Abstract
Hearing loss is a prevalent disorder that affects people of all ages. On top of the existing hearing aids and cochlear implants, there is a growing effort to regenerate functional tissues and restore hearing. However, studying and evaluating these regenerative medicine approaches in a big animal model (e.g. pigs) whose anatomy, physiology, and organ size are similar to a human is challenging. In big animal models, the cochlea is bulky, intricate, and veiled in a dense and craggy otic capsule. These facts complicate 3D microscopic analysis that is vital in the cochlea, where structure-function relation is time and again manifested. To allow 3D imaging of an intact cochlea of newborn and juvenile pigs with a volume up to ∼ 250 mm3, we adapted the BoneClear tissue clearing technique, which renders the bone transparent. The transparent cochleae were then imaged with cellular resolution and in a timely fashion, which prevented bubble formation and tissue degradation, using an adaptive custom-built light-sheet fluorescence microscope. The adaptive light-sheet microscope compensated for deflections of the illumination beam by changing the angles of the beam and translating the detection objective while acquiring images. Using this combination of techniques, macroscopic and microscopic properties of the cochlea were extracted, including the density of hair cells, frequency maps, and lower frequency limits. Consequently, the proposed platform could support the growing effort to regenerate cochlear tissues and assist with basic research to advance cures for hearing impairments.
1. Introduction
Hearing loss is a silently growing health disability that affects about 6% of the population worldwide [1]. The leading causes of hearing loss include ear infections, ototoxic medicines, loud noise exposure, and genetic diseases (e.g. Usher syndrome) [2,3]. With over one billion young people at risk of hearing loss [4], there is an ongoing effort to develop new regenerative therapeutics and to evaluate their success in preclinical studies. Consequently, a method for measuring subtle changes in the intricate 3D structure of the inner ear with cellular resolution is required. The 3D information is especially important when studying the inner ear and cochlea in particular, where the structure-function relation is repeatedly manifested; the cochlear structure directly correlates to the audible frequency range. However, due to the intricate structure of the inner ear, which is composed of a series of canals and cavities that are carved inside the dense temporal bone, it is difficult to image the cochlea with cellular resolution while maintaining the intact structure. Using classical histological techniques (e.g. 2D slices) to yield 3D information regarding cellular cartography, neuronal innervations, and vasculature of the cochlea is difficult, if not impossible, and requires extensive experience and practice. Thus, a fast and detailed 3D analysis method that enables the investigation of the entire cochlea as a whole is needed.
Recently, tissue clearing methods which render entire organs transparent have emerged. These tissue clearing methods and optical imaging techniques have enabled the acquisition of 3D structural and cellular information from intact organs [5–14], and even intact bones [7,15–18]. For comparison, without tissue clearing, a state-of-the-art confocal microscope can only image ∼ 100 µm deep in the uncleared tissues [19,20]. Given that tissue clearing methods work in osseous tissue, these methods were applied to characterize the 3D cochlear structure of rodents and provided insightful information that could not have been extracted previously [9,21–25]. Although rodent models are enormously useful both in basic science and drug development, there are many instances where big animal models (e.g. pigs and non-human primates) have proven superior in predicting the true translation potential of therapeutics. For the cochlea, these big animal models are considered more suitable for preclinical applications as their anatomy, physiology, and organ size are similar to the human cochlea. However, larger organ size creates several inherent challenges for 3D labeling and imaging of the cochlea: (i) The imaging volume is at least an order of magnitude larger than for rodents, which also results in more demanding quantification, e.g. more cells to count and more variable signal-to-noise ratios across the individual tissue. (ii) Antibody staining of hair cells, stem cells, neurons, and other cell populations through thick (mm scale) layers of dense bone is challenging. (iii) The unique architecture of the cochlea, with its canals and cavities, creates deflections and obstructions in the illumination path, which create various imaging artifacts and degrade the image quality.
To address these challenges, we show the advantages of imaging cochlea samples using an adaptive light-sheet fluorescence microscope (LSFM) that can continuously change the illumination angle and refocus the detection lens to overlap with the perpendicular illumination beam. Together with the BoneClear method [7], which facilitates antibody penetration through thick bones, intact cochleae of wild type pigs (up to 10 weeks old; ∼ 29.5 Kg) are imaged and anatomically characterized at both the macroscopic and microscopic level. To the best of our knowledge, this is the first high-resolution 3D reconstruction of the intact porcine cochlea. It should be noted that light-sheet microscopy was previously and extensively used to image intact cochleae of mice, gerbil, and guinea pigs [21,25–28], and interestingly, the first modern light-sheet microscope was developed to image the cochlea [27,29]. From the reconstruction, we extracted the frequency map of the porcine cochlea, extrapolated the lower frequency limit (at 60 dB), and mapped the density and diameter of inner and outer hair cells. This technology opens new avenues for evaluating therapeutics for hearing loss prevention and cell regeneration, as well as identification of developmental milestones in the auditory system.
2. Materials and methods
2.1. Animals
In this study, cochleae were extracted from seven transgenic leucine-rich repeat-containing G-protein coupled receptor 5 (LGR5-GFP) fetuses (Day 30, 50, and 80 mixed genders), six wild-type newborn (NB, mixed gender), and four wild-type Yorkshire 8-10 weeks old pigs (females). All animal protocols were approved by the Institutional Animal Care and Use Committee (IACUC) at North Carolina State University, following the standards of the National Institute of Health and Committee on Care and Use of Laboratory Animals.
2.2. Tissue clearing
Here, the BoneClear [7] procedure was optimized for the porcine cochlea; therefore, we will only outline significant steps and differences between our implementation versus the original protocol (Fig. 1). Given the size and complexity of pigs, perfusion of PBS and PFA was not conducted, and the specimen was typically obtained shortly after euthanasia (5–30 min). For ease of processing, the first step is to separate the head from the body and to cut a rectangular window on top of the skull, as shown in Fig. S1(a). A hacksaw was used to cut through the dense bone, and razor blades were used to cut through the skin and ligaments. The brain was removed to observe the location of the inner ear (Fig. S1(b)). Then, the skull was cut in half (Fig. S1(c)), and excess bone around the cochlea was removed. Overall, a cochlea extraction can take 10-20 minutes for a juvenile pig, and 5-10 minutes to a newborn pig.
Fig. 1.
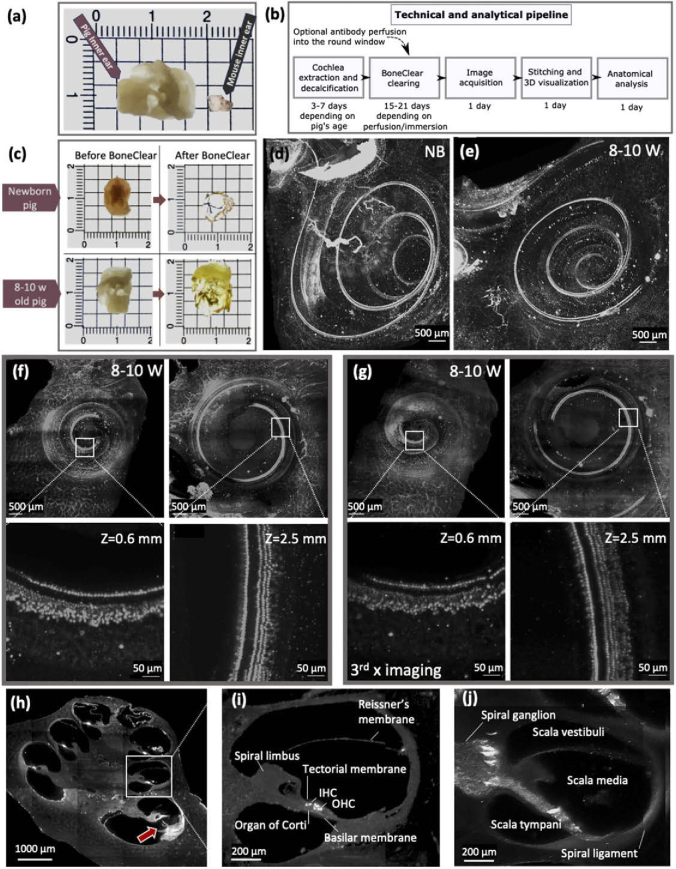
A 3D reconstruction of an intact cochlea of a pig. (a) Size comparison between pig and mouse inner ear; the mouse image was adapted from Ref. [22]. (b) A block diagram outlining the key steps of tissue preparation and imaging. (c) Micrographs of a newborn and 8-10 w old pigs’ cochleae before and after BoneClear. The samples were imaged in air. (d) Maximum intensity projection fluorescence image of a newborn pig’s cochlea and (e) an 8-10 w old pig’s cochlea stained with anti-MYOSIN VIIa for inner and outer hair cells. Using BoneClear protocol and customized light-sheet microscopy, cochleae were imaged from one end to another, which corresponds to ∼3-6.5 mm deep inside the tissue. (f) From left to right, digital slices (XY plane, 100 µm thick) that were taken at 0.6 mm and 2.5 mm deep inside the 8-10 w old pig’s cochlea, respectively. The zoomed-in regions show the inner and outer hair cells at different depths. (g) The corresponding digital slices (XY plane, 100 µm thick) that were taken at 0.6 and 2.5 mm deep inside the same 8-10 w old pig’s cochlea after imaging the sample three times. The zoomed-in regions show the inner and outer hair cells that can still be detected. Please note, the sample was remounted between the acquisitions of the overviews and the zoomed-in regions in (f) and (g). Consequently, the remounting caused a slight variation in the visible structure. (h) A transverse section (100 µm thick) at 1.6 mm deep inside the 8-10 w old pig’s cochlea. (i) A zoomed-in region from (h) that illustrates the anatomy of the cochlea duct. This sample was stained with anti-MYOSIN VIIa for inner and outer hair cells. (j) A similar region to (i) but stained with anti-PGP9.5 to label neurons. The images presented in (d-j) were captured using the custom LSFM (10× objective, 0.6 NA).
Then, the tissues were post-fixed in PBS/0.5% PFA/10% sucrose at room temperature for 2 h. The tissues were further fixed in PBS/0.5% PFA at 4 °C overnight and then washed with PBS at room temperature for 1 h three times. The tissues were decalcified in 350 mM EDTA-Na (pH 6.5) at 37 °C for 2-5 days, depending on the age of the pigs, with a fresh buffer change every 24 h. All the incubation steps were performed with gentle shaking. After decalcification, the cochlea was further separated from the bony structure, as illustrated in Visualization 1 (25.3MB, mp4) . For fetus cochlea, this step was skipped since the cochlea was extracted even before the decalcification step. The decalcified tissues were dehydrated at room temperature with the methanol gradient (diluted in ddH2O): 20% methanol for 2 h, 40% methanol for 2 h, 60% methanol for 2 h, 80% methanol for 2 h, and 100% methanol for 2 h twice. The tissues were decolorized at 4°C overnight with a mixture of 30% H2O2 and 100% methanol (v:v = 1:10). The tissues were rehydrated at room temperature with the inverse methanol gradient (diluted in ddH2O): 100% methanol for 2 h, 80% methanol for 2 h, 60% methanol for 2 h, 40% methanol for 2 h, 20% methanol for 2 h, and PBS for 2 h. The tissues were permeabilized with PBS/0.2% Triton X-100/0.1% Deoxycholate/10% DMSO/25 mM EDTA (pH 6.5) at 37 °C overnight, and then blocked with PBS/0.2% Triton X-100/10% DMSO/5% normal donkey serum/25 mM EDTA (pH 6.5) at 37 °C overnight. All the incubation steps were performed with gentle shaking. The tissues were immunolabeled with the primary antibodies diluted (1:250) in PBS/0.2% Tween-20/10 µg/mL heparin/5% normal donkey serum/25 mM EDTA (pH 6.5) at 37 °C for 5 days using immersion or 2 days using perfusion.
To accelerate the labeling, we used a perfusion pump (Instech; P720/37 K) with a highly accurate flow rate of 1.9 µL/min, when the inlet to the cochlea was the round window. The perfused antibody solution exited from the oval window to a 5 mL reservoir. The primary antibodies used in this study are listed in Table S1. The tissues were washed with PBS/0.2% Tween-20/10 µg/mL heparin/25 mM EDTA (pH 6.5) at 37 °C for 24 h, with the fresh buffer, changed every 12 h. The tissues were further immunolabeled with the secondary antibodies listed in Table S1 diluted (1:250) in PBS/0.2% Tween-20/10 µg/mL heparin/5% normal donkey serum/25 mM EDTA (pH 6.5) at 37 °C for 5 days using immersion or 2 days using perfusion. The tissues were washed with PBS/0.2% Tween-20/10 µg/mL heparin/25 mM EDTA (pH 6.5) at 37 °C for 48 h, with the fresh buffer, changed every 12 h. All the incubation steps were performed with gentle shaking. The tissues were dehydrated at room temperature with the methanol gradient (diluted in ddH2O): 20% methanol for 4 h, 40% methanol for 2 h, 60% methanol for 2 h, 80% methanol for 2 h, and 100% methanol for 2 h twice. The tissues were then incubated at room temperature with the mixture of dichloromethane and methanol (v:v = 2:1) for 2 h twice, followed by 100% dichloromethane for 30 min four times. The tissues were cleared at room temperature with 100% dibenzyl-ether (DBE) with a refractive index of 1.562 for 12 h three times. All the incubation steps were performed with gentle shaking. The imaging was performed while the samples were immersed in DBE.
2.3. Custom-build light-sheet microscopy
The adaptive LSFM was inspired by previous designs that were used for in-vivo imaging [30,31]. Figure 2 shows the custom light-sheet design, which is equipped with three continuous-wave lasers (488 nm, 561 nm, and 640 nm; Coherent OBIS series), each beam was expanded three times using an achromatic Galilean beam expander (Thorlabs; GBE03-A). The three beams were then combined using a set of dichroic mirrors (Thorlabs; DMLP505R and DMLP605R), and an iris was used to control the Gaussian beam waist on the sample (∼ 8 µm). The combined beam was then focused on a 2D pivot galvo system (Thorlabs; GVS202) using achromatic doublet (Thorlabs; AC254-125-A-ML). The role of the galvo system was to change the yaw and pitch angles of the illumination beam to compensate for beam deflections and obstructions. Then, the illumination beam was collimated (Thorlabs; AC254-125-A-ML) and aligned to the center of the scanning galvo system (Cambridge Technology; 6215H), which was controlled using a dual-channel function generator (Tektronix; AFG31022A). The scanning galvo system's primary role was to dither the Gaussian beam up and down (10k Hz) to create a light-sheet that illuminates the entire field of view. Potentially, the dual-axis galvo system could also change the roll angle, however, this degree of freedom was rarely used in our implementation. A combination of a scan lens (Thorlabs; CLS-SL) and a tube lens (Thorlabs; AC508-180-A-ML) was used to conjugate the galvo mirror with the back focal plane of the objective lens (Thorlabs; AC254-035-A-ML). The sample was mounted on a custom holder, which was connected to a precise XYZ theta stage (ASI; Stage-4D-50). The stage was used to move the sample across the light-sheet to achieve 3D reconstruction. A custom-made chamber (aluminum and glass) was used to immerse the sample and the detection objective (10×/numerical aperture (NA) 0.6, Olympus; XLPLN10XSVMP-2). The working distance of the detection objective was 8 mm, and a silicone membrane was employed to minimize potential damage to the objective lens when immersed in 100% DBE (Fig. S2). The detection objective was placed on a motorized linear translation stage (Newport; 561D-XYZ, and CONEX-TRB12CC motor) to correct for the index of refraction mismatches throughout the image acquisition. The detected light passed through a filter wheel with an appropriate emission filter (Thorlabs; FW102C, MF525-39; AVRO; FF01-593/40-25 and FF01-676/37-25) and was focused on the camera (Hamamatsu, C13440-20CU) using a tube lens (ASI; TL180-MMC). The field of view of the custom-built light-sheet microscope was 1.77 mm2. The entire control software and the graphical user interface (GUI) were written in MATLAB. A full list of light-sheet components, a description of our approach of immersing the objective lens in DBE, and a detailed diagram that contains pictures of the setup are provided in Supplementary Table S2, Fig. S2, and Fig. S3, respectively.
Fig. 2.
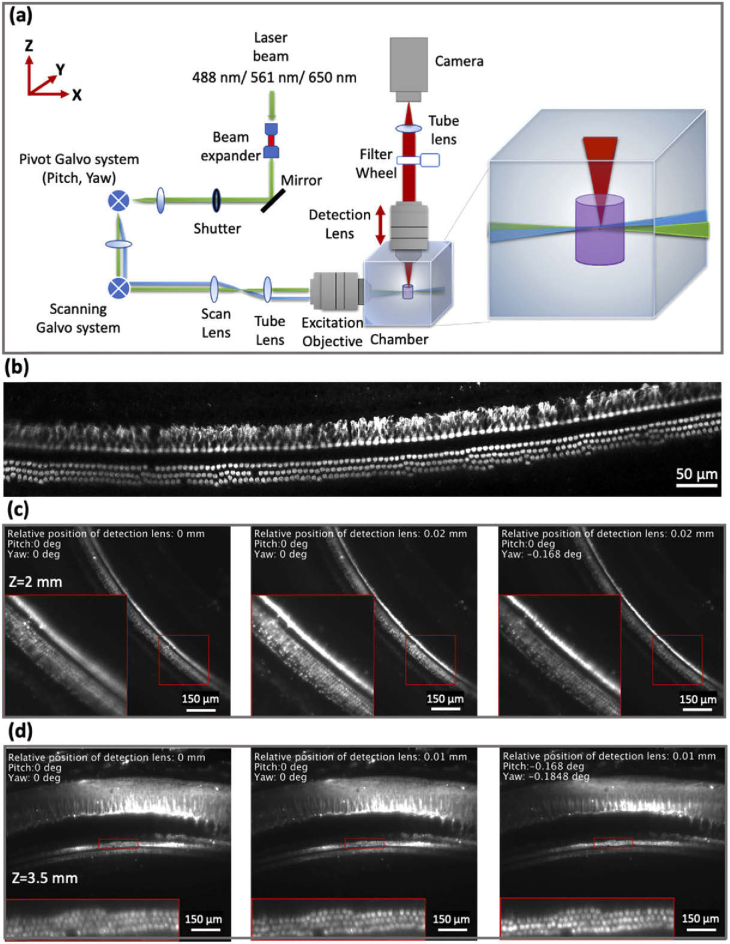
Degrees of freedom in light-sheet fluorescence microscopy. (a) A schematic diagram of the custom-built light-sheet microscope. The green beam corresponds to a normal illumination when no correction is applied to the illumination beam, while the blue beam corresponds to a change in the yaw angle of the illumination beam to compensate for light deflection by the sample. The detection objective movement is shown by a double arrow. (b) High-resolution images of IHC and OHC rows that were acquired by the custom-built LSFM next to the surface, where no corrections were used. (c and d) The images were taken at 2 and 3.5 mm respectively deep inside the tissue stained with anti-MYOSIN VIIa with no corrections, detection objective correction, and detection objective and pivot galvo system corrections, respectively. The red boxed regions in the images represent the zoomed-in area. The zoomed-in regions are shown with higher magnification, thus illustrating image quality enhancement utilizing various corrections.
Each cochlea was imaged with a voxel size of 0.65 × 0.65 ×10 µm3, and the resulting datasets were stitched using TeraStitcher [32]. For display in the figures and media, a gamma correction of 1.6 was applied to the raw data.
2.4. Confocal microscopy
For imaging small sections of tissues out of the entire intact cochlea, a laser scanning confocal microscope was used (Olympus FLUOVIEW FV3000). Two intact cochleae were imaged using the confocal microscope (NB and 8 - 10 weeks) to compare the acquisition time and image quality in terms of hair cell counting against the LSFM. The confocal microscope was equipped with the 4×/NA 0.16, 10×/NA 0.75, and 30× Si oil/NA 1.05 Plan Apo objectives. The whole tissue was placed in a holder and immersed in 100% dibenzyl-ether covered with a coverslip and imaged with either 10× or 4× magnification. Imaris (Oxford Instruments) and ImageJ were used to reconstruct the image stacks. For display in the figures and media, a gamma correction of 1.6 was applied to the raw data.
2.5. Spiral tracing
The data was imported into Imaris 9.5 software, and the associated voxel dimensions were set in the edit/image properties tab. We then used the “measurement points” tool under the “3D view” tab to assign points manually, approximately every 50 µm along the organ of Corti (where hair cells are located) from the apical to the basal turn. The intersect was set to “specific channel”, using the associated cochlea’s signal as input. To trace the spiral and to maintain consistency, points were placed at the center of the cochlear duct or on the inner portion of the outer hair cells as illustrated in Fig. S4. We then exported the assigned points, which include the 3D coordinate information, for further analysis (MATLAB).
2.6. Cell counting
A semi-automated pipeline was utilized to measure inner and outer hair cell densities from the obtained cochlea datasets. Partial maximum intensity projection (MIP) images were first generated for every 10-20 Z-planes. Because the entirety of the organ of Corti (OC) is located within a limited range of depths within a tile, all other depths that do not contain the OC were removed. This led to a substantial improvement in the signal-to-noise ratio, given the dimension reduction step ahead. This denoising procedure was performed in ImageJ using its selection brush tool [33]. Afterward, an overall MIP which contained all the hair cells in the cochlea was generated for each dataset. Given that the cell bodies of inner hair cells (IHCs) and outer hair cells (OHCs) were similar and hard to separate, IHC and OHC regions were segmented before counting. Masks for IHCs and OHCs were generated manually in ImageJ for each half-turn [33]. In the meantime, one image patch was extracted per half-turn for IHC and OHC, respectively. Such patches were used as quality controls to validate the subsequent counting process manually. Half-turn semi-automatic counting of IHCs and OHCs was performed with ilastik software [34], version 1.3.
ilastk is a software that provides an interface for interactive training of the cell counting algorithm [34]. Annotations for training are done manually by drawing dots on cell centers, while brushstrokes are painted over the background. The ilastik classification workflow utilizes the user’s annotations to assign labels to pixels based on pixel features. The training and counting can be done over the same image. Here, annotations were done over 5-10% of the sample, and for each half-turn, models were trained for IHCs and OHCs separately to minimize cell counting mistakes.
The ilastik counting model was evaluated by comparing ilastik’s results with manual counting results of quality control patches. For each counting event, a region on each half-turn was randomly selected for manual counting, and the manual counts were then used to validate the ilastik counting result. The error rate threshold was set to 5% during the training. If the ilastik counting error was more than 5% compared to the manual counting, more annotations were added for retraining and refining the ilastik results.
For small regions that could not be counted by ilastik due to noise or blur, manual counting was conducted instead. The length of OC per half-turn was measured by tracing the basilar membrane in Imaris.
A total of five datasets were subject to quantification analyses: three cochleae were taken from three NB pigs (NB-1, NB-2, and NB-3) and two cochleae were taken from two 8-10 w pigs (8-10 w-1 and 8-10 w-2). Among the images that were analyzed, NB-3 and 8-10 w-1 were captured using a confocal microscope and the remaining images were acquired using the custom-built light-sheet microscope.
2.7. Statistical analysis
The statistical analyses were performed using GraphPad Prism version 8.
3. Results and discussion
3.1. Extraction, clearing, and imaging of an intact porcine cochlea
Inner ear extraction from pigs can be challenging because the inner ear is entombed in a dense bony structure. We have demonstrated the extraction procedure on pigs ranging in age from newborn to 8-10-week-old (8-10 w) juveniles. The extraction process is explained in detail in the method section (Fig. S1 and Visualization 1 (25.3MB, mp4) ), and the extracted inner ear is at least an order of magnitude larger in volume in comparison with rodents (Fig. 1(a)). We cleared the pig cochlea to transparency and labeled hair cells and neurons through the surrounding bone. The critical steps of our technical and analytical pipeline alongside the timeline are outlined in Fig. 1(b). As an optional step, to speed up by six days the standard immunolabeling process that uses passive diffusion, antibodies were perfused through the round window. Qualitatively, the perfusion method provides similar results to passive diffusion. Figure 1(c) shows the inner ear of the newborn and juvenile pigs before and after tissue clearing, and MIPs of the cochleae of newborn and juvenile pigs are illustrated in Figs. 1(d) and 1(e), respectively. In these images, the anti-MYOSIN VIIa was used to label inner hair cells (IHCs) and outer hair cells (OHCs), the sensory receptors of the auditory system. Through mechanotransduction, IHCs sense the sound wave and send the signal to the auditory nerve, and the OHCs enhance the hearing performance in terms of sensitivity and selectivity [35].
Figure S5, Visualization 2 (27.7MB, mp4) , and Visualization 3 (40.7MB, mp4) illustrate that our light-sheet microscope can image through the entire cleared cochlea (∼ 6 mm deep), which corresponds to an imaging volume of ∼250 mm3. Figure S6 shows the image quality at the center of the volume (∼ 3 mm) and in the basal turn of the cochlea. Furthermore, by utilizing light-sheet microscopy that selectively illuminates only the plane being imaged, photobleaching was minimized [36], and the acquisition time was accelerated by a factor of ∼10 [36]. To image an 8-10 w old pig cochlea using a confocal microscope (Fig. S7), required 96 hours. This time was decreased to 10 hours using the light-sheet microscope. Furthermore, the acceleration in the acquisition time (several hours versus several days) proved paramount for cochlea imaging, as we frequently observed trapped air bubbles inside the cochlear duct that were generated during the last steps of the sample preparation. The air bubbles accumulated with time if the sample remained in the immersion media, and the bubbles became a significant obstruction. An example of a trapped air bubble is recorded in Visualization 4 (31.9MB, mp4) . Therefore, using LSFM, the air bubble issue was minimized as the acquisition time was minimized. Finally, the reduction of photobleaching was demonstrated by imaging the entire sample stained with anti-MYOSIN VIIa (the same tissue represented in Fig. 1(f)) two additional times (Fig. 1(g)). For a fair comparison, similar areas and depths are presented with identical display parameters (e.g. brightness and contrast). Quantitatively, we measured the intensity variations between the three different acquisitions of the same sample at five matching regions along the organ of Corti (Fig. S8). Please note that these corresponding regions might be imaged at different depths between the three acquisitions, as the sample was mounted with slightly different orientation each time. After the second and third imaging, the bleaching data showed 33 ± 26% and 73 ± 15% intensity reduction (mean ± standard deviation, n = 1; five regions), in comparison to the first imaging, respectively (Table S3). Please note that by using a confocal microscope, the samples are typically photobleached completely after the first imaging session (60 - 96 hr).
The 3D reconstruction reveals the cochlea’s structure from different viewpoints that highlight distinct anatomical substructures. Figure 1(h) shows a transverse cross-section of the 8-10 w cochlea, exposing the organ of Corti and spiral ganglion. The organ of Corti is the home to the hair cells that are innervated by nerve fibers and spiral ganglion cells (SGCs), which are auditory nerves that spiral around the modiolus [35]. The higher magnification of the cochlear duct cross-section at the second turn is presented in Figs. 1(i) and 1(j), which are labeled against MYOSIN VIIa and PGP9.5 (pan-neural marker), respectively. Visualization 5 (6.5MB, mp4) shows multiple cross-sections (z slices) of an 8-10 w porcine cochlea stained against MYOSIN VIIa and Fig. S9 shows a more detailed view of labeled neurons. The rupture that is observed in the basilar membrane at the beginning of a basal turn in Fig. 1(h) (marked by a red solid arrow) is generated due to the antibody perfusion. The perfusion tube is inserted through the round window of the cochlear duct, and if inserted too deep, it can create a rupture in the membrane and structure of the cochlea.
3.2. Cochlea imaging via a custom-built and adaptive light-sheet fluorescence microscopy
A custom LSFM was constructed [30,31] (Fig. 2(a) and methods section) to image the large volume of the pig cochlea (∼250 mm3). In LSFM, the illumination and detection paths are separated and perpendicular to each other, and the sample is typically illuminated by a thin sheet of light. In our implementation, the thin sheet of light is generated by fast dithering/scanning of a Gaussian beam (∼ 8 µm waist) up and down across the field of view by the scanning galvo system (Fig. 2(a)). Given that the illumination beam is selectively illuminating the part in the sample that is being imaged, optical sectioning is achieved. In various past LSFM implementations, the pivot galvo system is used to change the yaw and pitch angles of the illumination sheet of light [37], and to correct for deflections and obstructions in the passage of light through the specimen [38,39]. We followed this approach in order to address the unique labyrinthine structure of the cochlea. Figure 2(a) depicts two illumination beams; the green one is under normal illumination conditions, i.e., no tilt is added to the illumination beam by the pivot galvo, while the blue beam depicts the illumination beam in the case that a yaw angle is introduced by the pivot galvo. An additional degree of freedom (and the most important one when imaging the cochlea) that improves the image quality is the movement of the detection objective relative to the light-sheet position (red double arrow in Fig. 2(a)). This movement is used to compensate for cases where the illumination beam does not overlap with the focal plane of the detection objective; this typically happens while imaging deep into the tissue. Deep in the tissue, slight variations in the index of refraction of the tissue versus the immersion media (DBE) would minutely change the focal distance of the detection objective and would prevent it from overlapping with the illumination beam, thus blurring the acquired image. Figure 2(b) shows an LSFM image that was acquired at a relatively shallow depth, thus demonstrating the optimal image quality of the microscope when no corrections are required. The image shows that the morphology of the inner hair cells (upper row) is observed with sub-cellular resolution.
Figures 2(c) and 2(d) demonstrate two examples in which definite improvement in the image quality of hair cells was achieved by changing the detection lens position and/or illumination angles via the pivot galvo system (pitch and yaw angles). Therefore, for optimal imaging results, a calibration procedure is required before the imaging process. The whole volume is divided into tiles, and for each tile, calibration points were defined. The first calibration point was defined on the surface of the tile, and then additional calibration points were added every one mm in depth until reaching the end of the sample. At each calibration point, the optimal positions of the pivot galvo system, and the detection lens were defined manually (visual judgment of the user) and recorded. For instance, for a given tile, if the sample depth was 2.7 mm, we define 4 calibration points at 0, 1, 2, and 2.7 mm. These calibration points were defined for each sample and were not valid from one sample to another, since each sample had a unique structure that required customized calibration. The 1 mm distance between calibration points was subjectively determined to minimize the effort of the user while providing good imaging results to count all the cells in the volume. Later, in the imaging process, the LSFM adaptively changed (by linear interpolation) the pivot galvo system and objective lens positions, according to these calibration parameters. Visualization 6 (29.2MB, mp4) provides the correction values that were used to capture images at each depth throughout an 8-10 w porcine cochlea, which is labeled for PGP9.5. It is worth mentioning that the calibration settings that provided the best results, can include the zero angles for the pivot galvo mirrors as shown in Fig. 2(c).
To characterize the illumination beam waist, we removed the emission filter and imaged the light-sheet in DBE without any specimen. Then we calculated the beam waist (full width half maximum) in a few points along the illumination profile. In the center of the field of view, the beam waist is ∼8 µm and on the edge, the beam waist is ∼25 µm (Fig. S10).
The LSFM generates between 80 to 300 gigabytes (GB) of imaging data for each cochlea given a voxel size of 0.65×0.65×10 µm3. Using the adaptive LSFM, high-quality 3D reconstruction of the cochlea was obtained, and the anatomy and physical structure of the porcine cochlea were analyzed.
3.3. Physical properties of the cochlea: frequency maps
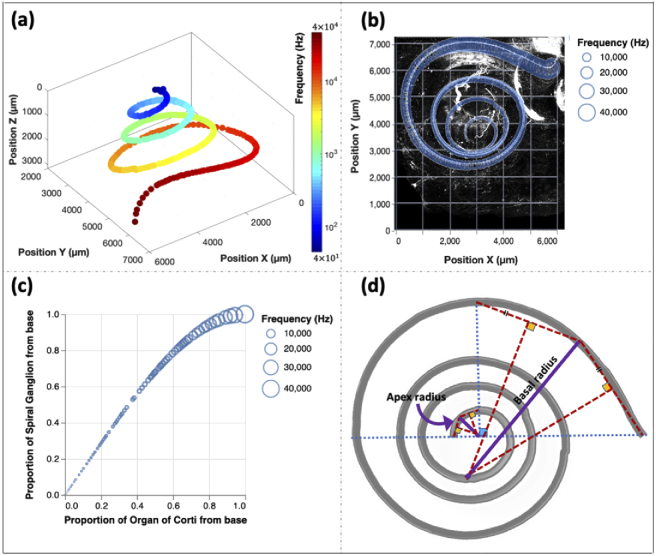
The frequency map [40] is a one-to-one mapping between a specific audible frequency and the corresponding location along the basilar membrane that strongly deforms in response to the onset of this frequency (Fig. 3(a)). Consequently, the local deformation in the membrane only activates specific hair cells in the vicinity of the deformation. To generate the frequency map, the hair cells coordinates were first extracted using Imaris by tracing hair cells from apex to base. The hair cells are residing in the organ of Corti. These coordinates were used to calculate the length of the basilar membrane and to correlate position to frequencies using the Greenwood function. The Greenwood equation is defined as , where F is the frequency (Hz), a is a constant equal to 2.1. The constant a (unitless) is an empirical constant obtained from the critical band function, and it is the slope of the straight portion of the log frequency-position function. The x (unitless) is a proportion of basilar membrane length with the apex being 0 and base being 1, k is an integration constant (unitless) which was defined originally as 1, but later it was replaced by a number ranging from 0.8 to 0.9 to set the lower frequency limit. The lower frequency limit is dictated by convention or best fit to data (requires slight adjustment for each species). Finally, constant A (Hz), which varies based on the species to best fit the data and determine the upper-frequency limit, is 165.4 Hz for humans [40]. Based on the results of this study and audiogram values reported for the hearing frequency range (40 Hz-40 kHz) of pigs [41,42], the constant A was calculated to be twice the value reported for humans [40]. The large value of A is logical as pigs have almost double the hearing range than humans [43]. Figure 3(b) illustrates the frequency map overlaid on the maximum intensity projection of an NB cochlea image.
Fig. 3.
Physical properties of the porcine cochlea were derived using a 3D reconstruction. (a) The 3D frequency map of a newborn (NB) pig. The map was derived using the Greenwood formula. (b) The frequency map overlaid on a MIP image of a NB cochlea stained with MYOSIN VIIa. (c) Frequency matched points along the relative lengths of the organ of Corti and spiral ganglion (NB pig). (d) A schematic diagram of radii that were measured to calculate the low-frequency limit of hearing for pigs. For all plots, the frequency range is between 40 Hz - 40 kHz.
Clinically, cochlear implants electrically stimulate auditory neurons located within the spiral ganglion (SG), these neurons are the first to fire action potentials from the auditory system and to supply auditory input to the brain [35]. Therefore, it is vital to correlate specific audible frequencies and the location of the SG neurons that are excited by these frequencies, i.e. the SG position-frequency function. A previously reported cubic function was used to derive the SG position-frequency function based on the Greenwood function values [40]. This approach requires tracing either the OC length or the SG length (NB pig cochlea stained with PGP9.5; Visualization 7 (68.5MB, mp4) ). Figure 3(c) shows the frequency-matched points on the OC and its corresponding position on the SG. The cubic function is presented as , where y is the percentage distance from the base of the SG, and x is the percentage distance from the base of the OC [44]. The length of the SG for a NB pig is calculated to be 27.5 mm using the tracing method, while for OC in pigs with similar age, it is calculated to be 33.5 mm. The length difference between OC and SG, which was previously reported for humans [44], seems to be smaller. This discrepancy could be related to the species or sample preparation techniques i.e. 2D versus 3D.
To validate the accuracy of the 3D reconstruction, we calculated the low-frequency limit of hearing, based on the ratio of the radii of curvature from the basal and apical turns [45,46], and compared it to pig audiogram results [41,42]. To calculate the radii of basal and apical turns, we considered the outermost and innermost quarter of the cochlea, respectively, as shown in Fig. 3(d). We calculated the lower frequency limit at 60 dB, using the following equation; where Hz, (unitless), and is the ratio of basal to apical turn radii [46]. Using our data, the lower frequency limit for newborn pigs (n = 3) is calculated to be 40.81 ± 0.39 Hz and for 8-10 weeks old pigs (n = 2), this value is calculated to be 38.75 ± 0.00 Hz (see Table S4; the values are reported as mean ± standard deviation). The reported values are in good agreement with the value derived from porcine audiogram results (See Table S5) [41,42]. This calculation further supports the hypothesis that the ratio of basal-to-apical turn radii determines the lower frequency range and not the size of the cochlea [45].
3.4. Physical properties of the cochlea: quantification of hair cells located in the organ of Corti
The high-resolution 3D reconstruction of the cochlea enables the characterization of both macroscopic (e.g. frequency maps) and microscopic properties (e.g. number of cells) of the intact cochlea. Here, we reconstructed the entire cochlea to count the number of IHCs and OHCs in the organ of Corti. The structure and number of these cell populations are indicative of the developmental stage of the hearing system, and a reduction in their number corresponds to hearing impairment due to aging, ear infections, ototoxic medicines, loud noise exposure, genetic diseases, and more [47].
A semi-automated cell detection pipeline simplified the analysis of the large volume of data acquired by the LSFM, and its key steps are outlined in Fig. 4(a). To count hair cells, we used images of the whole cochlea tissue labeled against MYOSIN VIIa. The Z-slices that did not contain hair cells were removed to reduce the background noise, and the dimensionality of the dataset was reduced to a single MIP, which contained the entire population of hair cells. IHC and OHC rows were segmented manually using ImageJ for each half-turn of the cochlea spiral. The masked area was then submitted to ilastik, open-source software that uses machine learning for image classification, and segmentation [34]. ilastik was trained to count hair cells in each half-turn independently. For quality control, we compared the ilastik counting result with manual counting at a randomly selected tile for each half-turn. If the error was higher than 5%, the algorithm was trained again, and the process repeated. At the end of the quality control stage, the cell counts were nearly equivalent to cell counts performed manually. An example of automated counting versus manual counting is illustrated in Fig. 4(b). The intensity values represent the cell density, hence, the integral of which over an imaged region provides an estimate to the number of cells in that region. The intensity values are lower at the edges since the density learned by the algorithm was approximated by a normalized Gaussian function placed on the center of each cell. To qualitatively compare and validate the performance of the pipeline on LSFM data, which has a variable signal-to-noise ratio, we also imaged one NB and one 8-10 w cochlea samples using confocal microscopy. With both imaging modalities, the number of counted hair cells using the pipeline was comparable. However, using the confocal microscope, it was challenging to count the disorganized hair cells captured on the apical turn (turn 3.5). Table S6 provides detailed information on the hair cell counts from samples that were captured either by the confocal microscope or by the light-sheet microscope. For hair cell counting purposes, the data from both imaging techniques are utilized to increase the validity of the results.
Fig. 4.
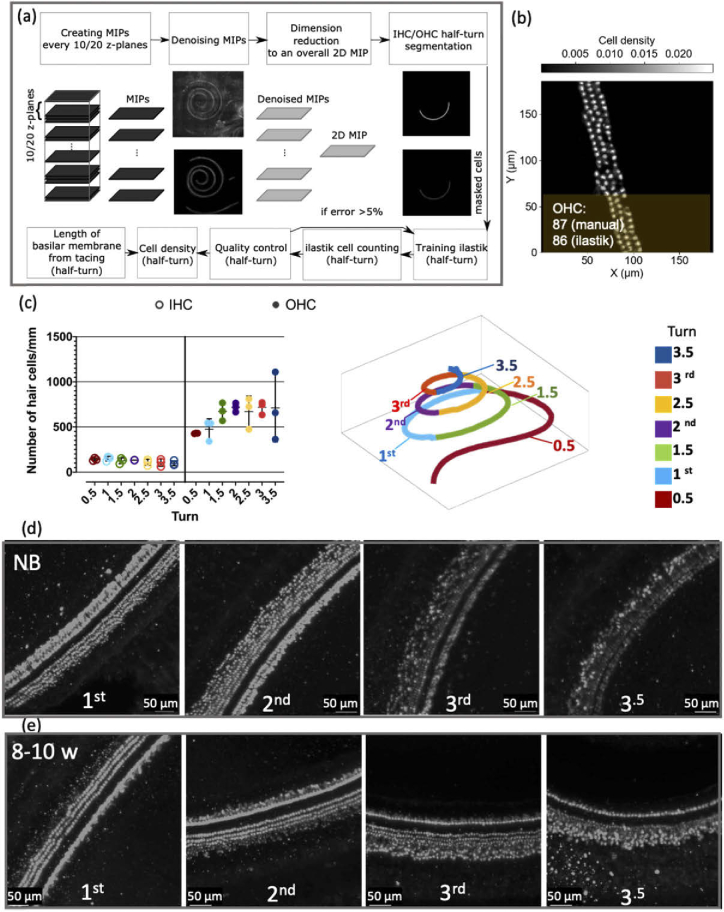
Characterization of hair cell numbers and organization in the developing porcine cochlea. (a) A block diagram of the semi-automated hair cell counting pipeline. (b) A heatmap, which is generated by ilastik, is used to count hair cells. The manual quality control for each half-turn is done and compared with ilastik counting. (c) The IHC and OHC density (number of hair cells/mm) of NB pigs is presented for each half-turn and color-coded based on their spatial location (see the schematic diagram of the cochlea on the right side). The IHC data points are illustrated by empty circles, and the OHC data points are depicted with full circles (n = 3, all values are mean ± SD). (d and e) The organization of IHCs and OHCs at each turn for newborn and 8-10 w old pigs, respectively. The basal turn (1st) has the most, and the apical turn (3.5) has the least organization.
The density of hair cells for each half-turn in NB pigs (n = 3) is reported in Fig. 4(c) and the densities are color-coded based on spatial position along the cochlea (see right side of Fig. 4(c)). The turn densities from basal to apical are decreasing for IHCs (significant linear trend, n = 3, NB pigs, F(1, 14) = 7.29, P-value < 0.05, Slope = -8.1, one-way ANOVA) and increasing for OHCs (significant linear trend, n = 3, NB pigs, F(1, 14) = 6.51, P-value < 0.05, Slope = 47.41, one-way ANOVA). The total number of IHCs for a NB pig is counted to be 4068 ± 901 and for OHCs is 18220 ± 2214, (the values are reported as mean ± standard deviation; n = 3, NB pigs). Both NB (n = 3) and 8-10 w old pigs’ (n = 2) cochleae hair cells density data are provided in Fig. S11, and in Table S6.
Typically, the ratio between IHCs and OHCs rows is reported to be 1:3; simply put, for each well-organized IHC row, there should be three organized rows of OHCs. However, in the developing cochlea, some reports have shown variability in the classical 1:3 IHC/OHC row ratio, with up to five rows of OHCs [35,47,48]. In both NB and 8-10 w pigs’ cochleae, the number of OHC rows in some regions is higher than three. We also observed that OHC rows are spatially disorganized especially toward the apex, and previous reports claim that only organized rows of OHC take part in the auditory process [47,48]. Therefore, the disorganization and increase in OHC row numbers are considered a transient event that ends with the maturation of the cochlea [47], and this can explain why our IHC/OHC row ratio deviates from 1:3.
Our observations also suggest an interesting developmental trend in the porcine cochlea: with the maturation of the pig, the OHC start to organize in a spatial order, starting from the basal turn (0.5 turns) toward the apex (3.5 turns), as illustrated in Figs. 4(d) and 4(e) respectively. The comparison between NB and 8-10 w old pigs in Figs. 4(e) and 4(f) suggests that OHCs organization in 8-10 w old pigs starts half a turn earlier than in the NB pig (see 2nd turn in both cases).
We further characterized the hair cells’ diameter along the cochlea turns and reported the values in Table S7, the comparison between IHC (Fig. S12) and OHC (Fig. S13) diameters in NB pigs in all the half-turns showed a significant difference with OHCs diameter being smaller (P-value < 0.05, two-tailed unpaired t-test, n = 3, newborn pigs).
Our imaging technique, combined with a semi-automated counting method, supplies a toolbox for future quantitative inner ear phenotyping in different developmental stages and a method for correlating structural changes with functional hearing impairment. This includes counting the missing hair cells in response to a stimulus (e.g. loud noise) which is a common technique to detect hearing impairment [49].
4. Summary and conclusions
Hearing loss is a substantial disorder that affects hundreds of millions of people of all ages. Consequently, there is a growing effort to develop new regenerative therapeutics for addressing hearing loss. Ideally, studying and evaluating new therapies should be done in animal models that mimic the physiology and scale of humans. However, given the big animal’s cochlea size, many challenges arise to study the cochlea in 2D, let alone in 3D, including large volumes to image and labeling through the osseous tissue. Here we have demonstrated that using a tissue clearing technique that was designed to allow antibody penetration through bones (BoneClear), an intact cochlea of NB and juvenile pigs can be imaged using a custom LSFM. The LSFM allows modifying the pitch and yaw angles of the illumination beam, as well as the position of the detection objective, to enhance the image quality through the complex structure of the cochlea. Based on a direct comparison with confocal microscopy, the LSFM can image the samples at least ten times faster while obtaining sufficient image quality to count the entire hair cell population inside the cochlea using a semi-automated pipeline. This technology can benefit hearing research by providing insights into the cochlear cartography, hearing loss pathology, physiological effects of drugs, and the design of hearing prosthetics.
The proposed technique has a few limitations: first, the cochlea samples need to be imaged relatively fast after the clearing process, as trapped air bubbles tend to accumulate with time and obstruct the illumination. We have tried multiple techniques to eliminate the bubbles including placing the sample in a vacuum chamber and perfusing clearing media directly to the cochlea, but we achieved limited success. Therefore, for optimal results, the samples should be imaged soon after they are placed in the clearing media. Second, not all antibodies are compatible with the methanol dehydration step, thus limiting the ability to study specific cellular populations. However, we have tried this technique with several mainstream antibodies that are used to study the cochlea, including, TH, VGLUT, SOX2, and PECAM, and they have shown compatibility with the clearing technique (Fig. S14). Third, before imaging, the LSFM user needs to set calibration points manually per tile, and the LSFM uses these points to interpolate the detection objective position and the illumination angles throughout the entire sample. Although the use of the multiple calibration points dramatically improves the imaging results, this process can be tedious. Therefore, the automation of this process will improve the user experience and the throughput of the system. In the past, the AutoPilot platform addressed this issue with living samples [30,31], but its direct integration with tissue clearing might be challenging, given the big differences in the optical properties of the imaged specimens. Therefore, the automation of the microscope control system will be the subject of future work. Lastly, in our implementation, the diameter of the Gaussian illumination beam is increasing towards the edges of the field of view (Fig. S10), thus resulting in increased background noise in comparison with the center of the field of view, where the illumination beam has a minimum diameter. This fact did not obstruct the counting of hair cells; however, in very demanding biological applications such as neuronal tracing, this issue will have to be addressed. To cope with this problem, complementary methods, which were devised to reject background noise (e.g.; HiLo [50], structured illumination [51], and focal point scanning [21]), could be implemented in parallel with our microscope’s corrections, thus providing a further enhancement to the imaging quality.
Acknowledgments
The authors would like to thank Dr. Liara Gonzalez and the NCSU Central Procedure Lab for their help with tissue collection.
Disclosures
The authors declare no conflicts of interest.
See Supplement 1 (11.7MB, pdf) for supporting content.
References
- 1.WHO, “WHO | Estimates,” https://www.who.int/pbd/deafness/estimates/en/.
- 2.Wilson B. S., Tucci D. L., Merson M. H., O’Donoghue G. M., “Global hearing health care: new findings and perspectives,” Lancet 390(10111), 2503–2515 (2017). 10.1016/S0140-6736(17)31073-5 [DOI] [PubMed] [Google Scholar]
- 3.Brigande J. V., “Hearing in the mouse of Usher,” Nat. Biotechnol. 35(3), 216–218 (2017). 10.1038/nbt.3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO , Hearing Loss Due to Recreational Exposure to Loud Sounds A Review World Health Organization (2015).
- 5.Richardson D. S., Lichtman J. W., “Clarifying Tissue Clearing,” Cell 162(2), 246–257 (2015). 10.1016/j.cell.2015.06.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ueda H. R., Ertürk A., Chung K., Gradinaru V., Chédotal A., Tomancak P., Keller P. J., “Tissue clearing and its applications in neuroscience,” Nat. Rev. Neurosci. 21(2), 61–79 (2020). 10.1038/s41583-019-0250-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Q., Liu K., Yang L., Wang H., Yang J., “BoneClear: whole-tissue immunolabeling of the intact mouse bones for 3D imaging of neural anatomy and pathology,” Cell Res. 29(10), 870–872 (2019). 10.1038/s41422-019-0217-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhau S., Todorov M. I., Cai R., Steinke H., Kemter E., Wolf E., Lipfert J., Bechmann I., Erturk A., “Cellular and Molecular Probing of Intact Transparent Human Organs,” bioRxiv 643908 (2019).
- 9.Urata S., Iida T., Yamamoto M., Mizushima Y., Fujimoto C., Matsumoto Y., Yamasoba T., Okabe S., “Cellular cartography of the organ of corti based on optical tissue clearing and machine learning,” eLife 8, e40946 (2019). 10.7554/eLife.40946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan C., Cai R., Quacquarelli F. P., Ghasemigharagoz A., Lourbopoulos A., Matryba P., Plesnila N., Dichgans M., Hellal F., Ertürk A., “Shrinkage-mediated imaging of entire organs and organisms using uDISCO,” Nat. Methods 13(10), 859–867 (2016). 10.1038/nmeth.3964 [DOI] [PubMed] [Google Scholar]
- 11.Chung K., Wallace J., Kim S. Y., Kalyanasundaram S., Andalman A. S., Davidson T. J., Mirzabekov J. J., Zalocusky K. A., Mattis J., Denisin A. K., Pak S., Bernstein H., Ramakrishnan C., Grosenick L., Gradinaru V., Deisseroth K., “Structural and molecular interrogation of intact biological systems,” Nature 497(7449), 332–337 (2013). 10.1038/nature12107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang B., Treweek J. B., Kulkarni R. P., Deverman B. E., Chen C. K., Lubeck E., Shah S., Cai L., Gradinaru V., “Single-cell phenotyping within transparent intact tissue through whole-body clearing,” Cell 158(4), 945–958 (2014). 10.1016/j.cell.2014.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Treweek J. B., Chan K. Y., Flytzanis N. C., Yang B., Deverman B. E., Greenbaum A., Lignell A., Xiao C., Cai L., Ladinsky M. S., Bjorkman P. J., Fowlkes C. C., Gradinaru V., “Whole-body tissue stabilization and selective extractions via tissue-hydrogel hybrids for high-resolution intact circuit mapping and phenotyping,” Nat. Protoc. 10(11), 1860–1896 (2015). 10.1038/nprot.2015.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Renier N., Wu Z., Simon D. J., Yang J., Ariel P., Tessier-Lavigne M., “IDISCO: A simple, rapid method to immunolabel large tissue samples for volume imaging,” Cell 159(4), 896–910 (2014). 10.1016/j.cell.2014.10.010 [DOI] [PubMed] [Google Scholar]
- 15.Jing D., Zhang S., Luo W., Gao X., Men Y., Ma C., Liu X., Yi Y., Bugde A., Zhou B. O., Zhao Z., Yuan Q., Feng J. Q., Gao L., Ge W. P., Zhao H., “Tissue clearing of both hard and soft tissue organs with the pegasos method,” Cell Res. 28(8), 803–818 (2018). 10.1038/s41422-018-0049-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenbaum A., Chan K. Y., Dobreva T., Brown D., Balani D. H., Boyce R., Kronenberg H. M., Mcbride H. J., Gradinaru V., “Bone CLARITY: Clearing, imaging, and computational analysis of osteoprogenitors within intact bone marrow,” Sci. Transl. Med. 9(387), eaah6518 (2017). 10.1126/scitranslmed.aah6518 [DOI] [PubMed] [Google Scholar]
- 17.Jing D., Yi Y., Luo W., Zhang S., Yuan Q., Wang J., Lachika E., Zhao Z., Zhao H., “Tissue Clearing and Its Application to Bone and Dental Tissues,” J. Dent. Res. 98(6), 621–631 (2019). 10.1177/0022034519844510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grüneboom A., Hawwari I., Weidner D., Culemann S., Müller S., Henneberg S., Brenzel A., Merz S., Bornemann L., Zec K., Wuelling M., Kling L., Hasenberg M., Voortmann S., Lang S., Baum W., Ohs A., Kraff O., Quick H. H., Jäger M., Landgraeber S., Dudda M., Danuser R., Stein J. V., Rohde M., Gelse K., Garbe A. I., Adamczyk A., Westendorf A. M., Hoffmann D., Christiansen S., Engel D. R., Vortkamp A., Krönke G., Herrmann M., Kamradt T., Schett G., Hasenberg A., Gunzer M., “A network of trans-cortical capillaries as mainstay for blood circulation in long bones,” Nat. Metab. 1(2), 236–250 (2019). 10.1038/s42255-018-0016-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graf B. W., Boppart S. A., “Imaging and analysis of three-dimensional cell culture models,” Methods Mol. Biol. 591, 211–227 (2010). 10.1007/978-1-60761-404-3_13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pawley J. B., Handbook of Biological Confocal Microscopy: Third Edition (Springer US, 2006). [Google Scholar]
- 21.Santi P. A., Johnson S. B., Hillenbrand M., GrandPre P. Z., Glass T. J., Leger J. R., “Thin-sheet laser imaging microscopy for optical sectioning of thick tissues,” BioTechniques 46(4), 287–294 (2009). 10.2144/000113087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.MacDonald G. H., Rubel E. W., “Three-dimensional imaging of the intact mouse cochlea by fluorescent laser scanning confocal microscopy,” Hear. Res. 243(1-2), 1–10 (2008). 10.1016/j.heares.2008.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tinne N., Antonopoulos G. C., Mohebbi S., Andrade J., Nolte L., Meyer H., Heisterkamp A., Majdani O., Ripken T., “Three-dimensional hard and soft tissue imaging of the human cochlea by scanning laser optical tomography (SLOT),” PLoS One 12(9), e0184069 (2017). 10.1371/journal.pone.0184069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nolte L., Tinne N., Schulze J., Heinemann D., Antonopoulos G. C., Meyer H., Nothwang H. G., Lenarz T., Heisterkamp A., Warnecke A., Ripken T., “Scanning laser optical tomography for in toto imaging of the murine cochlea,” PLoS One 12(4), e0175431 (2017). 10.1371/journal.pone.0175431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hutson K. A., Pulver S. H., Ariel P., Naso C., Fitzpatrick D. C., “Light sheet microscopy of the gerbil cochlea,” J. Comp. Neurol. 2020, cne.24977 (2020). 10.1002/cne.24977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kopecky B., Johnson S., Schmitz H., Santi P., Fritzsch B., “Scanning thin-sheet laser imaging microscopy elucidates details on mouse ear development,” Dev. Dyn. 241(3), 465–480 (2012). 10.1002/dvdy.23736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Voie A. H., Burns D. H., Spelman F. A., “Orthogonal-plane fluorescence optical sectioning: Three-dimensional imaging of macroscopic biological specimens,” J. Microsc. 170(3), 229–236 (1993). 10.1111/j.1365-2818.1993.tb03346.x [DOI] [PubMed] [Google Scholar]
- 28.Voie A. H., “Imaging the intact guinea pig tympanic bulla by orthogonal-plane fluorescence optical sectioning microscopy,” Hear. Res. 171(1-2), 119–128 (2002). 10.1016/S0378-5955(02)00493-8 [DOI] [PubMed] [Google Scholar]
- 29.Santi P. A., “Light sheet fluorescence microscopy: A review,” J. Histochem. Cytochem. 59(2), 129–138 (2011). 10.1369/0022155410394857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Royer L. A., Lemon W. C., Chhetri R. K., Keller P. J., “A practical guide to adaptive light-sheet microscopy,” Nat. Protoc. 13(11), 2462–2500 (2018). 10.1038/s41596-018-0043-4 [DOI] [PubMed] [Google Scholar]
- 31.Royer L. A., Lemon W. C., Chhetri R. K., Wan Y., Coleman M., Myers E. W., Keller P. J., “Adaptive light-sheet microscopy for long-term, high-resolution imaging in living organisms,” Nat. Biotechnol. 34(12), 1267–1278 (2016). 10.1038/nbt.3708 [DOI] [PubMed] [Google Scholar]
- 32.Bria A., Iannello G., “TeraStitcher - A tool for fast automatic 3D-stitching of teravoxel-sized microscopy images,” BMC Bioinformatics 13(1), 316 (2012). 10.1186/1471-2105-13-316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A., “Fiji: An open-source platform for biological-image analysis,” Nat. Methods 9(7), 676–682 (2012). 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berg S., Kutra D., Kroeger T., Straehle C. N., Kausler B. X., Haubold C., Schiegg M., Ales J., Beier T., Rudy M., Eren K., Cervantes J. I., Xu B., Beuttenmueller F., Wolny A., Zhang C., Koethe U., Hamprecht F. A., Kreshuk A., “ilastik: interactive machine learning for (bio)image analysis,” Nat. Methods 16(12), 1226–1232 (2019). 10.1038/s41592-019-0582-9 [DOI] [PubMed] [Google Scholar]
- 35.Raphael Y., Altschuler R. A., “Structure and innervation of the cochlea,” Brain Res. Bull. 60(5-6), 397–422 (2003). 10.1016/S0361-9230(03)00047-9 [DOI] [PubMed] [Google Scholar]
- 36.Tomer R., Ye L., Hsueh B., Deisseroth K., “Advanced CLARITY for rapid and high-resolution imaging of intact tissues,” Nat. Protoc. 9(7), 1682–1697 (2014). 10.1038/nprot.2014.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huisken J., Stainier D. Y. R., “Even fluorescence excitation by multidirectional selective plane illumination microscopy (mSPIM),” Opt. Lett. 32(17), 2608 (2007). 10.1364/OL.32.002608 [DOI] [PubMed] [Google Scholar]
- 38.Ji N., Freeman J., Smith S. L., “Technologies for imaging neural activity in large volumes,” Nat. Neurosci. 19(9), 1154–1164 (2016). 10.1038/nn.4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gualda E. J., Pereira H., Martins G. G., Gardner R., Moreno N., “Three-dimensional imaging flow cytometry through light-sheet fluorescence microscopy,” Cytometry, Part A 91(2), 144–151 (2017). 10.1002/cyto.a.23046 [DOI] [PubMed] [Google Scholar]
- 40.Greenwood D. D., “A cochlear frequency-position function for several species—29 years later,” J. Acoust. Soc. Am. 87(6), 2592–2605 (1990). 10.1121/1.399052 [DOI] [PubMed] [Google Scholar]
- 41.Heffner R. S., Heffner H. E., “Hearing in domestic pigs (Sus scrofa) and goats (Capra hircus),” Hear. Res. 48(3), 231–240 (1990). 10.1016/0378-5955(90)90063-U [DOI] [PubMed] [Google Scholar]
- 42.Lovell J. M., Harper G. M., “The morphology of the inner ear from the domestic pig (Sus scrofa),” J. Microsc. 228(3), 345–357 (2007). 10.1111/j.1365-2818.2007.01852.x [DOI] [PubMed] [Google Scholar]
- 43.Heffner H. E., Heffner R. S., “Auditory perception,” in Farm Animals and the Environment, Phillips C., Piggins D., eds. (CAB International, 1992), pp. 159–184. [Google Scholar]
- 44.Sridhar D., Stakhovskaya O., Leake P. A., “A frequency-position function for the human cochlear spiral ganglion,” Audiol. Neuro-otol. 11(1), 16–20 (2006). 10.1159/000095609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manoussaki D., Dimitriadis E. K., Chadwick R. S., “Cochlea’s graded curvature effect on low frequency waves,” Phys. Rev. Lett. 96(8), 088701 (2006). 10.1103/PhysRevLett.96.088701 [DOI] [PubMed] [Google Scholar]
- 46.Manoussaki D., Chadwick R. S., Ketten D. R., Arruda J., Dimitriadis E. K., O’Malley J. T., “The influence of cochlear shape on low-frequency hearing,” Proc. Natl. Acad. Sci. U. S. A. 105(16), 6162–6166 (2008). 10.1073/pnas.0710037105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iii J. W. H., “Development of the ear and hearing background science,” J. Perinatol. 20, 11–19 (2000). [DOI] [PubMed] [Google Scholar]
- 48.Raphael Y., Lenoir M., Wroblewski R., Pujol R., “The sensory epithelium and its innervation in the mole rat cochlea,” J. Comp. Neurol. 314(2), 367–382 (1991). 10.1002/cne.903140211 [DOI] [PubMed] [Google Scholar]
- 49.Schmitz H. M., Johnson S. B., Santi P. A., “Kanamycin-furosemide ototoxicity in the mouse cochlea: A 3-dimensional analysis,” Otolaryngol.--Head Neck Surg. 150(4), 666–672 (2014). 10.1177/0194599813519071 [DOI] [PubMed] [Google Scholar]
- 50.Schröter T. J., Johnson S. B., John K., Santi P. A., “Scanning thin-sheet laser imaging microscopy (sTSLIM) with structured illumination and HiLo background rejection,” Biomed. Opt. Express 3(1), 170 (2012). 10.1364/BOE.3.000170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schacht P., Johnson S. B., Santi P. A., “Implementation of a continuous scanning procedure and a line scan camera for thin-sheet laser imaging microscopy,” Biomed. Opt. Express 1(2), 598 (2010). 10.1364/BOE.1.000598 [DOI] [PMC free article] [PubMed] [Google Scholar]