Suaeda salsa. Seed Sci Res. 25:335–344. [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Liu Y, Chen M, Song YP, Song J, Wang BS, Feng G. 2012. Relationships between ion and chlorophyll accumulation in seeds and adaptation to saline environments in Suaeda salsa populations. Plant Biosyst. 146:142–149. [Google Scholar]
- Liu F, Jin Z, Wang Y, Bi YP, Melton JT. 2017. Plastid genome of Dictyopteris divaricata (Dictyotales, Phaeophyceae): understanding the evolution of plastid genomes in brown algae. Mar Biotechnol. 19:627–637. [DOI] [PubMed] [Google Scholar]
- Liu QQ, Liu RR, Ma YC, Song J. 2018. Physiological and molecular evidence for Na+ and Cl− exclusion in the roots of two Suaeda salsa populations. Aquat Bot. 146:1–7. [Google Scholar]
- Qu X-J. 2019. Complete plastome sequence of an endangered species, Calocedrus rupestris (Cupressaceae). Mitochondrial DNA B. 4:762–763. [Google Scholar]
- Qu X-J, Moore MJ Li D-Z, Yi T-S. 2019. PGA: a software package for rapid, accurate, andflexible batch annotation of plastomes. Plant Methods. 15:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Shi G, Gao B, Fan H, Wang B. 2011. Waterlogging and salinity effects on two Suaeda salsa populations. Physiol Plant. 141:343–351. [DOI] [PubMed] [Google Scholar]
- Song J, Shi WW, Liu RR, Xu YG, Sui N, Zhou JC, Feng G. 2017. The role of the seed coat in adaptation of dimorphic seeds of the euhalophyte Suaeda salsa to salinity. Plant Species Biol. 32:107–114. [Google Scholar]
- Song J, Wang B. 2015. Using euhalophytes to understand salt tolerance and to develop saline agriculture: Suaeda salsa as a promising model. Ann Bot. 115:541–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Zhou JC, Zhao WW, Xu HL, Wang FX, Xu YG, Wang L, Tian CY. 2016. Effects of salinity and nitrate on production and germination of dimorphic seeds applied both through the mother plant and exogenously during germination in Suaeda salsa. Plant Species Biol. 31:19–28. [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui N, Li M, Li K, Song J, Wang BS. 2010. Increase in unsaturated fatty acids in membrane lipids of Suaeda salsa L. enhances protection of photosystem II under high salinity. Photosynthetica. 48:623–629. [Google Scholar]
- Wang HY, Jiang DF, Huang YH, Wang PM, Li T. 2013. Study on the phylogeny of Nephroma helveticum and allied species. Mycotaxon. 125:263–275. [Google Scholar]
- Wang F, Xu YG, Wang S, Shi W, Liu R, Feng G, Song J. 2015. Salinity affects production and salt tolerance of dimorphic seeds of Suaeda salsa. Plant Physiol Biochem. 95:41–48. [DOI] [PubMed] [Google Scholar]
- Yang MF, Song J, Wang BS. 2010. Organ-specific responses of vacuolar H-ATPase in the shoots and roots of C halophyte Suaeda salsa to NaCl. J Integr Plant Biol. 52:308–314. [DOI] [PubMed] [Google Scholar]
- Zhou JC, Fu TT, Sui N, Guo JR, Feng G, Fan JL, Song J. 2016. The role of salinity in seed maturation of the euhalophyte Suaeda salsa. Plant Biosystems. 150:83–90. [Google Scholar]