Abstract
The COVID-19 pandemic, caused by SARS-CoV-2, is a pressing public health emergency garnering a rapid response from scientists across the globe. Host cell invasion is initiated through direct binding of the viral spike protein to the host receptor angiotensin-converting enzyme 2 (ACE2). Disrupting the spike protein–ACE2 interaction is a potential therapeutic target for treating COVID-19. We have developed a proximity-based AlphaLISA assay to measure the binding of SARS-CoV-2 spike protein receptor binding domain (RBD) to ACE2. Utilizing this assay platform, a drug-repurposing screen against 3384 small-molecule drugs and preclinical compounds was carried out, yielding 25 high-quality primary hits, of which only corilagin was validated in cherry-picking. This established AlphaLISA RBD–ACE2 platform can facilitate evaluation of biologics or small molecules that can perturb this essential viral–host interaction to further the development of interventions to address the global health pandemic.
Keywords: COVID19, ACE2, drug repurposing, AlphaLISA, high-throughput screening, assay development
The SARS-CoV-2 pandemic has driven an urgent need to understand the molecular basis of infection and the consequent COVID-19 disease as well as a need to rapidly identify pharmacologic interventions for both preventing infection and treating infected patients. Given that no approved therapeutics for treating any coronaviruses existed at the time SARS-CoV-2 emerged (late 2019), early attention has focused on drug repurposing opportunities.1,2 Drug repurposing is an attractive approach to treating SARS-CoV-2, as active FDA-approved drugs or unapproved drug candidates previously shown to be safe in human clinical trials can be fast-tracked to the clinic. One such example is remdesivir for treatment of COVID-19 (GS-5734, Gilead Sciences, Inc.), a virus-RNA-dependent RNA polymerase inhibitor that was in Phase II clinical trials for treating Ebola virus at the time SARS-CoV-2 emerged. Remdesivir was rapidly shown to be active against SARS-CoV-2 in vitro and progressed to clinical trials, leading to the FDA granting emergency use authorization.3 The understanding of each target and process critical for SARS-CoV-2 infection and replication will allow assays to be developed for both high-through drug-repurposing screening and to support new therapeutic development.4
One therapeutic target receiving significant attention is the interaction between the SARS-CoV-2 spike protein (Uniprot: P0DTC2) and the host protein angiotensin-converting enzyme 2 (ACE2, Uniprot: Q9BYF1).5 ACE2 is anchored to the extracellular surface of the cell and is responsible for catalyzing the conversion of angiotensin II into angiotensin 1–7 part of the renin–angiotensin system. The spike protein is exposed on the outer surface of the coronavirus particle and has evolved to bind to ACE2 with high affinity.6 This binding between SARS-CoV-2 spike protein and ACE2 is the first step in viral infection.7,8 The spike protein comprises two functional regions: The S1 region contains the ACE2 receptor binding domain (RBD), and the S2 region is responsible for membrane fusion. Because ACE2 binding by RBD is the first step in virus infection, preventing the interaction between the spike RBD and ACE2 is considered a viable therapeutic strategy, and work with prior zoonotic coronaviruses SARS and MERS has demonstrated proof-of-concept for this approach.9−11 The primary therapeutic strategy being pursued for inhibiting the spike–ACE2 interaction is the development of selective antibodies against the spike RBD. Other approaches include antibodies that bind to ACE2 (though these may produce side effects),12 development of soluble recombinant ACE2 to compete for spike binding and limit cell entry,13 or small molecules that directly bind to ACE2 and biophysically reduce its affinity for binding to spike protein.
At National Center for Advancing Translational Sciences (NCATS), we are developing both protein/biochemical and cell-based assays to interrogate a number of biological targets to enable identification of potential therapeutic leads. Our initial focus is on performing drug repurposing screening for each assay and rapidly sharing the data through the NCATS OpenData portal for COVID-19 (https://opendata.ncats.nih.gov/covid19).14 As part of this effort, we sought to develop a proximity assay for measuring the interaction between ACE2 and SARS-CoV-2 spike RBD to facilitate the identification of small molecules and biologics that disrupt this essential process.
AlphaLISA is a proximity-based assay that uses a pair of donor and acceptor beads to measure interaction (or disruption) of two tagged proteins/targets of interest.15 When the donor bead is excited by light at 680 nm, it converts ambient oxygen to singlet oxygen. If an acceptor bead is in close proximity to the singlet oxygen, then it emits a luminescent signal at 615 nm (and when this proximity is perturbed, there is a loss of signal). As AlphaLISA is amenable to quantitative high-throughput sceening (qHTS) and can leverage multiple tagged ACE2 and spike RBD constructs that are commercially available, this methodology could prove useful as a discovery approach for small or large molecules that inhibit infection.15,16
Herein we describe the development of an AlphaLISA assay using a recombinant SARS-CoV-2 spike protein RBD fused to an Fc tag and recombinant soluble human ACE2 fused to a biotinylated AviTag to model a simplified spike RBD–ACE2 interaction system. The ratio of proteins in the assay was optimized, and proof-of-concept protein–protein disruption was demonstrated using untagged ACE2 or RBD protein as competitive inhibitors. The assay was used for a drug-repurposing screen of >3000 compounds, using a library of approved small-molecule drugs (the NCATS Pharmaceutical Collection, NPC) and a library of compounds with demonstrated anti-infective activity.17,18 We also screened all compounds in parallel against the TruHits counterassay to identify and rule out false-positive hits.
Methods
Reagents
ACE2-His-Avi (human ACE2 residues 18–740, C-terminal His-Avi tags; catalog no. AC2H82E6) was acquired from ACROBiosystems (Newark, DE). RBD-Fc (SARS-CoV-2 spike protein residues 319–541, C-terminal Fc tag; catalog no.40592-V02H), ACE2-His (ACE2 residues 1–740, C-terminal His tag; catalog no. 10108-H08H), and S1-His (spike protein residues 1–667, C-terminal His tag; catalog no. 40150-V08B1) were acquired from Sino Biological (Wayne, PA). All proteins were reconstituted in 1× PBS at pH 7.4 supplemented with 0.05 mg/mL bovine serum albumin (BSA). Streptavidin (strep) donor beads (catalog no. 6760002) and protein A acceptor beads (catalog no. AL101) were acquired from PerkinElmer (Waltham, MA).
Cross-Titration Experiment
Concentrations of ACE2-His-Avi and RBD-Fc in the AlphaLISA assay were selected by cross-titrating ACE2-His-Avi (300–0.1 nM) against RBD-Fc (300–0.1 nM) in 1× PBS (pH 7.4) supplemented with 0.05 mg/mL BSA. Each concentration combination of ACE2-His-Avi and RBD-Fc were mixed in a 384-well plate (square-well, high-base, white, medium-binding; catalog no. EWB010000A, Aurora Microplates, Whitefish, MT) and incubated at 25 °C for 30 min. Streptavidin donor beads and protein A acceptor beads were then added to the wells using a multichannel pipet to a final concentration of 10 μg/mL each. AlphaLISA signal was read using a PheraSTAR (BMG Labtech, Cary, NC) plate reader with a 384-well format focal lens equipped with an AlphaLISA optical module (BMG Labtech, Cary, NC). The optimal concentration combination of ACE2-His-Avi and RBD-Fc was identified by the combination that returned the highest signal in the presence of 10 μg/mL streptavidin donor beads and 10 μg/mL protein A acceptor beads.
Competition ACE2- and S1-Binding Assay
A competition assay was performed to confirm that the AlphaLISA assay was sensitive the ACE2-His-Avi interactions with RBD-Fc. ACE2-His (Sino Biological, Wayne, PA) and S1-His (Sino Biological, Wayne, PA) are not recognized by either the streptavidin donor bead nor the protein A acceptor bead. A gradient of ACE2-His concentrations (1 μM to 0.01 nM) were mixed with RBD-Fc (4 nM). After 30 min of preincubation at 25 °C ACE2-His-Avi (4 nM) was added to the mixture and allowed to incubate at 25 °C for another 30 min. A mixture of streptavidin donor beads and protein A acceptor beads were then added to a final concentration of 10 μg/mL each. The resulting mixture was incubated at 25 °C for 40 min before reading the AlphaLISA luminescent signal with a PheraSTAR plate reader. S1-His competition assay was performed in the same manner with some changes S1-His (300 nM to 0.1 nM) was mixed with ACE2-His-Avi (4 nM) and preincubated for 30 min at 25 °C, and RBD-Fc was then introduced to the mixture before adding beads.
AlphaLISA Screening Protocol
The AlphaLISA screening assay was performed according to the protocol in Table 1.
Table 1. Detailed SARS-CoV-2 Spike–ACE2 AlphaLISA qHTS Protocol.
| step no. | process | notes |
|---|---|---|
| 1 | Predispense 20 nL of compounds and controls into 1536-well plates. | Compound transfer was performed using an ECHO 650 acoustic dispenser (LabCyte). Compounds (20 nL) in dose response were transferred to columns 5–48. Positive control and DMSO (neutral control) were dispensed into columns 1–4 using 1536-well plates (square-well, high-base white plate, solid bottom, nonsterile, nontreated; catalog no. EWB010000A, Aurora Microplates). |
| 2 | Dispense 2 μL of ACE2-His-Avi to wells except column 2. | 4× ACE2-His-Avi (16 nM, catalog no. AC2-H82E6, Acro Biosystems) in PBS (catalog no. 10010–031, ThermoFisher) + 0.05 mg/mL BSA (catalog no. BP1600-1, Fisher Scientific) |
| 3 | Incubate at RT for 30 min and 200 rpm shaking. | preincubation of ACE2 with compound step |
| 4 | Dispense 2 μL of RBD-Fc to wells, except column 3. | 4× RBD-Fc (16 nM, catalog no. 40592-V02H, Sino Biological) in PBS + 0.05 mg/mL BSA |
| 5 | Incubate at RT for 30 min and 200 rpm shaking. | equilibration of RBD-Fc and ACE2-Avi complexes |
| 6 | Dispense 4 μL of AlphaLISA donor/acceptor beads, except for column 1. | 2× protein A acceptor beads (20 μg/mL, catalog no. AL101M, PerkinElmer), 2× streptavidin donor beads (20 μg/mL, catalog no. 6760002, PerkinElmer) in PBS + 0.05 mg/mL BSA |
| 7 | Incubate at RT for 30 min and 200 rpm of shaking. | Final assay conditions: 4 nM ACE2-His-Avi, 4 nM S1-RBD-Fc, 1× PBS, 0.05 mg/mL BSA, 10 μg/mL protein A acceptor beads, and 10 μg/mL streptavidin donor beads |
| 8 | Read plates on PHERAstar FSX (BMG Labtech). | Fastest read settings, AlphaLISA Module (680 nm excitation, 615 nm emission; catalog no. 1802H1, BMG Labtech) |
TruHits Counter-Assay
The TruHits counterassay was performed according to the protocol Table 2.
Table 2. Detailed TruHits AlphaLISA Counterassay Protocol.
| step no. | process | notes |
|---|---|---|
| 1 | Compounds and controls (20 nL) were predispensed into 1536-well plates. | Compound transfer was performed using an ECHO 650 acoustic dispenser (LabCyte). Compounds in doses were dispensed to columns 5–48, and controls were dispensedto columns 1 using 1536-well plates (catalog no. EWB010000A, Aurora Microplates). |
| 2 | Dispensed 3 μL of biotin-BSA acceptor beads to all wells, except column 2. | 2× biotin-BSA acceptor beads (0.02 mg/mL, TruHits kit catalog no. 6760627, PerkinElmer) in PBS (catalog no. 10010–031, ThermoFisher) + 0.05 mg/mL BSA (catalog no. BP1600-1, Fisher Scientific) |
| 3 | Incubated at RT for 30 min and 200 rpm shaking. | incubation of Biotin-BSA acceptor beads with test compounds |
| 4 | Dispensed 3 μL of streptavidin donor beads to wells, except column 3. | 2× streptavidin donor beads (0.02 mg/mL, TruHits kit catalog no. 6760627, PerkinElmer) in PBS (catalog no. 10010–031, ThermoFisher) in PBS + 0.05 mg/mL BSA |
| 5 | Incubated at RT for 30 min and 200 rpm shaking. | Streptavidin–biotin complex formation. Final assay conditions: 10 μg/mL streptavidin donor beads, 10 μg/mL Biotin-BSA acceptor beads in PBS + 0.05 mg/mL BSA |
| 6 | Read on PHERAstar FSX (BMG Labtech). | Fastest read settings, AlphaLISA Module (680 nm excitation, 615 nm emission) (catalog no. 1802H1, BMG Labtech) |
Hit Confirmation Assays
Cherry-picked compounds were tested in triplicate dose–response assay against the AlphaLISA assay (see above) and a biotin-IgG counterassay to confirm their activity and specificity. Compounds were prepared as a triplicate 11-point, 1:3 dilution series using an ECHO 550 (LabCyte) acoustic dispenser. The ACE2 Alphalisa was performed on these compounds as described above. We also designed a biotin-IgG counterassay to test compounds for their ability to interfere with the Fc-protein A and streptavidin–biotin interactions, resulting in a false positive. For the biotin-IgG counter assay, biotinylated rabbit IgG was dispensed to each well to a final concentration of 1.6 nM in PBS + 0.05 mg/mL BSA. This mixture was incubated at room temperature for 30 min before adding the streptavidin donor beads and protein A acceptor beads to a final concentration of 10 μg/mL in PBS + 0.05 mg/mL. The resulting mixture was incubated at room temperature for another 40 min before reading AlphaLISA signal with a PHERAstar FSX plate reader (BMG Labtech).
Data Processing and Analysis
To determine compound activity in the qHTS assay, the concentration–response data for each sample was plotted and modeled by a four-parameter logistic fit yielding IC50 and efficacy (maximal response) values. Raw plate reads for each titration point were first normalized relative to positive control (−100% activity, full inhibition) and DMSO-only wells (basal, 0% activity). Data normalization and curve fitting were performed using in-house informatics tools. Compounds were designated as classes 1–4 according to the type of concentration–response curve (CRC) observed.19 In brief, classes 1.1 and 1.2 were the highest-confidence complete CRCs containing upper and lower asymptotes with efficacies ≥80% and <80%, respectively. Classes 2.1 and 2.2 were incomplete CRCs having only one asymptote with efficacy ≥80% and <80%, respectively. Class 3 CRCs showed activity at only the highest concentration or were poorly fit. Class 4 CRCs were inactive having a curve-fit of insufficient efficacy or lacking a fit altogether. Data visualization was performed using Prism (GraphPad, San Diego, CA) and Spotfire (PerkinElmer) software programs. Scheme figures were all prepared using Illustrator (Adobe, San Jose, CA).
Results
Assay Design
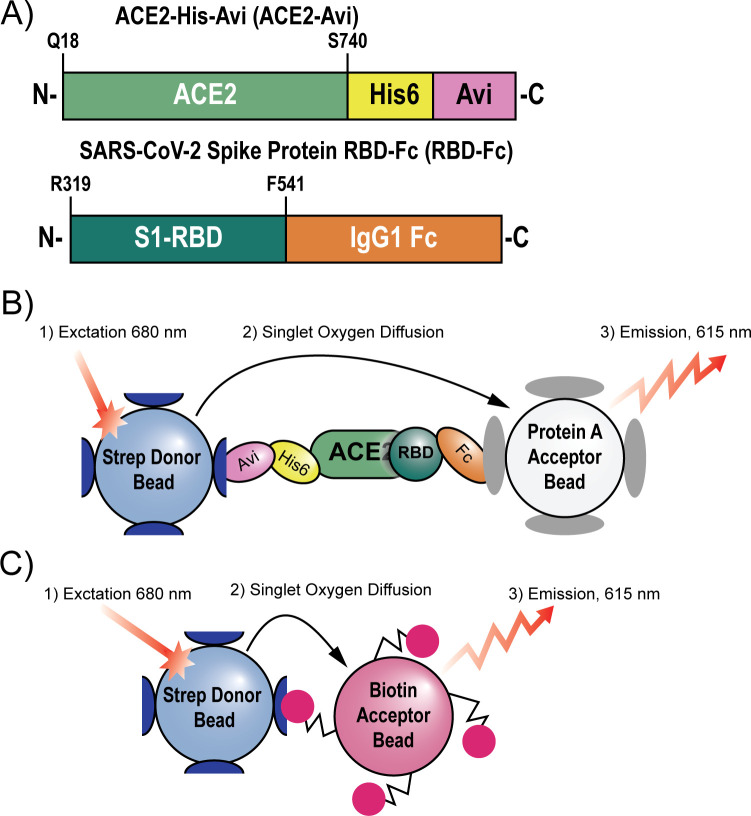
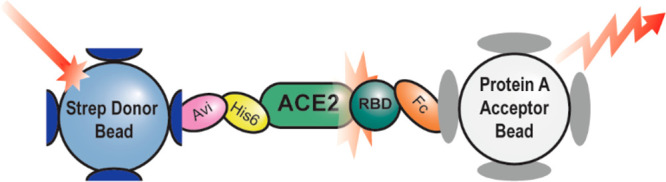
We set out to develop a 1536-well plate assay capable of measuring SARS-CoV-2 spike RBD binding to the ACE2 receptor. Two recombinant protein constructs were selected to model the RBD–ACE2 interaction in vitro: A truncated spike protein RBD (residues 319–541) was fused to a C-terminal Fc tag, and a truncated soluble ACE2 (residues 18–740) was fused to a C-terminal poly-His tag followed by an AviTag (Figure 1A). These constructs were selected because the Fc tag and AviTag form tight and specific interactions with protein A and streptavidin, respectively, making them ideal handles for AlphaLISA particles. When the RBD binds to ACE2, the Fc tag and AviTag provide platforms for the protein A acceptor beads and streptavidin donor beads, respectively, and bring them into close proximity (Figure 1B). Once this bead–protein supercomplex forms, the donor bead can be excited with light at 680 nm, causing it to generate a superoxide which diffuses to the acceptor bead, generating a luminescent signal at 615 nm.
Figure 1.
Scheme describing the assays employed in this paper. (A) The recombinant protein constructs ACE2-His-Avi (ACE2-Avi) (Acro Biosystems) and SARS-CoV-2 spike protein receptor binding domain-Fc (RBD-Fc) (Sino Biological) were used to model ACE2–RBD binding. (B) AlphaLISA assay system used to monitor ACE2–RBD interacts. Streptavidin donor beads recognize the Avi tag on ACE2. The protein A acceptor beads recognize the Fc tag on RBD. When in proximity the donor beads can be excited with light at 680 nm. This generates singlet oxygen which diffuses to the acceptor beads, causing the acceptor beads to luminesce at 615 nm. (C) The TruHits counterscreen uses streptavidindonor beads which directly interact with biotin acceptor beads. Because no intermediary molecule is needed to bring the donor and acceptor beads in proximity, the TruHits assay can identify compounds which directly interfere with the AlphaLISA readout.
Defining Assay Parameters
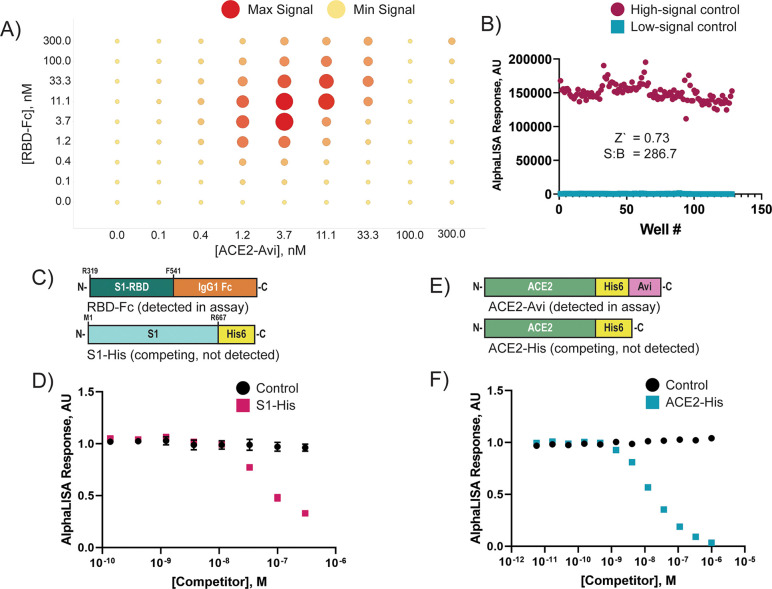
We first had to determine the working concentrations of RBD-Fc and ACE2-Avi that maximize the AlphaLISA signal. We selected 10 μg/mL for the concentration of donor and acceptor beads because it gave good signal with minimal well-to-well cross-talk (data not shown). To determine the optimal protein concentrations for the assay, we performed a cross-titration of RBD-Fc and ACE2-Avi varying each from 300 to 0.1 nM (Figure 2A). The maximum signal was achieved at 3.7 nM of both RBD-Fc and ACE2-Avi. Next, we tested the signal consistency of a high-signal control (ACE2-Avi + RBD-Fc + both AlphaLISA beads) and a low-signal control (ACE2-Avi + RBD-Fc + streptavidin donor beads) in a 1536-well format (Figure 2B). At 4 nM ACE2-Avi, 4 nM RBD-Fc, and 10 μg/mL streptavidin donor beads and protein A acceptor beads, we determined a plate Z′ value of 0.73 and signal-to-background ratio of 286.7, indicating satisfactory assay performance.
Figure 2.
AlphaLISA assay monitors ACE2–RBD interactions. (A) ACE2-Avi and RBD-Fc were titrated against each other from 300–0.1 nM in matrix format to determine the optimal protein concentrations for the AlphaLISA assay. ACE2-Avi and RBD-Fc were mixed at the concentrations indicated and allowed to equilibrate at 25 °C for 30 min. Streptavidin donor beads and protein A acceptor beads were then introduced to the solutions to a final concentration of 5 μg/mL of each bead. After 40 min of incubation at 25 °C the signal intensity was read using a PheraStar plate reader. (B) Assay suitability to 1536-well format was determined by combining 4 nM ACE2-Avi with 4 nM RBD-Fc in PBS + 0.05 mg/mL BSA. The mixture was incubated at 25 °C for 30 min. Streptavidin donor beads (10 μg/mL) were added to columns 1–8 of a 1536-well plate. The protein A acceptor beads (10 μg/mL) were added to columns 5–8. The entire mixture was incubated at 25 °C for 40 min before reading the low-signal control (columns 1–4) and high-signal control (columns 5–8) using a PheraSTAR plate reader with an AlphaLISA module. Z′ and signal-to-background (S/B) were calculated from these sample measurements. (C) Spike protein constructs used for this competition assay were RBD-Fc, which is recognized by the AlphaLISA beads, and S1-His, which competes for ACE2 binding and is not recognized by the AlphaLISA beads. (D) His-S1 was mixed in dose–response with 4 nM ACE2-Avi and allowed to incubate at 25 °C for 30 min before adding 4 nM RBD-Fc. Data points indicate the mean ± standard deviation, n = 3. (E) ACE2 protein constructs used for this competition assay were ACE2-Avi, which is biotinylated and recognized by the AlphaLISA beads, and ACE2-His, which competes for RBD binding but is not detected by the AlphaLISA beads. (F) ACE2-His was preincubated in dose–response with 4 nM RBD-Fc and allowed to incubate at 25 °C for 30 min before adding 4 nM ACE2-Avi. Data points indicate the mean ± standard deviation, n = 3. Both His-S1 and ACE2-His showed dose-dependent signal loss, indicating the AlphaLISA signal is mediated by RBD-Fc binding to ACE2-Avi.
Next we wanted to confirm the AlphaLISA signal measured was dependent on the RBD-Fc interaction with ACE2-Avi (i.e., the assay is “inhibitable”). We tested the ability of a truncated SARS-CoV-2 spike protein with a C-terminal poly-His tag to compete with RBD-Fc for ACE2 binding and a soluble ACE2 with a C-terminal poly-His tag to compete with ACE2-Avi for RBD binding (Figure 2C,E). These constructs were selected because both the S1-His and ACE2-His constructs should recognize their respective binding partner, ACE2-Avi and RBD-Fc, but will not be engaged by the AlphaLISA beads. Indeed, both S1-His and ACE2 were able to compete with their respective binding partners in a dose-dependent manner (Figure 2D,F), indicating that the AlphaLISA assay is dependent on RBD–ACE2 interaction and therefore is “inhibitable”.
qHTS Screen of NPC and AIL Compounds Sets
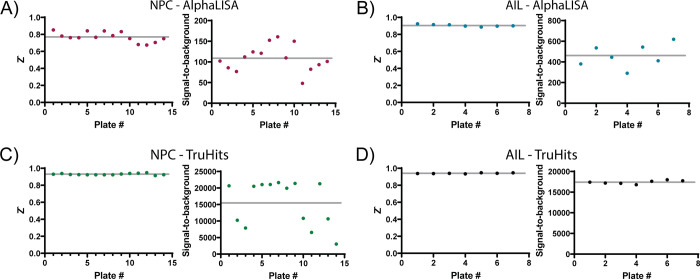
Having established the conditions and suitable performance for the RBD–ACE2 AlphaLISA assay, we employed it for qHTS. To this end, we selected two libraries: the NCATS Pharmaceutical Collection (NPC, library of approved small-molecule drugs in the USA, Europe, and Japan; 2678 compounds) and an in-house anti-infectives library (AIL, compounds shown to be active against infectious diseases, 739 compounds) to screen against our AlphaLISA RBD–ACE2 binding assay (HTS assay protocol shown in Table 1). When screening the NPC library, the average plate Z′ was 0.77 with an average signal-to-background ratio of 108.5 (Figure 3A). The AIL collection screen had an average plate Z′ of 0.90 and signal-to-background ratio of 460.7 (Figure 3B). Both NPC and AIL were also screened in parallel against the TruHits counterassay platform to rule out false-positive hits (Figure 1C, TruHits HTS protocol displayed in Table 2). For NPC, the TruHits counterassay performed with an average Z′ of 0.93 and average signal-to-background ratio of 15 477 (Figure 3C), and the AIL had an average Z′ of 0.94 with an average signal-to-background ratio of 17 410 (Figure 3D). All HTS data is available for download at https://opendata.ncats.nih.gov/covid19.
Figure 3.
Assay performance for compound libraries tested in the primary AlphaLISA assay and the TruHits counterassay. Z′ scores and signal-to-background values are plotted for each plate in the NPC AlphaLISA assay (A), the AIL AlphaLISA assay (B), the NPC TruHits counterscreen (C), and the AIL TruHits counterscreen (D). Data points in all plots indicate the Z′ or signal-to-background for individual plates. Gray lines in all plots represent the mean values.
Hit Identification
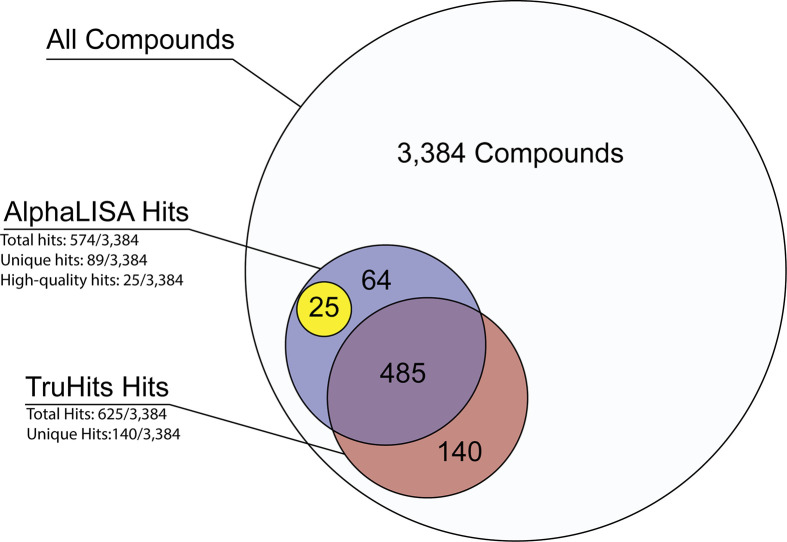
Hit compounds for both the AlphaLISA primary assay and the TruHits counter screen were identified as those with an IC50 < 50 μM and a maximum efficacy ≥ −50%, effectively removing all class 4 and some class 3 compounds (see the “Methods” section for curve class designations). A total of 574 initial hits were identified for the AlphaLISA assay (16.9% hit rate), and a total of 624 initial hits were identified for the TruHits counterassay (18.4% hit rate, Figure 4, Table S1). Out of these initial hits, 89 were unique to the AlphaLISA assay forming our first pool of potential active compounds. Twenty-five compounds were identified as capable high-quality hits and selected for cherry-picking and hit confirmation (Table S2).
Figure 4.
Hit summary from NPC and AIL compound sets. AlphaLISA and TruHits assays were performed against the NPC and AIL compound sets (3384 compounds total). Potential hits in both the AlphaLISA and TruHits assays were defined as having an IC50 ≥ 50 μM and a maximum response less than or equal to −50%. Of those hits, 89 were found to be unique to the AlphaLISA assay, and those curves were visually inspected to identify 25 high-quality hits for cherry-picking. Low-quality hits have TruHits response curves that satisfy the conditions for a “potential hit” but do not significantly diverge from the AlphaLISA data. High-quality hits have minimal TruHits response which do significantly diverge from the AlphaLISA response curve.
Hit Confirmation
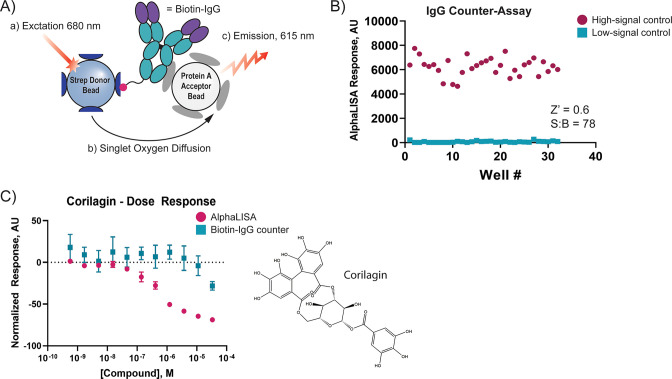
To confirm that the 25 high-quality hits disrupted ACE2–RBD interactions we performed cherry-pick hit confirmation assays. First, we repeated the ACE2 AlphaLISA assay with each of the 25 hit compounds re-plated in a triplicate dose–response assay to validate their activity from the primary screen. We also developed a counterassay using biotinylated rabbit IgG, which could replicate the interactions between the AlphaLISA beads and the Fc- and biotin-tags, without the ACE2–RBD complex (Figure 5A,B). Corilagin was the only compound which showed activity against ACE2–RBD with an IC50 of 5.5 μM (Figure 5C).
Figure 5.
Hit confirmation assays to confirm cherry-picked compound activity. (A) Cartoon depicting biotin-IgG counter assay used for cherry-picking. Biotinylated rabbit IgG acts as a platform for both the streptavidin donor bead and protein A acceptor bead. Compounds which disrupt either the streptavidin–IgG or protein A–Fc interaction will prevent the generation of a luminescent signal at 615 nm, indicating assay interference. (B) Biotin-IgG counterassay performance was evaluated in the 1536 format by incubating 1.6 nM biotinylated rabbit IgG in PBS + 0.05 mg/mL BSA with 10 μg/mL each of protein A acceptor beads and streptavidin donor beads. The mixture was incubated for 40 min at room temperature before measuring AlphaLISA luminescence on a PHERAstar FSX (BMG Labtech) plate reader. The high signal control represents the AlphaLISA signal in the vehicle control wells (DMSO). The low signal control represents the signal generated in the absence of biontinylated rabbit IgG. Data points indicate signal for individual replicants. (C) Corilagin showed activity in the AlphaLISA assay but not the biotin-IgG counter assay, indicating it inhibits the ACE2–RBD interaction in this proximity assay (IC50 = 5.5 μM). The structure of corilagin is presented to the right of the dose–response curve. Data points and error bars represent the mean ± standard deviation, n = 3.
Discussion
We developed the first well-based proximity assay for measuring COVID-19 RBD–ACE2 interactions using AlphaLISA technology. The AlphaLISA approach has the advantages of high signal-to-background ratios and robust assay performance (Z′ values) and is widely applicable to a variety of therapeutic types (small molecules, nanobodies, peptides, etc.). Molecules which interfere with the generation of or those that scavenge singlet oxygen present a problem as do those which absorb or emit light in the 615–680 nm wavelengths.20 However, the ease of use and ability to rapidly deploy an assay using this platform positions it as a strong primary assay for therapeutic screenings. Furthermore, the availability of the TruHits counterscreen simplifies the identification of most false-positive compounds.
While TruHits cannot account for the protein A–Fc interaction used in our AlphaLISA assay, it does aid in funneling down the potential hits to numbers that are more manageable for lower-throughput or more costly orthogonal approaches. In this case, we developed a supplemental biotin-IgG counterassay which fully replicated the bead–ligand interactions used in the primary assay, but material limitations made it more suitable to testing cherry-picked hits. Our screen identified 25 high-quality hits that were not active in the TruHits counterassay screen. These hits were cherry-picked, but only corilagin was validated in the ACE2–RBD AlphaLISA assay and the biotin-IgG counterassay. The original set of 3384 compounds included ACE2 inhibitors such as enalapril, moexipril, and lisinopril, but none showed ACE2–RBD binding inhibition in the AlphaLISA assay (Table S1). Similarly, compounds such as remdesivir and ceftazidime, which have gained notoriety over the course of the COVID-19 pandemic, showed no significant activity in the primary AlphaLISA screen nor the TruHits counterscreen.
While there was no expectation of a clinical drug repurposing opportunity emerging from the screen, the initial 25 hit compounds include several compounds approved for treating patients. A number of molecules containing PAINS moieties such as phenolics, including the confirmed hit corilagin (a polyphenolic natural product), and careful follow-up is required to understand mechanism. Nevertheless, the assay can serve to support the identification (HTS) or optimization of small-molecule and protein-based therapeutic candidate molecules directed to specifically inhibit the ACE2–spike interaction. Several assays have been developed at NCATS, and the data are available via the NCATS OpenData Portal (https://opendata.ncats.nih.gov/covid19/index.html), with regular updates. This portal contextualizes the data generated by our AlphaLISA assay, and we are continually working to ensure compounds are tested across a range of assays, from ACE2 enzyme activity to pseudoparticle entry assays. At current writing, corilagin shows activity in this AlphaLISA assay, weak activity in the ACE2 enzymatic assay, and no activity in blocking cytopathic effects (CPE).
SARS-CoV-2 follows its predecessor, SARS-CoV, by using its spike protein to engage host ACE2 receptors to invade host cells. In response to the SARS-CoV outbreak in 2003, efforts targeting the ACE2–spike interaction emphasized antibody-based therapeutics.21,22 Phage display libraries10,11 and surface plasmon resonance23 were used to develop neutralizing peptides. However, reported small-molecule screens for SARS-CoV at the time focused on target-agnostic cellular approaches.24 Focused protein–protein interaction (PPI, proximity) assays were not widely available for screening in 2003. Advances in screening technology over the past decades, such as the AlphaLISA technology used in this work, enable us to address specific aspects of COVID-19 biology on a scale that was inaccessible during previous outbreaks.
Currently, protein–protein interaction assays have been used to determine vital information about ACE2–spike interactions,7,8 but the methods used (ITC and SPR) are low-throughput. At the time of disclosure, neither protein–protein interaction assays for ACE2–spike binding nor any HTS with ACE2–spike proximity assay approaches have yet been published. The commercial availability of orthogonal PPI methods using fluorescence resonance energy transfer (FRET), saturation transfer difference (STD) NMR, co-immunoprecipitation, and microscale thermophoresis support the need for further development of such assays and broad interest of the scientific community.
The AlphaLISA assay we developed presents a quick, simple, and qHTS-amenable approach to ACE2–RBD therapeutic development. Although the drug-repurposing data presented herein yielded no actionable therapeutic candidates, the assay we developed can be leveraged for structure–activity relationship studies and support novel drug development targeting ACE2–RBD interactions. Multimerization of ACE2 and spike have been demonstrated to enhance the interaction between SARS-CoV-2 spike and host ACE2.25 This presents an opportunity for ACE2–RBD assays to develop as the field grows. The advantage to AlphaLISA is that we can continue to use the methods developed herein toward more sophisticated qHTS assay design to suit our evolving knowledge of SARS-CoV-2 biology.
Acknowledgments
This work was supported by the Intramural Research Program of the National Center for Advancing Translational Sciences, National Institutes of Health.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsptsci.0c00161.
Author Contributions
Q.M.H. and M.D.H. developed and designed the experiments. Q.M.H. conducted the experiments. Q.M.H., K.M.W., and M.S. conducted data analysis. Z.I., R.T.E., and P.S. prepared and plated compound collections. All authors contributed to writing and editing the manuscript.
The authors declare no competing financial interest.
Notes
Assay data are available via the NCATS OpenData Portal https://opendata.ncats.nih.gov/covid19/index.html, with regular updates.
This article is made available via the ACS COVID-19 subset for unrestricted RESEARCH re-use and analyses in any form or by any means with acknowledgement of the original source. These permissions are granted for the duration of the World Health Organization (WHO) declaration of COVID-19 as a global pandemic.
Supplementary Material
References
- Guy R. K.; DiPaola R. S.; Romanelli F.; Dutch R. E. (2020) Rapid repurposing of drugs for COVID-19. Science 368 (6493), 829–830. 10.1126/science.abb9332. [DOI] [PubMed] [Google Scholar]
- Tu Y. F.; Chien C. S.; Yarmishyn A. A.; Lin Y. Y.; Luo Y. H.; Lin Y. T.; Lai W. Y.; Yang D. M.; Chou S. J.; Yang Y. P. (2020) A Review of SARS-CoV-2 and the Ongoing Clinical Trials. Int. J. Mol. Sci. 21 (7), 2657. 10.3390/ijms21072657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman R. T.; Roth J. S.; Brimacombe K. R.; Simeonov A.; Shen M.; Patnaik S.; Hall M. D. (2020) Remdesivir: A Review of Its Discovery and Development Leading to Emergency Use Authorization for Treatment of COVID-19. ACS Cent. Sci. 6 (5), 672–683. 10.1021/acscentsci.0c00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G. D.; De Clercq E. (2020) Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nat. Rev. Drug Discovery 19 (3), 149–150. 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- Lei C.; Qian K.; Li T.; Zhang S.; Fu W.; Ding M.; Hu S. (2020) Neutralization of SARS-CoV-2 spike pseudotyped virus by recombinant ACE2-Ig. Nat. Commun. 11 (1), 2070. 10.1038/s41467-020-16048-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. H.; Moore M. J.; Vasilieva N.; Sui J. H.; Wong S. K.; Berne M. A.; Somasundaran M.; Sullivan J. L.; Luzuriaga K.; Greenough T. C.; Choe H.; Farzan M. (2003) Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426 (6965), 450–454. 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J.; Ye G.; Shi K.; Wan Y. S.; Luo C. M.; Aihara H.; Geng Q. B.; Auerbach A.; Li F. (2020) Structural basis of receptor recognition by SARS-CoV-2. Nature 581, 221. 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R. H.; Zhang Y. Y.; Li Y. N.; Xia L.; Guo Y. Y.; Zhou Q. (2020) Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367 (6485), 1444. 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho T. Y.; Wu S. L.; Chen J. C.; Wei Y. C.; Cheng S. E.; Chang Y. H.; Liu H. J.; Hsiang C. Y. (2006) Design and biological activities of novel inhibitory peptides for SARS-CoV spike protein and angiotensin-converting enzyme 2 interaction. Antiviral Res. 69 (2), 70–6. 10.1016/j.antiviral.2005.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung K. S.; Meanwell N. A. (2007) Recent developments in the virology and antiviral research of severe acute respiratory syndrome coronavirus. Infect. Disord.: Drug Targets 7 (1), 29–41. 10.2174/187152607780090739. [DOI] [PubMed] [Google Scholar]
- Prabakaran P.; Zhu Z.; Xiao X.; Biragyn A.; Dimitrov A. S.; Broder C. C.; Dimitrov D. S. (2009) Potent human monoclonal antibodies against SARS CoV, Nipah and Hendra viruses. Expert Opin. Biol. Ther. 9 (3), 355–68. 10.1517/14712590902763755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B.; Zhang Q.; Ge J.; Wang R.; Sun J.; Ge X.; Yu J.; Shan S.; Zhou B.; Song S. (2020) Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 584, 115. 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Monteil V.; Kwon H.; Prado P.; Hagelkruys A.; Wimmer R. A.; Stahl M.; Leopoldi A.; Garreta E.; Hurtado del Pozo C.; Prosper F.; et al. (2020) Inhibition of SARS-CoV-2 Infections in Engineered Human Tissues Using Clinical-Grade Soluble Human ACE2. Cell 181 (4), 905. 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brimacombe K. R., Zhao T., Eastman R. T., Hu X., Wang K., Backus M., Baljinnyam B., Chen C. Z., Chen L., and Eicher T.. et al. (June 5, 2020) An Open Data portal to share COVID-19 drug repurposing data in real time. bioRxiv (Microbiology), DOI: 10.1101/2020.06.04.135046.
- Yasgar A.; Jadhav A.; Simeonov A.; Coussens N. P. (2016) Alpha Screen-Based Assays: Ultra-High-Throughput Screening for Small-Molecule Inhibitors of Challenging Enzymes and Protein-Protein Interactions. Methods Mol. Biol. 1439, 77–98. 10.1007/978-1-4939-3673-1_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L.; Li L.; Tien C.; LaBarbera D. V.; Chen C. (2019) Targeting HIV-1 Protease Autoprocessing for High-throughput Drug Discovery and Drug Resistance Assessment. Sci. Rep. 9 (1), 301. 10.1038/s41598-018-36730-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R.; Southall N.; Wang Y.; Yasgar A.; Shinn P.; Jadhav A.; Nguyen D. T.; Austin C. P. (2011) The NCGC pharmaceutical collection: a comprehensive resource of clinically approved drugs enabling repurposing and chemical genomics. Sci. Transl. Med. 3 (80), 80ps16 10.1126/scitranslmed.3001862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R.; Zhu H.; Shinn P.; Ngan D.; Ye L.; Thakur A.; Grewal G.; Zhao T.; Southall N.; Hall M. D.; Simeonov A.; Austin C. P. (2019) The NCATS Pharmaceutical Collection: a 10-year update. Drug Discovery Today 24 (12), 2341–2349. 10.1016/j.drudis.2019.09.019. [DOI] [PubMed] [Google Scholar]
- Inglese J.; Auld D. S.; Jadhav A.; Johnson R. L.; Simeonov A.; Yasgar A.; Zheng W.; Austin C. P. (2006) Quantitative high-throughput screening: A titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc. Natl. Acad. Sci. U. S. A. 103 (31), 11473–11478. 10.1073/pnas.0604348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussens N. P., Auld D., Roby P., Walsh J., Baell J. B., Kales S., Hadian K., and Dahlin J. L. (2004) Compound-Mediated Assay Interferences in Homogenous Proximity Assays, in Assay Guidance Manual (Sittampalam G. S., Grossman A., Brimacombe K., Arkin M., Auld D., Austin C. P., Baell J., Bejcek B., Caaveiro J. M. M., and Chung T. D. Y., et al. , Eds.), Eli Lilly & Company and the National Center for Advancing Translational Sciences, Bethesda, MD. [PubMed] [Google Scholar]
- Sui J. H.; Li W. H.; Murakami A.; Tamin A.; Matthews L. J.; Wong S. K.; Moore M. J.; Tallarico A. S. C.; Olurinde M.; Choe H.; Anderson L. J.; Bellini W. J.; Farzan M.; Marasco W. A. (2004) Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc. Natl. Acad. Sci. U. S. A. 101 (8), 2536–2541. 10.1073/pnas.0307140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M. (2004) SARS Immunity and Vaccination. Cell Mol. Immunol 1 (3), 193–198. [PubMed] [Google Scholar]
- Struck A. W.; Axmann M.; Pfefferle S.; Drosten C.; Meyer B. (2012) A hexapeptide of the receptor-binding domain of SARS corona virus spike protein blocks viral entry into host cells via the human receptor ACE2. Antiviral Res. 94 (3), 288–296. 10.1016/j.antiviral.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao R. Y.; Tsui W. H.; Lee T. S.; Tanner J. A.; Watt R. M.; Huang J. D.; Hu L.; Chen G.; Chen Z.; Zhang L.; He T.; Chan K. H.; Tse H.; To A. P.; Ng L. W.; Wong B. C.; Tsoi H. W.; Yang D.; Ho D. D.; Yuen K. Y. (2004) Identification of novel small-molecule inhibitors of severe acute respiratory syndrome-associated coronavirus by chemical genetics. Chem. Biol. 11 (9), 1293–9. 10.1016/j.chembiol.2004.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui I., Zhou X. X., Lim S. A., Elledge S. K., Solomon P., Rettko N. J., Zha B. S., Kirkemo L. L., Gramespacher J. A., and Liu J.. et al. (May 21, 2020) Trimeric SARS-CoV-2 Spike interacts with dimeric ACE2 with limited intra-Spike avidity. bioRxiv (Biochemistry), DOI: 10.1101/2020.05.21.109157
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.