Abstract
Liriodendron tulifipera L. belongs to Magnoliaceae which is one of the basal angiosperm families. To understand intra-species variations on chloroplast genome in Liriodendron genus, we presented complete chloroplast genome of L. tulifipera, which is 156,387 bp long and has four subregions: 85,606 bp of large single copy (LSC) and 18,778 bp of small single copy (SSC) regions are separated by 26,002 bp of inverted repeat (IR) regions including 129 genes (84 coding genes, 8 rRNAs, and 37 tRNAs). The overall GC content of the chloroplast genome is 37.0% and those in the LSC, SSC, and IR regions are 34.9%, 30.5%, and 42.8%, respectively. Twelve single nucleotide polymorphisms (SNPs) located in one region and one insertion and deletion are found between two L. tulifipera genomes. INDEL Phylogenetic trees show that two L. tulifipera chloroplasts are clustered together and are sister to Magnolia species.
Keywords: Liriodendron tulifipera, intra-species variations, chloroplast genome, Liriodendron, Magnoliaceae
Genus Liriodendron is one of the genera belonging to Magnoliaceae, which is basal angiosperm family (Chase et al. 2016). Due to this importance, two Liriodendron chloroplast genomes were analyzed (Cai et al. 2006; Li et al. 2016). In addition, mitochondrial genomes of Liriodendron tulifipera have also been sequenced to show its phylogenetic position (Richardson et al. 2013; Park, Kim , Kwon, under review).
Intraspecies variations on chloroplast genome have been used for distinguishing its origins or cultivars (Selvaraj et al. 2008; Whittall et al. 2010; Huang et al. 2014; Ishizuka et al. 2017). Owing to the development of rapid development of sequencing technologies, organelle genomes, especially chloroplast, can be sequenced and assembled with simple processes and low costs. To understand intra-species variations on chloroplast of L. tulifipera, we completed second chloroplast genome of L. tulifipera.
Total DNA of L. tulifipera collected in the forest managed by Kookmin University in Korea was extracted from fresh leaves using DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Genome sequencing was performed using HiSeq2000 at Macrogen Inc., Korea. Chloroplast genome was assembled and confirmed by Velvet 1.2.10 (Zerbino and Birney 2008), SOAPGapCloser 1.12 (Zhao et al. 2011), BWA 0.7.17 (Li 2013) and SAMtools 1.9 (Li et al. 2009). Geneious R11 11.0.5 (Biomatters Ltd, Auckland, New Zealand) was used for chloroplast genome annotation based on L. tulifipera chloroplast genome (NC_008326).
The chloroplast genome of Korean L. tulifipera (Genbank accession is MK477550) is 159,885 bp long (GC ratio is 39.2%) and has four subregions: 85,606 bp of large single copy (LSC; 34.9%) and 18,778 bp of small single copy (SSC; 30.5%) regions are separated by 26,002 bp of inverted repeat (IR; 42.8%), which is longer than that of L. tulifipera by 1 bp (Cai et al. 2006). It contains 129 genes (84 protein-coding genes, 8 rRNAs, and 37 tRNAs); 18 genes (7 protein-coding genes, 4 rRNAs, and 7 tRNAs) are duplicated in IR regions.
Based on pair-wise alignment of both Liriodendron chloroplasts, 12 single nucleotide polymorphisms (SNPs) and one insertion and deletion (INDEL) are identified. This amount of intra-species sequence variations is larger than those of Coffea (Park, Xi et al., under review; Park, Kim, Xi, Nho et al., under review) and Marchantia (Kwon et al. under review), similar to that of Nymphaea (Park, Kim, Kwon et al. under review) and smaller than those of Duchesnea (Park, Kim, Lee, under review), Pseudostellaria (Kim et al., under review), Camellia (Park, Kim, Xi, et al. under review), Rehmannia (Jeon et al. 2019), and Illicium (Park, Kim, Xi, under review) . Interestingly, all 12 SNPs are found continuously at 67,837 bp to 67,848 bp, which is different from normal SNPs. One INDEL is located in IR-SSC boundary.
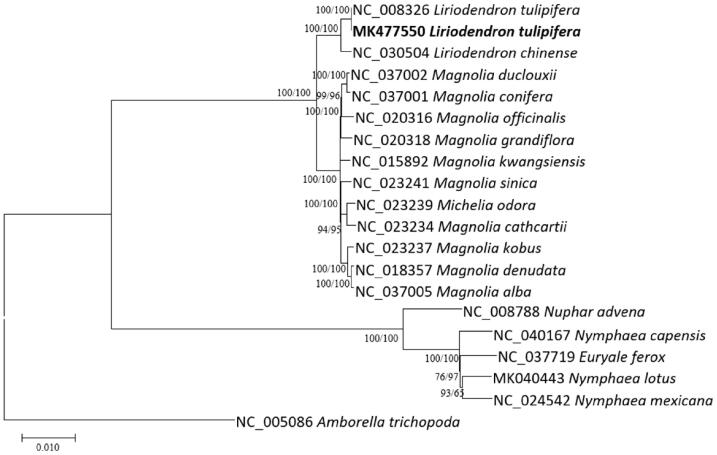
Fourteen Magnoliaceae containing two L. tulifipera chloroplasts, four Nymphaceae, and one Amborella chloroplast genomes as an outgroup were used for drawing neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1000) trees using MAFFT 7.388 (Katoh and Standley 2013) and MEGA X (Kumar et al. 2018). Phylogenetic trees show that two L. tulifipera chloroplasts are clustered together and are sister to Magnolia species (Figure 1).
Figure 1.
Neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) phylogenetic trees of 14 Magnoliaceae, 4 Nymphaceae, and 1 Amborella complete chloroplast genomes: two Liriodendron tulifipera (MK477550 in this study and NC_008326), Liriodendron chinense (NC_030504), Magnolia alba (NC_037005), Magnolia cathcartii (NC_023234), Magnolia conifera (NC_037001), Magnolia denudata (NC_018357), Magnolia duclouxii (NC_37002), Magnolia grandiflora (NC_020318), Magnolia kobus (NC_023237), Magnolia kwangsiensis (NC_015892), Magnolia officinalis (NC_020316), Magnolia sinica (NC_023241), Michelia odora (NC_023239), Euryale ferox (NC_037719), Nuphar advena (NC_008788), Nymphaea capensis (NC_040167), Nymphaea lotus (MK040443), Nymphaea mexicana (NC_024542), and Amborella trichopoda (NC_005086). The numbers above branches indicate bootstrap support values of maximum likelihood and neighbor joining phylogenetic trees, respectively.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Cai Z, Penaflor C, Kuehl JV, Leebens-Mack J, Carlson JE, dePamphilis CW, Boore JL, Jansen RK. 2006. Complete plastid genome sequences of Drimys, Liriodendron, and Piper: implications for the phylogenetic relationships of magnoliids. BMC Evol Biol. 6:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase M, Christenhusz M, Fay M, Byng J, Judd W, Soltis D, Mabberley D, Sennikov A, Soltis P, Stevens P. 2016. An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG IV. Bot J Linnean Soc. 181:1–20. [Google Scholar]
- Huang H, Shi C, Liu Y, Mao S-Y, Gao L-Z. 2014. Thirteen Camellia chloroplast genome sequences determined by high-throughput sequencing: genome structure and phylogenetic relationships. BMC Evol Biol. 14:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka W, Tabata A, Ono K, Fukuda Y, Hara T. 2017. Draft chloroplast genome of Larix gmelinii var. japonica: insight into intraspecific divergence. J Forest Res. 22:393–398. [Google Scholar]
- Jeon J-H, Park H-S, Park JY, Kang TS, Kwon K, Kim YB, Han J-W, Kim SH, Sung SH, Yang T-J. 2019. Two complete chloroplast genome sequences and intra-species diversity for Rehmannia glutinosa (Orobanchaceae). Mitochondrial DNA B. 4:176–177. [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability . Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Heo K-I, Park J. under review. The second complete chloroplast genome sequence of Pseudostellaria palibiniana (Takeda) Ohwi (Caryophyllaceae): intraspecies variations based on geographical distribution. doi: 10.1080/23802359.2019.1591179 [DOI] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon W, Kim Y, Park J. under review. The complete mitochondrial genome of Korean Marchantia polymorpha subsp. ruderalis Bischl. & Boisselier: inverted repeats on mitochondrial genome between Korean and Japanese isolates. doi: 10.1080/23802359.2019.1565975 [DOI] [Google Scholar]
- Li B, Li Y, Cai Q, Lin F, Meng Q, Zheng Y. 2016. The complete chloroplast genome of a Tertiary relict species Liriodendron chinense (Magnoliaceae). Conserv Genet Resour. 8:279–281. [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv Preprint arXiv. 13033997. [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Kim Y, Kwon M. under review. The complete mitochondrial genome of tulip tree, Liriodendron tulifipera L. (Magnoliaceae): intra-species variations on mitochondrial genome.doi: 10.1080/23802359.2019.1591242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Kim Y, Xi H, Nho M, Woo J, Seo Y. under review. The complete chloroplast genome of high production individual tree of Coffea arabica L. (Rubiaceae).doi: 10.1080/23802359.2019.1600386 [DOI] [Google Scholar]
- Park J, Xi H, Kim Y, Heo K-I, Nho M, Woo J, Seo Y, Yang JH. under review. The complete chloroplast genome of cold hardiness individual of Coffea arabica L. (Rubiaceae).doi: 10.1080/23802359.2019.1586472 [DOI] [Google Scholar]
- Park J, Kim Y, Kwon W, Nam S, Song MJ.. under review. The second complete chloroplast genome sequence of Nymphaea alba L. (Nymphaeaceae) to investigate inner-species variations.doi: 10.1080/23802359.2019.1565968 [DOI] [Google Scholar]
- Park J, Kim Y, Lee K. under review. The complete chloroplast genome of Korean mock strawberry, Duchesnea chrysantha (Zoll. & Moritzi) Miq. (Rosoideae).doi: 10.1080/23802359.2019.1573114 [DOI] [Google Scholar]
- Park J, Kim Y, Xi H, Oh YJ, Hahm KM, Ko J. under review. The complete chloroplast genome of common camellia tree, Camellia japonica L. (Theaceae), adapted to cold environment in Korea.doi: 10.1080/23802359.2019.1580164 [DOI] [Google Scholar]
- Park J, Kim Y, Xi H. under review. The complete chloroplast genome of aniseed tree, Illicium anisatum L. (Schisandraceae).doi: 10.1080/23802359.2019.1584062 [DOI] [Google Scholar]
- Richardson AO, Rice DW, Young GJ, Alverson AJ, Palmer JD. 2013. The “fossilized” mitochondrial genome of Liriodendron tulipifera: ancestral gene content and order, ancestral editing sites, and extraordinarily low mutation rate. BMC Biol. 11:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaraj D, Sarma RK, Sathishkumar R. 2008. Phylogenetic analysis of chloroplast matK gene from Zingiberaceae for plant DNA barcoding. Bioinformation. 3:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittall JB, Syring J, Parks M, Buenrostro J, Dick C, Liston A, Cronn R. 2010. Finding a (pine) needle in a haystack: chloroplast genome sequence divergence in rare and widespread pines. Mol Ecol. 19:100–114. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12:S2. [DOI] [PMC free article] [PubMed] [Google Scholar]