NRPs form a class of secondary metabolites with biocontrol and pharmaceutical potential. This work describes the identification of novel bogorol variants and succinylated bogorols (namely, succilins) and further investigates their biosynthetic pathway and mode of action. Adenylation domain-mediated lipoinitiation of bogorols represents a novel pathway by which NRPs incorporate fatty acid tails. This pathway provides the possibility to engineer the lipid tail of NRPs without identifying a fatty acid coenzyme ligase, which is usually not present in the biosynthetic gene cluster. The terminal reductase domain (TD) and BogI-mediated valinol formation and their effect on the biological activity of bogorols are revealed. Succinylation, which is rarely reported in NRPs, was discovered in the bogorol family of peptides. We demonstrate that bogorols combat bacterial pathogens by forming pores in the cell membrane. We also report the synergistic effect of two natural products (relacidine B and bogorol K) produced by the same strain, which is relevant for competition for a niche.
KEYWORDS: bogorol variants, succilins, Brevibacillus laterosporus, biosynthesis, lipoinitiation, reduction, succinylation, mode of action, synergy
ABSTRACT
Nonribosomal peptides (NRPs) are a class of secondary metabolites usually produced by microorganisms. They are of paramount importance in different applications, including biocontrol and pharmacy. Brevibacillus spp. are a rich source of NRPs yet have received little attention. In this study, we characterize four novel bogorol variants (bogorols I to L, cationic linear lipopeptides) and four succilins (succilins I to L, containing a succinyl group that is attached to the Orn3/Lys3 in bogorols I to L) from the biocontrol strain Brevibacillus laterosporus MG64. Further investigation revealed that the bogorol family of peptides employs an adenylation pathway for lipoinitiation, different from the usual pattern, which is based on an external ligase and coenzyme A. Moreover, the formation of valinol was proven to be mediated by a terminal reductase domain and a reductase encoded by the bogI gene. Furthermore, succinylation, which is a novel type of modification in the family of bogorols, was discovered. Its occurrence requires a high concentration of the substrate (bogorols), but its responsible enzyme remains unknown. Bogorols display potent activity against both Gram-positive and Gram-negative bacteria. Investigation of their mode of action reveals that bogorols form pores in the cell membrane of both Gram-positive and Gram-negative bacteria. The combination of bogorols and relacidines, another class of NRPs produced by B. laterosporus MG64, displays a synergistic effect on different pathogens, suggesting the great potential of both peptides as well as their producer B. laterosporus MG64 for broad applications. Our study provides a further understanding of the bogorol family of peptides as well as their applications.
IMPORTANCE NRPs form a class of secondary metabolites with biocontrol and pharmaceutical potential. This work describes the identification of novel bogorol variants and succinylated bogorols (namely, succilins) and further investigates their biosynthetic pathway and mode of action. Adenylation domain-mediated lipoinitiation of bogorols represents a novel pathway by which NRPs incorporate fatty acid tails. This pathway provides the possibility to engineer the lipid tail of NRPs without identifying a fatty acid coenzyme ligase, which is usually not present in the biosynthetic gene cluster. The terminal reductase domain (TD) and BogI-mediated valinol formation and their effect on the biological activity of bogorols are revealed. Succinylation, which is rarely reported in NRPs, was discovered in the bogorol family of peptides. We demonstrate that bogorols combat bacterial pathogens by forming pores in the cell membrane. We also report the synergistic effect of two natural products (relacidine B and bogorol K) produced by the same strain, which is relevant for competition for a niche.
INTRODUCTION
Nonribosomal peptides (NRPs) are a class of secondary metabolites synthesized by megaenzymes named nonribosomal peptide synthetases (NRPSs). NRPs display diverse properties, some of which can help the producer to survive in natural environments. Surfactins and viscosins are essential for the motility of Bacillus subtilis and Pseudomonas putida, respectively (1–4). Massetolide A and rhamnolipid are involved in biofilm formation by different Pseudomonas species (5–7). Bacillibactin and petrobactin are siderophores that can chelate iron and make it available for Bacillus species (8, 9). Many NRPs display antimicrobial activity against a broad range of microorganisms, including bacteria, fungi, oomycetes, and viruses, etc. Because of this important property, some NRPs, such as surfactins and iturins, are used as biocontrol agents to defend against plant diseases (10), while others, such as daptomycin and polymyxin, are used as antibiotics to treat animal and human diseases caused by pathogenic bacteria (11, 12).
Brevibacillus is a genus belonging to the Firmicutes, one of the phyla with a great abundance of NRPS enzymes (13). Brevibacillus spp. are a rich source of NRPs, which has been known for many decades. Tyrocidine and gramicidin S, two cyclic lipopeptides with potent antibacterial activity, were discovered from Brevibacillus brevis in the 1940s (14, 15). A class of linear cationic lipopeptides, including bogorols, BT peptide, BL-A60, and brevibacillins, was discovered from Brevibacillus laterosporus in the past 2 decades (16–20). Brevicidine, laterocidine, and relacidines, a class of cationic lipopeptides that selectively combat Gram-negative bacteria, were characterized from B. laterosporus in the past 2 years (21, 22). Even so, we believe that the biosynthetic potential of Brevibacillus spp., especially B. laterosporus, has been underestimated. On the one hand, genome mining of B. laterosporus MG64, a plant-growth-promoting rhizobacterium from perennial ryegrass, revealed that around 80% of the biosynthetic gene clusters (BGCs) have not been assigned to known products yet (23). On the other hand, some structurally similar NRPs produced by Brevibacillus spp. were overlooked (24). For example, bogorols, BT peptide, and brevibacillins share similar structures but have different amino acid residues in the second, fourth, fifth, or ninth positions. A minor difference in the amino acid residues has been shown to affect the biological properties of the bogorols: a replacement of isoleucine by valine at the fourth position decreases the antibacterial activity severely (17). Therefore, more attention should be paid to the characterization of the rich variety of NRPs from Brevibacillus spp.
In this study, we aimed to identify and characterize novel NRPs produced by the biocontrol strain B. laterosporus MG64, which showed very good activity against a variety of pathogens, including plant pathogens and mammalian pathogens (23). Furthermore, we investigate the biosynthesis and mode of action of the compounds to have a better understanding of their potential for application.
RESULTS
Purification and characterization of bioactive peptides from B. laterosporus MG64.
We previously isolated a biocontrol strain, B. laterosporus MG64, that can inhibit a wide range of plant pathogens and mammalian pathogens (23). To identify and characterize the bioactive compounds, B. laterosporus MG64 was inoculated in Lennox broth (LB) for 24 h, and the supernatant was used to extract the metabolites with a column filled with C18 silica gel. The crude extract was further purified by high-performance liquid chromatography (HPLC), and the different peaks that were collected were screened for bioactivity against Xanthomonas campestris pv. campestris, a Gram-negative pathogen that causes bacterial wilt in perennial ryegrass, and Bacillus cereus, a Gram-positive bacterium that causes food spoilage. The major peaks showed bioactivity against both strains and were selected for further characterization.
Four different compounds were characterized by liquid chromatography-tandem mass spectrometry (LC-MS/MS) (see Fig. S1 in the supplemental material). Compounds 1 and 2 have the same mass of 1,555.06 Da (m/z 778.53 [M + 2H]2+). The clear signals of y and b ions indicated the presence of Val (5), Orn (1), Ile (1), Lys (2), and Tyr (1) (with the numbers in parentheses indicating the amounts of amino acid residues) (Fig. S1A). The difference between compounds 1 and 2 are the amino acid residues at positions 4 and 5, where compound 1 has Ile4-Val5 and compound 2 has these inverted. The total masses and amino acid compositions of both compounds are similar to those of bogorols, which form a class of linear peptides that are composed of a fatty acid tail (2-hydroxy-3-methylvaleric acid) and 13 amino acid residues (including a valinol at the last position) (16, 17). The bogorol biosynthetic gene cluster identified by antiSMASH also supports this hypothesis (Fig. 1A). With bogorol as a reference structure, we successfully identified the components at both termini, namely, C6H11O2-dehydrobutyrine (Dhb) for the N terminus (b2 = 198.11) (Fig. S1A) and Leu-valinol for the C terminus (y2 = 217.19) (Fig. S1A). However, Leu9 in the reported bogorols is replaced by Val9 in compounds 1 and 2. Therefore, we propose them to be novel variants of bogorol. Similarly, compound 3 (m/z 785.54 [M + 2H]2+) and compound 4 (m/z 792.54 [M + 2H]2+) were identified to be novel variants of bogorol as well. Compared to compounds 1 and 2, compound 3 has a 14-Da increase in mass, which is a result of two consecutive Ile residues at positions 4 and 5 (Fig. S1B). Compound 4 has Lys instead of Orn at position 3, which is the only difference from compound 3 (Fig. S1C). Compounds 1 to 4 were therefore named bogorols I to L, respectively (Fig. 1B).
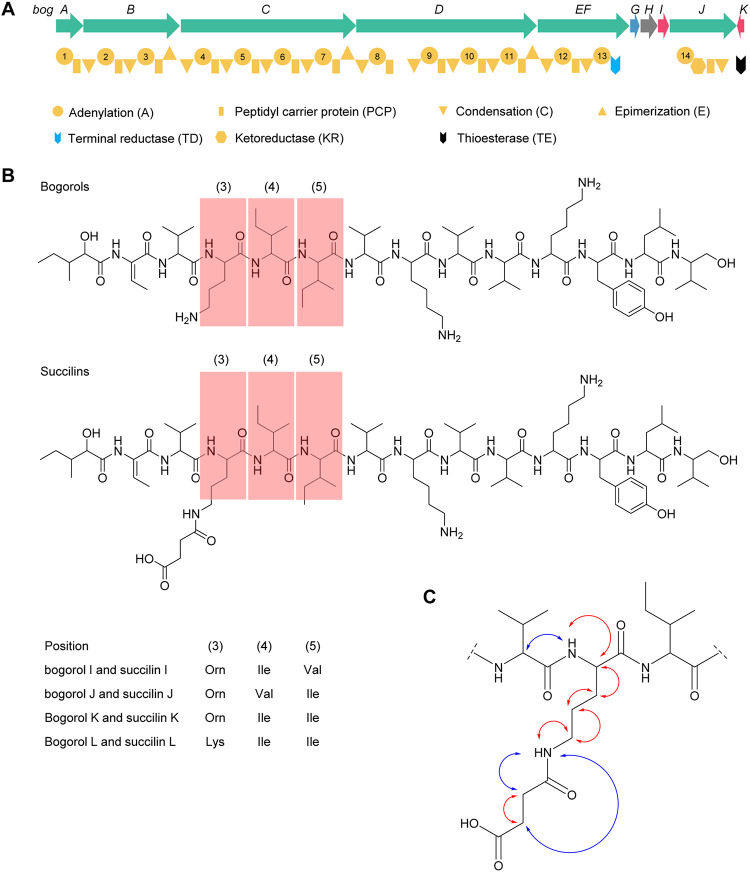
FIG 1.
Bogorols and succilins produced by B. laterosporus MG64. (A) Gene cluster of bogorols harbored by B. laterosporus MG64. Green, core biosynthetic gene; blue, transport-related gene; gray, unknown gene; red, additional biosynthetic gene. (B) Structures of bogorols and succilins characterized by LC-MS/MS and NMR. Amino acid residues at positions 3 to 5 (red shaded) of different peptides are shown in the table below the structures. (C) Two-dimensional (2D) NMR correlations (red, 1H-1H TOCSY; blue, 1H-1H NOESY) confirming succinylation at the third amino acid residue (Lys) of succilin K (see also Fig. S3 in the supplemental material).
The structures of bogorol K and bogorol L were further confirmed by 1H nuclear magnetic resonance (NMR), 1H-1H total correlation spectroscopy (TOCSY) NMR, and 1H-1H nuclear Overhauser effect spectroscopy (NOESY) NMR (Fig. S1D and E). Their chemical shift (parts per million) assignments (partial) are shown in Tables S1 and S2, respectively. A Marfey-type analysis (25) was also conducted to detect the configuration of amino acids in bogorol K and bogorol L. Both compounds have an l-configuration of all the Val, Leu, Ile, and valinol residues and a d-configuration of Tyr at position 11 (Table S3). The Orn residue in position 3 of bogorol K is in the d-configuration, which suggests that the Lys residue in bogorol L should also be in a d-configuration. The other two Lys residues at positions 7 and 10 of both compounds displayed either a d- or l-configuration, which could not be distinguished from each other. This result is consistent with the prediction from antiSMASH where epimerization domains were predicted at the 3rd, 7th, and 11th modules (Fig. 1A).
Fortuitously, we found another group of peptides (compounds 5 to 8) that have masses of 1,655.10 Da (compounds 5 and 6), 1,669.11 Da (compound 7), and 1,683.10 Da (compound 8), which are 100 Da higher than those of bogorols I to L, respectively. They are eluted by HPLC in the same order as bogorols I to L but with a longer elution time (data not shown). Further analysis of tandem MS data revealed that these peptides are likely the products of a succinyl group attached to Orn3 or Lys3 of bogorols I to L with a peptide bond (Fig. 1B; Fig. S2A to C). In the NMR analysis, the succinylation at Orn3 in peptide 7 was supported by the appearance of 2 methylene signals at 2.28 and 2.36 ppm, which showed a strong correlation by 1H-1H TOCSY NMR (Fig. 1C; Fig. S3A). Both signals showed NOESY cross-peaks with an amide N-H signal at 7.79 ppm, which in turn showed a correlation with the signal of the δ-CH2 of Orn3 at 2.98 ppm by both TOCSY and NOESY NMR (Fig. 1C; Fig. S3B and C). The downfield shift of this δ-H from 2.76 ppm (in bogorol K) to 2.98 ppm is consistent with the acetylation of the Orn3 amine resulting in a more electron-withdrawing amide (Table S4). This result confirms the attachment of a succinyl group at the end of the side chain of the Orn3. Therefore, we name compounds 5 to 8 as succilins I to L, which are probably modification products arising from bogorols I to L (Fig. 1B).
Adenylation domain-mediated lipoinitiation.
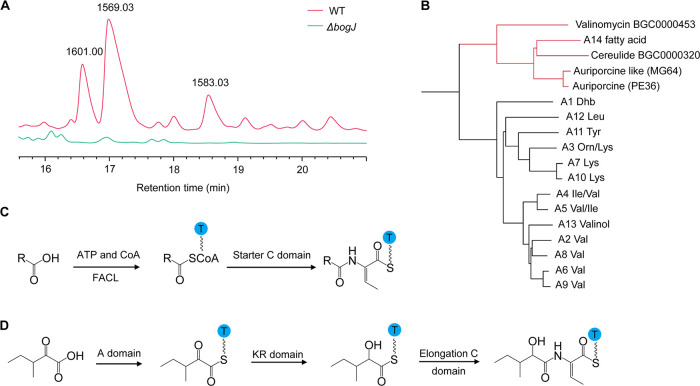
Members of the bogorol family of peptides contain a fatty acid tail at the N terminus, which is a representative characteristic of lipopeptides. The lipid tails of lipopeptides are usually incorporated into the peptides through a starter condensation (C) domain (26). However, such a domain was not found in the gene cluster of bogorols (Fig. 1A), suggesting a potentially novel lipidation of bogorols. Inspection of the biosynthetic modules reveals an additional module that is encoded by the bogJ gene. The knockout mutant of bogJ in B. laterosporus LMG15441, a transformable bogorol producer (27), impaired the production of bogorols completely (Fig. 2A; Fig. S4A), suggesting that the bogJ gene is essential for the biosynthesis of bogorols. To investigate the potential substrate activation by this module, a phylogenetic tree involving the 14th adenylation (A) domain and the other A domains from the bogorol gene cluster, as well as other A domains that are known to activate alpha-hydroxy acid, was constructed. The result revealed that A14 is phylogenetically far away from other A domains in the bogorol gene cluster. Instead, it was clustered together with A domains that activate alpha-hydroxy acid (Fig. 2B). Further inspection revealed that such A domains are normally followed by a ketoreductase (KR) domain. They form a special module together with a peptidyl carrier protein (PCP) domain. A similar module was found in another gene cluster from B. laterosporus MG64 (Fig. S4B). This gene cluster was identified to be auriporcine, a lipopeptide originally identified from B. laterosporus PE36 (Fig. S4B) (28). Interestingly, the fatty acid tail of auriporcine, which is incorporated by the A-KR-PCP module, is the same as that of bogorols (Fig. S4C). All these results suggest that the lipoinitiation of bogorols is mediated by the module encoded by bogJ. This lipoinitiation is different from the usual pattern employed by most of the NRPs, in which the fatty acid is activated by ligation to coenzyme A in the presence of a fatty acid coenzyme ligase (FACL) and the activated fatty acid is then incorporated into the assembly line by a starter C domain (Fig. 2C) (26). In the case of bogorols, a 3-methyl-2-oxopentanoic acid was first activated by an A domain, which usually activates amino acid residues. After activation, the α-keto acid was reduced into 2-hydroxy-3-methylvaleric acid by the KR domain and entered the next module under the guidance of a C domain (Fig. 2D) (29).
FIG 2.
Adenylation domain-mediated lipoinitiation of bogorol peptides. (A) Production of bogorols by the wild-type (WT) and ΔbogJ mutant strains of B. laterosporus LMG15441. Strains were grown in MEM broth for 48 h and extracted with a C18 column filled with silica gel. The extracted products were subsequently subjected to HPLC. (B) Phylogenetic tree of A domains from the bogorol gene cluster harbored by B. laterosporus MG64 and reference A domains that incorporate hydroxy acid. Auriporcine was retrieved from B. laterosporus PE36, while cereulide and valinomycin were retrieved from the miBIG database. The A domains activating alpha-hydroxy acids are indicated in red. (C) Usual pattern of fatty acid incorporation in NRPs. CoA, coenzyme A; FACL, fatty acid coenzyme ligase. (D) Proposed pattern of bogorols incorporating fatty acid.
The formation of valinol is mediated by two reductases.
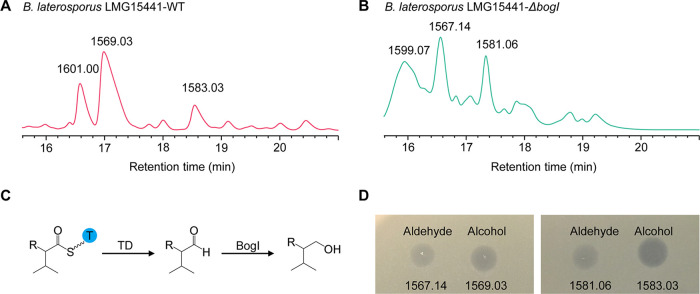
An outstanding characteristic of bogorol family peptides is the presence of a valinol (an alcohol form of valine) at the C terminus. Its formation is mediated by two possible mechanisms: (i) valinol is incorporated by the corresponding module, or (ii) valine is incorporated and then reduced to valinol. The first mechanism was considered more straightforward and energy-saving. However, analysis of A domains indicates that A13 shows very high similarity to other A domains that incorporate valine (Fig. 2B). Therefore, the second mechanism is more likely to be employed. Investigation of the gene cluster led us to the discovery of two reductases: the terminal reductase domain (TD) in BogF and an aldo/keto reductase, BogI. The TD is responsible for the release of the peptide chain from the PCP domain in a reduction pathway (30), while the function of BogI was unknown. To investigate the latter, the bogI gene in B. laterosporus LMG15441 was knocked out, resulting in the formation of a new group of compounds. These new peptides have masses that are 2 Da lower than those of the wild type (Fig. 3A and B; Fig. S4D). This mass reduction corresponds to the change of the alcohol form of valine to the aldehyde form, which indicates that valine was first reduced to the aldehyde form when released by the TD and then further reduced to the alcohol form by BogI (Fig. 3C). A comparison of the antibacterial activities of bogorols in the two different forms revealed that the alcohol form has better activity, underlining the importance of the reduction for biological activity (Fig. 3D).
FIG 3.
Formation of valinol in bogorol peptides. (A and B) Bogorols and their intermediates (aldehyde form) produced by wild-type (A) and ΔbogI mutant (B) strains of B. laterosporus LMG15441. The masses (daltons, based on MALDI-TOF analysis) of the three major compounds produced by each strain are indicated. (C) Two-step formation of alcohol-form amino acids in the bogorol family of peptides. (D) Spot-on-lawn assay of aldehyde-form and alcohol-form bogorols. X. campestris pv. campestris NCCB92058 was used as an indicator. Each compound was added at the same amount, namely, 10 μl of a 200-μg/ml stock.
Succinylation occurring in lipopeptides.
In this study, a novel class of peptides with an additional succinyl group attached to the Orn3/Lys3 residue of bogorols was identified as succilins. This modification is not observed in B. laterosporus LMG15441 (Fig. S5A), suggesting that succilins are not always formed when bogorols are produced. A comparison of the flanking genes of the bogorol BGC reveals some specific genes in each strain (Fig. S5B). However, the annotation of these genes did not result in a potential candidate, which was expected to be a transferase based on the similar modification, mannosylation, of ramoplanin (31, 32). Further investigation of the gene cluster revealed two transferases, bogN and bogR, which are present in both strains (Fig. S5B). However, the expression of bogN and bogR from B. laterosporus MG64 in B. laterosporus LMG15441 did not result in the production of succilins, suggesting that they are not responsible for succinylation (Fig. S5C). Interestingly, we found that bogorols were detected in B. laterosporus MG64 after a 14-h incubation in our experiment, whereas succilins were detected only after 16 h by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry and only after 18 h by HPLC (Fig. 4; Fig. S5D). This suggests that succinylation may require a high substrate concentration and is likely mediated by enzymes. However, the responsible gene is still unclear, and further research using transcriptomics and/or heterologous expression is needed.
FIG 4.
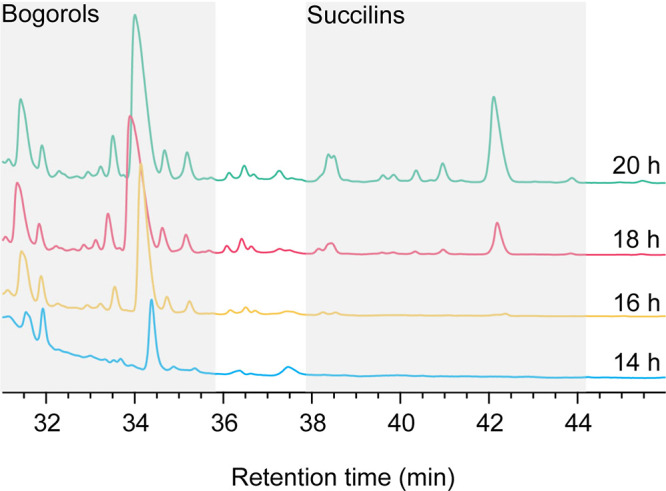
Production of bogorols and succilins by B. laterosporus MG64 at different time points. Fresh cells (10-μl culture with an OD600 of 1.0) were inoculated in 50 ml MEM broth and grown at 28°C for 14 h, 16 h, 18 h, and 20 h. The compounds were extracted from the supernatant at each time point and analyzed by HPLC and MALDI-TOF mass spectrometry.
Antibacterial activity and mechanism of action.
To investigate the potential of the newly identified bogorols and succilins for different applications, the antibacterial activity of bogorol K, bogorol L, and succilin K was assessed using plant pathogens (Xanthomonas species, Pseudomonas species, Pectobacterium carotovorum, and Ralstonia syzygii), a food pathogen (Bacillus cereus), and human pathogens (Pseudomonas aeruginosa, Klebsiella pneumoniae, Escherichia coli, and Staphylococcus aureus) as indicators. As shown in Table 1, bogorol K and bogorol L displayed potent activity against the Gram-positive pathogens tested (MIC values ranging from 1 to 4 μg/ml). Bogorols K and L were also active against various Gram-negative pathogens, except for the human pathogen P. aeruginosa and the plant pathogens P. carotovorum and R. syzygii, which are not inhibited by both compounds until their concentrations reach 32 μg/ml. Their antibacterial potency is similar to that of brevibacillin, a structurally similar lipopeptide. Succilin K, which is derived from bogorol K by succinylation at Orn3, showed only weak activity against Xanthomonas spp. and the Gram-positive pathogens tested, and the MIC values are at least 4 times higher than that of bogorol K.
TABLE 1.
MIC values of the identified compounds against different bacteria
| Pathogen | MIC (μg/ml)a
|
|||
|---|---|---|---|---|
| Bogorol K (compound 3) | Bogorol L (compound 4) | Succilin K (compound 7) | Brevibacillin | |
| Xanthomonas campestris pv. campestris NCCB92058 | 4 | 2 | 32 | 2 |
| Xanthomonas translucens pv. graminis LMG587 | 8 | 4 | 16 | 2 |
| Pseudomonas syringae pv. antirrhini LMG2131 | 16 | 8 | >32 | 8 |
| Pseudomonas syringae pv. tomato DC3000 | 16 | 8 | >32 | 16 |
| Pseudomonas aeruginosa PAO1 | 32 | 32 | >32 | 32 |
| Klebsiella pneumoniae LMG20218 | 16 | 32 | >32 | 16 |
| Escherichia coli ET8 | 8 | 8 | >32 | 8 |
| E. coli MG1665 | 16 | 16 | >32 | 16 |
| Pectobacterium carotovorum LMG5863 | >32 | >32 | >32 | ND |
| Ralstonia syzygii subsp. syzygii LMG6969 | >32 | 32 | >32 | ND |
| Staphylococcus aureus subsp. aureus 533 R4 | 2 | 2 | 16 | 1–2 |
| Bacillus cereus ATCC 14579 | 2 | 2 | 32 | 2 |
| Enterococcus faecium LMG16003 | 4 | 2 | 16 | 1–2 |
ND, not determined.
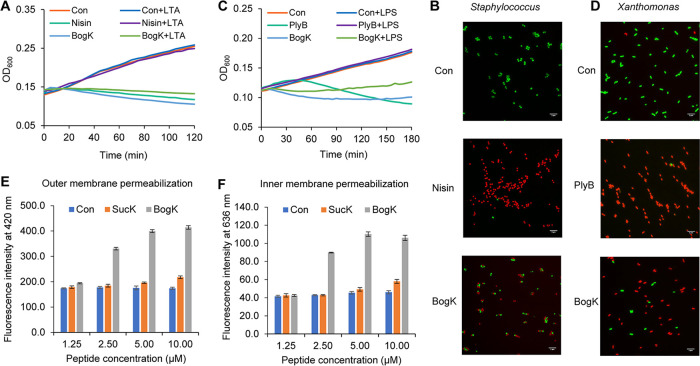
The Gram-positive human pathogen S. aureus subsp. aureus 533 R4 and the Gram-negative plant pathogen X. campestris pv. campestris NCCB92058 were used to investigate the mode of action of bogorol K. As shown in Fig. 5A, bogorol K inhibited the growth of Staphylococcus within 15 min, which is comparable to the positive control nisin. The addition of lipoteichoic acid (LTA), an important constituent of the cell wall of Gram-positive bacteria, did not affect the activity of bogorol K against Staphylococcus, suggesting that bogorol K does not bind to LTA, in contrast to nisin (Fig. 5A). Moreover, the addition of lipid II originating from Gram-positive bacteria did not decrease the inhibition zone of bogorol K toward Staphylococcus (Fig. S6A), suggesting that bogorol K does not bind to Gram-positive-type lipid II. However, the membrane permeability assay showed that bogorol K forms pores in the cell membrane of Staphylococcus, similar to the positive control nisin (Fig. 5B). Bogorol K inhibited the growth of the Gram-negative bacterium X. campestris within 15 min and showed weak binding to the lipopolysaccharide (LPS) compared to polymyxin B (Fig. 5C). Besides, bogorol K does not bind to lipid II originating from E. coli (Fig. S6B). Membrane permeability assays revealed that bogorol K disrupts the cellular membrane of X. campestris as it does to those of Gram-positive bacteria (Fig. 5D). Collectively, the results show that bogorol K employs the same mechanism for killing Gram-negative bacteria as that for Gram-positive bacteria by forming holes in the cellular membrane.
FIG 5.
Mode of action employed by the bogorol family of peptides. (A) Growth curves of S. aureus in the presence of bogorol K and LTA. (B) Permeation of bogorol K into the cell membrane of S. aureus. (C) Growth curve of X. campestris in the presence of bogorol K and LPS. (D) Permeation of bogorol K into the cell membrane of X. campestris. (E and F) Permeation of bogorol K and succilin K into the outer membrane (E) and inner membrane (F) of X. campestris. The permeabilization of the outer membrane is indicated by the staining of NPN in the hydrophobic part, while PI staining indicates inner membrane permeabilization. The MIC values of bogorol K and succilin K are around 2.5 μM and 20 μM, respectively. Con, control; BogK, bogorol K; PlyB, polymyxin B; SucK, succilin K.
We also investigated the cause of the difference in antibacterial activities between bogorol K and succilin K using two different dyes, 1-N-phenylnaphthylamine (NPN) and propidium iodide (PI). The hydrophobic dye NPN cannot penetrate the intact outer membrane of Gram-negative bacteria. However, once this membrane is damaged, NPN will bind to the phospholipid layer and become fluorescent. PI binds to DNA and emits fluorescence only when the inner membrane is damaged. Consequently, a combination of both dyes can be used to show the difference between bogorol K and succilin K in the penetration of the different layers of the cell membrane. It was observed that bogorol K can penetrate both the outer membrane and inner membrane of X. campestris when applied at concentrations above 2.5 μM (Fig. 5E and F). In contrast, succilin K barely showed penetration of either the outer membrane or the inner membrane at the same concentration as that of bogorol K (Fig. 5E and F). This result suggests that the outer membrane is the first barrier to succilins and that Orn3 in bogorols plays an important role in penetrating the outer membrane.
Synergistic effect of relacidine and bogorol.
Apart from discovering novel antibiotics, combining existing antibiotics is another strategy to combat the increasing number of drug-resistant bacteria (33). Here, we investigated the synergistic effect of relacidine, a compound that affects oxidative phosphorylation in cells (22), and bogorol, a compound that was proven to cause damage to the cell membranes in this study. Both compounds are produced by B. laterosporus MG64 but employ different modes of action. A study of their synergistic effect could lead to a better understanding of the antibacterial mechanism of B. laterosporus MG64 as well as its potential in different applications. As shown in Table 2, relacidine B and bogorol K displayed a synergistic effect on all the pathogens tested, including the Gram-negative plant pathogens X. campestris, P. syringae pv. antirrhini, and P. syringae pv. tomato and the Gram-positive human pathogen S. aureus subsp. aureus. This result suggests that B. laterosporus MG64 may employ a combination of compounds to combat other bacteria, thus making it better capable of occupying desirable niches in the natural environment.
TABLE 2.
Synergistic effect of relacidine B and bogorol Ka
| Pathogen | MICa (μg/ml) | MICb (μg/ml) | MICab (μg/ml) | MICba (μg/ml) | FICI |
|---|---|---|---|---|---|
| Xanthomonas campestris pv. campestris NCCB92058 | 4 | 0.5 | 0.5 | 0.125 | 0.375 |
| Pseudomonas syringae pv. antirrhini LMG2131 | 16 | 0.5 | 2 | 0.0625 | 0.25 |
| Pseudomonas syringae pv. tomato DC3000 | 16 | 0.5 | 2 | 0.0312 | 0.1875 |
| Staphylococcus aureus subsp. aureus 533 R4 | 2 | 64b | 0.5 | 2 | 0.2812 |
MICa, MIC value of bogorol K alone; MICab, MIC value of bogorol K when relacidine B is present; MICb, MIC value of relacidine B alone; MICba, MIC value of relacidine B when bogorol K is present. An FICI value below 0.5 indicates a synergistic effect of the two peptides.
The MIC is >32 μg/ml; here, we use 64 μg/ml for calculation.
DISCUSSION
In this study, we characterized four novel bogorol variants (bogorols I to L) from the biocontrol strain B. laterosporus MG64. The bogorol variants show a difference of at least one amino acid residue relative to structurally similar peptides (see Table S5 in the supplemental material) (16–20). Leu9 in bogorol B is replaced by Val9 in bogorol I, while Leu2 in brevibacillin is replaced by Val2 in bogorol K. Bogorol J has different amino acids at positions 5 and 9 compared to bogorol C, while bogorol L is different at positions 2 and 3 compared to brevibacillin. Nevertheless, the newly characterized peptides share many characteristic features with the reported ones. First of all, they are linear lipopeptides containing three positively charged amino acid residues (Orn or Lys), which contribute to the hydrophilicity of the peptides. Moreover, the bogorol family peptides have highly conserved fragments, and residues at position 1, positions 6 to 8, and positions 10 to 13 show no difference between variants. Moreover, the C termini of all the peptides contain valinol, the alcohol form of valine.
We also characterized four novel succilins (succilins I to L) from the same strain, B. laterosporus MG64. Succilins are similar to bogorols except that an additional succinyl group was attached to the side chain of Orn3/Lys3. This modification confers different chemical properties to succilins compared to bogorols. First of all, the masses of succilins are 100 Da higher than those of their corresponding bogorols. Moreover, the polarity of the peptides is decreased. As a result, they are eluted later than bogorols and have a lower solubility in water (data not shown). Besides, succilins retain only one positive charge, compared to the three in bogorols. These changes, especially the polarity and charges, result in a negative effect on the bioactivity of the peptides (Table 1). This is considered reasonable because (i) the phospholipid of the cell membrane in bacteria is negatively charged and has an electrostatic attraction to the cationic peptides (34), and a decrease of charges therefore causes weaker penetration into the membrane (Fig. 5E and F), and (ii) the amphipathic property of peptides is important for bioactivity. Hydrophobicity that is too high or too low could dramatically decrease the antimicrobial activity (35). Our study reports the succinylation of the bogorol family of peptides. Similar modified peptides were not detected in B. laterosporus LMG15441 (Fig. S5A), which also produces bogorols. The biological function of succilins remains unknown, but the higher level of production of succilins on agar plates using bogorols as a reference (data not shown) suggests that they are possibly involved in motility or biofilm formation, which needs further investigation.
Although several variants of bogorol have been reported (16–20), their biosynthesis was unclear to date. In this study, we successfully unraveled the biosynthesis process of bogorols and succilins with both in silico analysis and experimental methods (Fig. S7). We reveal that the lipoinitiation of bogorols does not follow the usual pattern observed in other lipopeptides, which is mediated by FACL, coenzyme A, and a starter C domain (26). Instead, the fatty acid of bogorols is incorporated by a special module, A-KR-PCP. Such modules are reported to incorporate α-keto acids and are normally found in depsipeptides, a group of compounds containing both hydroxy acid residues and amino acid residues (29). Modules that incorporate α-keto acid are found in natural products, including cereulide (36, 37), valinomycin (38, 39), cryptophycin (40), kutzneride (41), and hectochlorin (42). However, these modules were located between other modules that incorporate amino acids, which suggests that they are not responsible for lipoinitiation. In contrast, such modules in bogorols are encoded by an independent gene, bogJ. A C domain following the A-KR-PCP module and a TD at the end of the last module encoded by bogEF suggest that bogJ is responsible for lipoinitiation (Fig. S7). To the best of our knowledge, only bogorols and auriporcine employ such a novel lipoinitiation pathway. This expands our knowledge of lipoinitiation in natural products and provides a possible pathway for the engineering of lipid tails in NRPs.
We also reveal the pathway leading to the formation of valinol in bogorols. The aldehyde form of bogorols is released from the PCP domain by the TD and further reduced to the alcohol form by a reductase encoded by the bogI gene (Fig. S7). A similar process in gramicidin A was reported to happen in the presence of a TD, the aldoreductase LgrE, and the electron donor NADPH in vitro (43). A similar reduction in myxochelin A was reported to be mediated by the TD alone (44), which is different from the two-step process found in bogorols and gramicidin A. A previous study on BT peptides revealed that the valinol at the C terminus confers protease resistance to the peptides (20). Our results also reveal that the aldehyde form of bogorols is less active against pathogens than the alcohol form. Therefore, the formation of valinol is of vital importance for the biological activity of these peptides.
Succinylation is commonly found in proteins of diverse organisms, including bacteria, yeasts, and animals (45, 46). Studies have shown that succinylation is possibly relevant to numerous human diseases associated with mitochondria (47). However, reports on the succinylation of secondary metabolites are relatively rare. One of the reports is on the ribosomal peptides subtilin and entianin, which are produced by Bacillus subtilis (48). The succinylation of subtilin and entianin occurs at a tryptophan residue at position 1 and is affected by the concentration of glucose in the growth medium as well as the transition state regulator AbrB (48). Another report is on cerexins E1, F1, and F2, in which succinylation occurs at the ε-amino group of lysine or hydroxylysine (49). In this study, we report succinylation occurring in the bogorol family of peptides. Compared to other modifications, such as halogenation, glycosylation, and sulfation, etc., in NRPs (50), succinylation makes a profound difference in the chemical properties of peptides, and its occurrence likely demands a high concentration of the substrate (Fig. 4). The mechanism of succinylation in succilins remained unknown, and further research will be directed toward revealing the responsible enzymes.
The bogorol family of peptides was initially reported to have potent bioactivity against Gram-positive bacteria and weak bioactivity toward Gram-negative bacteria (16–19). However, we show that bogorol K and bogorol L also have potent activity against Gram-negative bacteria, especially Xanthomonas species. We also show that bogorol K targets the cell membrane of both Gram-positive and Gram-negative bacteria. This result is similar to those of a study on the mode of action of brevibacillin on Gram-positive bacteria (51). The membrane-targeting mechanism makes bogorol a promising candidate for studies on the synergistic effect of antimicrobials. Bogorols damage the cell membranes and remove the barrier to other compounds that target intracellular components. This is supported by the synergistic effect of bogorols and relacidines (Table 2), a class of compounds affecting the oxidative phosphorylation of cells (22). Further research will be directed to further evaluate its potential in different applications.
MATERIALS AND METHODS
Extraction and purification of antimicrobial compounds from B. laterosporus MG64.
A fresh colony of B. laterosporus MG64 was inoculated in 3 ml Lennox broth (LB) and incubated at 28°C overnight with shaking. The culture grown overnight was then diluted with fresh LB to an optical density at 600 nm (OD600) of 1.0 as the inoculum. One milliliter of the inoculum was inoculated into 100 ml of fresh LB and incubated at 28°C with shaking at 220 rpm for 24 h. After centrifugation at 10,000 × g for 10 min, the supernatant was collected and applied to a column filled with 10 g C18 silica gel (catalog no. 97727-U; Sigma-Aldrich). After washing with 20 ml of 20% acetonitrile plus 0.1% trifluoroacetic acid (TFA), the crude extract was eluted with 20 ml 95% acetonitrile plus 0.1% TFA. The crude extract was lyophilized and redissolved in Milli-Q water. After filtering with a 0.45-μm cellulose acetate membrane, an aliquot of the crude extract was applied to a high-performance liquid chromatography (HPLC) system equipped with an analytical C18 column (3.6-μm particles, 250 by 4.6 mm; Phenomenex) for purification. A linear gradient of water with 0.1% TFA (solvent A) and acetonitrile with 0.1% TFA (solvent B) was used to separate compounds. In each run, solvent B was linearly increased from 15% to 60% within 45 min at a flow rate of 0.5 ml/min. The effluent was monitored with a UV detector at a wavelength of 214 nm. Every single peak was collected and concentrated with lyophilization before testing activity against X. campestris pv. campestris and B. cereus.
Structure elucidation.
(i) LC-MS/MS. The antimicrobial compounds were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) to determine the amino acid sequences. An Ultimate 3000 ultrahigh-performance liquid chromatography (UHPLC) system equipped with a Kinetex WVO-C18 column (2.6-μm particles, 100 by 2.1 mm; Phenomenex) was used. A Q-Exactive Orbitrap-based mass spectrometer (Thermo Scientific, San Jose, CA, USA) equipped with a heated-electrospray ionization (HESI-II) electrospray source was coupled to the UHPLC system. Each sample (10 μl) was injected into the LC system and separated with water (with 0.1% formic acid) and acetonitrile (with 0.1% formic acid) at a flow rate of 500 μl min−1. A spray voltage of 3.5 kV (positive mode) and a capillary temperature of 275°C were used. The samples were measured in positive mode from m/z 300 to 2,000 at a resolution of 70,000. MS/MS data were recorded using either a DDA-top10 (data-dependent acquisition with the 10 highest-intensity eligible precursors) setup or targeted MS/MS using parallel reaction monitoring (PRM) mode at a resolution of 17,500.
(ii) NMR. Prior to NMR analysis, the peptides were purified by reverse-phase HPLC (RP-HPLC) as described above, and the pure fractions were pooled and lyophilized. The peptides were obtained as white solids (0.5 to 1 mg) and were dissolved in 0.5 ml deuterated dimethyl sulfoxide (DMSO-d6). 1H NMR, 1H-1H TOCSY NMR, and 1H-1H NOESY NMR spectra were recorded on a Bruker Ascend 600-MHz spectrometer. Chemical shifts in 1H NMR spectra were internally referenced to solvent signals (DMSO-d6 at δ-H = 2.50 ppm).
Identification and analysis of BGCs.
The Web-based tool antiSMASH (52) was used to identify and analyze the BGCs encoding bogorol and auriporcine. The genomic sequences of B. laterosporus MG64 (GenBank accession no. QJJD00000000), B. laterosporus LMG15441 (GenBank accession no. NZ_CP007806.1), and B. laterosporus PE36 (GenBank accession no. NZ_AXBT00000000) were used as inputs, and the gene clusters of bogorol and auriporcine were predicted. The flanking genes of the bogorol BGCs from MG64 (producer of succilin) and LMG15441 (nonproducer of succilin) were compared in order to identify the succinylation gene. The A-KR-PCP domains of auriporcine and bogorol (both compounds have the same fatty acid side chain) from both MG64 and PE36 were compared in order to investigate their lipoinitiation pathway.
Genome editing with the CRISPR-Cas9 system.
To knock out the bogI or bogJ gene in B. laterosporus LMG15441, vector pJOE8999 harboring the clustered regularly interspaced short palindromic repeat (CRISPR)-associated (Cas) system was employed (53). Single guide RNA (sgRNA) fragments were ligated into Eco31I sites of pJOE8999 after annealing. Short fragments located upstream and downstream of the sgRNA were PCR amplified with the primers listed in Table 3. The template for homologous repair was achieved by overlap PCR of the upstream and downstream fragments and ligated into the SfiI sites of pJOE8999 to make the final construct.
TABLE 3.
Primers used in this study
| Primer | Sequencea | Purpose |
|---|---|---|
| bogJ-sgRNA-F | TACGATTAGTAGAAAGTGTTTACG | Knockout of bogJ |
| bogJ-sgRNA-R | AAACCGTAAACACTTTCTACTAAT | Knockout of bogJ |
| bogJ-up-SfiI-F | AAGGCCAACGAGGCCGTAGCTCCGCTTCACAGTAG | Knockout of bogJ |
| bogJ-up-R | GGCGTTCTTCCTTGTCTCCTCCTGCTCTATG | Knockout of bogJ |
| bogJ-down-F | GGAGGAGACAAGGAAGAACGCCCTATTTCTG | Knockout of bogJ |
| bogJ-down-SfiI-R | AAGGCCTTATTGGCCGCTTGTAACGCTTGCAATG | Knockout of bogJ |
| bogI-sgRNA-F | TACGGAAGCCGTCTCAGTTAGACG | Knockout of bogI |
| bogI-sgRNA-R | AAACCGTCTAACTGAGACGGCTTC | Knockout of bogI |
| bogI-up-SfiI-F | AAGGCCAACGAGGCCTATGTAGCGGCTGTAAAGGG | Knockout of bogI |
| bogI-up-R | AGTAGGTGGCCACGGGAGCACTTTAACTCAG | Knockout of bogI |
| bogI-down-F | AGTGCTCCCGTGGCCACCTACTAAGGAAGAG | Knockout of bogI |
| bogI-down-SfiI-R | AAGGCCTTATTGGCCTAACCCGATCTTGAGGCTTG | Knockout of bogI |
| pJOE8999-check-F | CGTGTGATGCGAATTCTTGAC | Confirmation of cloning |
| pJOE8999-check-R | GATGCCACTCTTATCCATCAATCC | Confirmation of cloning |
The restriction sites are indicated in boldface type, while base-pairing regions in the sgRNA primers are indicated by underlining.
To prepare competent cells, a single colony of B. laterosporus LMG15441 was inoculated in LBS medium (LB medium with 0.5 M sorbitol) and incubated at 37°C with shaking (220 rpm) overnight. The culture grown overnight was diluted 100 times into fresh LBS medium and grown until the OD600 reached 0.8 to 0.9. Cells (100 ml) were cooled on ice for 5 min and centrifuged at 5,000 × g at 4°C for 5 min to collect the cells. The pellet was washed four times with electroporation buffer (0.5 M sorbitol, 0.5 M mannitol, and 10% glycerol) and finally resuspended in the same buffer. One microgram of the reconstructed vector was mixed with 100 μl of competent cells in an electroporation cuvette (1 mm) and shocked with a 2.0-kV voltage, 25-μF capacitance, and 200-Ω resistance. One milliliter of recovery medium (LB medium with 0.5 M sorbitol and 0.38 M mannitol) was added immediately, and the cells were incubated at 28°C with shaking at 150 rpm for 3 to 5 h. The cells were plated on LB plates with 30 μg/ml kanamycin and 0.2% mannose for inducing the expression of the Cas9 protein. The plate was incubated at 28°C for 24 h, and colonies were checked by colony PCR.
Inspection of bogorol production in mutants.
The mutants and wild-type strain of B. laterosporus LMG15441 were grown in minimal expression medium (MEM) (54) at 28°C overnight (16 h). The culture grown overnight was diluted with MEM broth to an OD600 of 1.0. Each 20-μl diluted culture was inoculated in 50 ml of MEM broth, and the cells were incubated at 28°C for 48 h. The supernatant was collected by centrifugation at 10,000 × g for 2 min. The compounds were extracted as described above and dissolved in Milli-Q water before filtering through a 0.45-μm cellulose membrane. The bogorol peptides were purified by HPLC as described above and identified by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry.
Production time of bogorols and succilins.
To investigate the production time of bogorols and succilins, the producer was grown in MEM broth overnight. The culture grown overnight was diluted to an OD600 of 1.0. For each 50 ml of MEM broth, 10 μl of the diluted bacterial culture was added. The cells were grown at 28°C for 14 h, 16 h, 18 h, and 20 h. At each time point, the supernatant was collected, and the compounds were extracted as described above. The extracts were lyophilized and dissolved in Milli-Q water before analysis by HPLC and MALDI-TOF mass spectrometry.
Antibacterial assays.
The MICs of the peptides were determined using the broth dilution method (55). Mueller-Hinton II broth (MHB) medium (BD BBL, catalog no. BD 212322) was used for all the indicator bacteria. The concentration of bacteria was adjusted to 5.0 × 105 CFU/ml in each well. The compounds (32 μg/ml) were 2-fold serially diluted in the 96-well plate. The OD600 was measured using the Tecan Infinite F200 Pro luminometer after incubation at 28°C for 36 h, and the MIC values were defined as the lowest concentration that completely inhibited the growth of the indicator bacteria.
Growth curve and LPS/LTA binding assay.
Cultures of X. campestris pv. campestris and Staphylococcus aureus subsp. aureus grown overnight were diluted to an OD600 of 0.05 in a 96-well plate and incubated in a microplate spectrophotometer (1,000 rpm at 28°C). When the OD600 reached 0.1, bogorol K (dissolved in DMSO) was added to each well at a final concentration of 8 μg/ml (2× MIC), and the same volume of DMSO was used as a negative control. Polymyxin B (1 μg/ml) was used as a positive control for X. campestris pv. campestris, while nisin (2 μg/ml) was used as a positive control for S. aureus subsp. aureus. Exogenous lipopolysaccharide (LPS) (0.1 mg/ml) or lipoteichoic acid (LTA) (0.1 mg/ml) was added to investigate the association with peptides. The growth curve in the following 3 h was recorded using the same microplate spectrophotometer under the same conditions, and four replicates were used for each treatment. The release of growing pressure indicates the binding of peptides to LPS or LTA.
Membrane permeability assay.
The membrane integrity of X. campestris pv. campestris and S. aureus subsp. aureus after treatment with bogorol was tested with the commercial Live/Dead BacLight bacterial viability kit (Invitrogen). Briefly, the culture of each strain grown overnight was diluted to an OD600 of 0.2. Bogorol K (dissolved in DMSO) was added to a concentration of 8 μg/ml (2× MIC), and the same volume of DMSO was added to the negative control. Polymyxin B was used as a positive control for X. campestris pv. campestris, while nisin was used as a positive control for S. aureus subsp. aureus. After treatment at room temperature for 15 min, cells were harvested (10,000 × g for 2 min) and washed with a 0.9% saline solution. Cells were finally suspended in 200 μl of a 0.9% saline solution, and two different dyes (3.34 mM SYTO9 and 20 mM propidium iodide) were added at a ratio of 1:1 (vol/vol). After staining in the dark for 15 min, cells were mounted on 1% agarose pads and analyzed with a Nikon Ti-E microscope (Japan).
NPN and PI staining assay.
X. campestris pv. campestris cells were grown in LB broth at 28°C overnight. The culture grown overnight was diluted 100 times with fresh LB broth and grown until the OD600 reached 1.0. The cells were collected (5,000 × g for 3 min) and washed with HEPES buffer twice. The cells were resuspended in HEPES buffer, and the OD600 was adjusted to 1.0. Both NPN and PI were added to the cells at a final concentration of 30 μM. The mixture was distributed into a 96-well plate, in which DMSO, bogorol K, and succilin K were added before. The concentration range of each compound was 1.25 mM, 2.5 mM, 5 mM, and 10 mM. The fluorescence signal at 350/420 nm was recorded every 10 min during a period of 1 h and normalized to the OD600. Three replicates were used in each treatment.
Synergy test.
The synergistic effect of bogorol K and relacidine B was tested using a checkerboard method (56). Briefly, bogorol K was 2-fold serially diluted in each column, while relacidine B was diluted using the same method in each row in a 96-well plate. Fresh cells were added to each well at a final concentration of 5 × 105 CFU/ml. The plate was incubated at 28°C for 24 h, and the OD600 was recorded. The fractional inhibitory concentration index (FICI) was calculated using the formula FICI = MICab/MICa + MICba/MICb, where MICa and MICb are the MIC values of bogorol K and relacidine B alone, respectively, and MICab is the MIC value of relacidine B in the presence of relacidine B, while MICba is the other way around. The FICI value suggests a synergistic effect (≤0.5), an additive effect (>0.5 to 1), no interaction (1 to 4), or an antagonistic effect (>4) of the two peptides (56).
Supplementary Material
ACKNOWLEDGMENTS
We thank Johan Kemmink for the help and discussion about the NMR analysis.
Z.L. and X.Z. are financially supported by the Chinese Scholarship Council (CSC). R.H.D.V. and G.R. acknowledge financial support from the Netherlands Organisation for Scientific Research (NWO) (Vici grant 724.013.003) and the Ministry of Education, Culture, and Science (Gravitation program no. 024.001.035).
Z.L. and O.P.K. designed the study. Z.L. performed most of the experiments and prepared the manuscript. R.H.D.V. performed the NMR analysis. P.C. and X.Z. contributed to the mode-of-action assays. All authors were involved in analyzing and discussing the results and revising the manuscript.
We declare we have no conflict of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Kinsinger RF, Shirk MC, Fall R. 2003. Rapid surface motility in Bacillus subtilis is dependent on extracellular surfactin and potassium ion. J Bacteriol 185:5627–5631. doi: 10.1128/jb.185.18.5627-5631.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kearns DB, Chu F, Rudner R, Losick R. 2004. Genes governing swarming in Bacillus subtilis and evidence for a phase variation mechanism controlling surface motility. Mol Microbiol 52:357–369. doi: 10.1111/j.1365-2958.2004.03996.x. [DOI] [PubMed] [Google Scholar]
- 3.Hamze K, Julkowska D, Autret S, Hinc K, Nagorska K, Sekowska A, Holland IB, Seror SJ. 2009. Identification of genes required for different stages of dendritic swarming in Bacillus subtilis, with a novel role for phrC. Microbiology 155:398–412. doi: 10.1099/mic.0.021477-0. [DOI] [PubMed] [Google Scholar]
- 4.de Bruijn I, de Kock MJ, Yang M, de Waard P, van Beek TA, Raaijmakers JM. 2007. Genome-based discovery, structure prediction and functional analysis of cyclic lipopeptide antibiotics in Pseudomonas species. Mol Microbiol 63:417–428. doi: 10.1111/j.1365-2958.2006.05525.x. [DOI] [PubMed] [Google Scholar]
- 5.de Bruijn I, de Kock MJ, de Waard P, van Beek TA, Raaijmakers JM. 2008. Massetolide A biosynthesis in Pseudomonas fluorescens. J Bacteriol 190:2777–2789. doi: 10.1128/JB.01563-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boles BR, Thoendel M, Singh PK. 2005. Rhamnolipids mediate detachment of Pseudomonas aeruginosa from biofilms. Mol Microbiol 57:1210–1223. doi: 10.1111/j.1365-2958.2005.04743.x. [DOI] [PubMed] [Google Scholar]
- 7.Davey ME, Caiazza NC, O’Toole GA. 2003. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J Bacteriol 185:1027–1036. doi: 10.1128/jb.185.3.1027-1036.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hotta K, Kim CY, Fox DT, Koppisch AT. 2010. Siderophore-mediated iron acquisition in Bacillus anthracis and related strains. Microbiology 156:1918–1925. doi: 10.1099/mic.0.039404-0. [DOI] [PubMed] [Google Scholar]
- 9.Abergel RJ, Wilson MK, Arceneaux JEL, Hoette TM, Strong RK, Byers BR, Raymond KN. 2006. Anthrax pathogen evades the mammalian immune system through stealth siderophore production. Proc Natl Acad Sci U S A 103:18499–18503. doi: 10.1073/pnas.0607055103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ongena M, Jacques P. 2008. Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol 16:115–125. doi: 10.1016/j.tim.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 11.Steenbergen JN, Alder J, Thorne GM, Tally FP. 2005. Daptomycin: a lipopeptide antibiotic for the treatment of serious Gram-positive infections. J Antimicrob Chemother 55:283–288. doi: 10.1093/jac/dkh546. [DOI] [PubMed] [Google Scholar]
- 12.Velkov T, Roberts KD, Nation RL, Thompson PE, Li J. 2013. Pharmacology of polymyxins: new insights into an ‘old’ class of antibiotics. Future Microbiol 8:711–724. doi: 10.2217/fmb.13.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H, Fewer DP, Holm L, Rouhiainen L, Sivonen K. 2014. Atlas of nonribosomal peptide and polyketide biosynthetic pathways reveals common occurrence of nonmodular enzymes. Proc Natl Acad Sci U S A 111:9259–9264. doi: 10.1073/pnas.1401734111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hotchkiss RD, Dubos RJ. 1941. The isolation of bactericidal substances from cultures of Bacillus brevis. J Biol Chem 141:155–162. [Google Scholar]
- 15.Gause GF, Brazhnikova MG. 1944. Gramicidin S and its use in the treatment of infected wounds. Nature 154:703. doi: 10.1038/154703a0. [DOI] [Google Scholar]
- 16.Barsby T, Kelly MT, Gagné SM, Andersen RJ. 2001. Bogorol A produced in culture by a marine Bacillus sp. reveals a novel template for cationic peptide antibiotics. Org Lett 3:437–440. doi: 10.1021/ol006942q. [DOI] [PubMed] [Google Scholar]
- 17.Barsby T, Warabi K, Sorensen D, Zimmerman WT, Kelly MT, Andersen RJ. 2006. The bogorol family of antibiotics: template-based structure elucidation and a new approach to positioning enantiomeric pairs of amino acids. J Org Chem 71:6031–6037. doi: 10.1021/jo060667p. [DOI] [PubMed] [Google Scholar]
- 18.Yang X, Huang E, Yuan C, Zhang L, Yousef AE. 2016. Isolation and structural elucidation of brevibacillin, an antimicrobial lipopeptide from Brevibacillus laterosporus that combats drug-resistant Gram-positive bacteria. Appl Environ Microbiol 82:2763–2772. doi: 10.1128/AEM.00315-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu Y, Zhou L, Lu F, Bie X, Zhao H, Zhang C, Lu Z, Lu Y. 2019. Discovery of a novel antimicrobial lipopeptide, brevibacillin V, from Brevibacillus laterosporus fmb70 and its application on the preservation of skim milk. J Agric Food Chem 67:12452–12460. doi: 10.1021/acs.jafc.9b04113. [DOI] [PubMed] [Google Scholar]
- 20.Wu X, Ballard J, Jiang YW. 2005. Structure and biosynthesis of the BT peptide antibiotic from Brevibacillus texasporus. Appl Environ Microbiol 71:8519–8530. doi: 10.1128/AEM.71.12.8519-8530.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li YX, Zhong Z, Zhang WP, Qian PY. 2018. Discovery of cationic nonribosomal peptides as Gram-negative antibiotics through global genome mining. Nat Commun 9:3273. doi: 10.1038/s41467-018-05781-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Z, Chakraborty P, de Vries RH, Song C, Zhao X, Roelfes G, Scheffers D-J, Kuipers OP. 30 June 2020. Characterization of two relacidines belonging to a novel class of circular lipopeptides that act against Gram-negative bacterial pathogens. Environ Microbiol doi: 10.1111/1462-2920.15145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Z, Song C, Yi Y, Kuipers OP. 2020. Characterization of plant growth-promoting rhizobacteria from perennial ryegrass and genome mining of novel antimicrobial gene clusters. BMC Genomics 21:157. doi: 10.1186/s12864-020-6563-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang X, Yousef AE. 2018. Antimicrobial peptides produced by Brevibacillus spp.: structure, classification and bioactivity: a mini review. World J Microbiol Biotechnol 34:57. doi: 10.1007/s11274-018-2437-4. [DOI] [PubMed] [Google Scholar]
- 25.Marfey P. 1984. Determination of D-amino acids. II. Use of a bifunctional reagent, 1,5-difluoro-2,4-dinitrobenzene. Carlsberg Res Commun 49:591–596. doi: 10.1007/BF02908688. [DOI] [Google Scholar]
- 26.Kraas FI, Helmetag V, Wittmann M, Strieker M, Marahiel MA. 2010. Functional dissection of surfactin synthetase initiation module reveals insights into the mechanism of lipoinitiation. Chem Biol 17:872–880. doi: 10.1016/j.chembiol.2010.06.015. [DOI] [PubMed] [Google Scholar]
- 27.Li YX, Zhong Z, Hou P, Zhang WP, Qian PY. 2018. Resistance to nonribosomal peptide antibiotics mediated by D-stereospecific peptidases. Nat Chem Biol 14:381–387. doi: 10.1038/s41589-018-0009-4. [DOI] [PubMed] [Google Scholar]
- 28.Theodore CM, Stamps BW, King JB, Price LS, Powell DR, Stevenson BS, Cichewicz RH. 2014. Genomic and metabolomic insights into the natural product biosynthetic diversity of a feral-hog-associated Brevibacillus laterosporus strain. PLoS One 9:e90124. doi: 10.1371/journal.pone.0090124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alonzo DA, Chiche-Lapierre C, Tarry MJ, Wang J, Schmeing TM. 2020. Structural basis of keto acid utilization in nonribosomal depsipeptide synthesis. Nat Chem Biol 16:493–496. doi: 10.1038/s41589-020-0481-5. [DOI] [PubMed] [Google Scholar]
- 30.Du L, Lou L. 2010. PKS and NRPS release mechanisms. Nat Prod Rep 27:255–278. doi: 10.1039/b912037h. [DOI] [PubMed] [Google Scholar]
- 31.Chen JS, Wang YX, Shao L, Pan HX, Li JA, Lin HM, Dong XJ, Chen DJ. 2013. Functional identification of the gene encoding the enzyme involved in mannosylation in ramoplanin biosynthesis in Actinoplanes sp. Biotechnol Lett 35:1501–1508. doi: 10.1007/s10529-013-1233-3. [DOI] [PubMed] [Google Scholar]
- 32.Wu MC, Styles MQ, Law BJ, Struck AW, Nunns L, Micklefield J. 2015. Engineered biosynthesis of enduracidin lipoglycopeptide antibiotics using the ramoplanin mannosyltransferase Ram29. Microbiology 161:1338–1347. doi: 10.1099/mic.0.000095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brochado AR, Telzerow A, Bobonis J, Banzhaf M, Mateus A, Selkrig J, Huth E, Bassler S, Zamarreno Beas J, Zietek M, Ng N, Foerster S, Ezraty B, Py B, Barras F, Savitski MM, Bork P, Gottig S, Typas A. 2018. Species-specific activity of antibacterial drug combinations. Nature 559:259–263. doi: 10.1038/s41586-018-0278-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ntwasa M. 2012. Cationic peptide interactions with biological macromolecules, p 139–164. In Abdelmohsen K. (ed), Binding protein. IntechOpen, London, England. [Google Scholar]
- 35.Chen Y, Guarnieri MT, Vasil AI, Vasil ML, Mant CT, Hodges RS. 2007. Role of peptide hydrophobicity in the mechanism of action of alpha-helical antimicrobial peptides. Antimicrob Agents Chemother 51:1398–1406. doi: 10.1128/AAC.00925-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ekman JV, Kruglov A, Andersson MA, Mikkola R, Raulio M, Salkinoja-Salonen MJM. 2012. Cereulide produced by Bacillus cereus increases the fitness of the producer organism in low-potassium environments. Microbiology 158:1106–1116. doi: 10.1099/mic.0.053520-0. [DOI] [PubMed] [Google Scholar]
- 37.Alonzo DA, Magarvey NA, Schmeing TM. 2015. Characterization of cereulide synthetase, a toxin-producing macromolecular machine. PLoS One 10:e0128569. doi: 10.1371/journal.pone.0128569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jaitzig J, Li J, Süssmuth RD, Neubauer P. 2014. Reconstituted biosynthesis of the nonribosomal macrolactone antibiotic valinomycin in Escherichia coli. ACS Synth Biol 3:432–438. doi: 10.1021/sb400082j. [DOI] [PubMed] [Google Scholar]
- 39.Huguenin-Dezot N, Alonzo DA, Heberlig GW, Mahesh M, Nguyen DP, Dornan MH, Boddy CN, Schmeing TM, Chin JW. 2019. Trapping biosynthetic acyl-enzyme intermediates with encoded 2,3-diaminopropionic acid. Nature 565:112–117. doi: 10.1038/s41586-018-0781-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding Y, Rath CM, Bolduc KL, Håkansson K, Sherman DH. 2011. Chemoenzymatic synthesis of cryptophycin anticancer agents by an ester bond-forming non-ribosomal peptide synthetase module. J Am Chem Soc 133:14492–14495. doi: 10.1021/ja204716f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fujimori DG, Hrvatin S, Neumann CS, Strieker M, Marahiel MA, Walsh CT. 2007. Cloning and characterization of the biosynthetic gene cluster for kutznerides. Proc Natl Acad Sci U S A 104:16498–16503. doi: 10.1073/pnas.0708242104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramaswamy AV, Sorrels CM, Gerwick WH. 2007. Cloning and biochemical characterization of the hectochlorin biosynthetic gene cluster from the marine cyanobacterium Lyngbya majuscula. J Nat Prod 70:1977–1986. doi: 10.1021/np0704250. [DOI] [PubMed] [Google Scholar]
- 43.Schracke N, Linne U, Mahlert C, Marahiel MA. 2005. Synthesis of linear gramicidin requires the cooperation of two independent reductases. Biochemistry 44:8507–8513. doi: 10.1021/bi050074t. [DOI] [PubMed] [Google Scholar]
- 44.Gaitatzis N, Kunze B, Müller R. 2001. In vitro reconstitution of the myxochelin biosynthetic machinery of Stigmatella aurantiaca Sg a15: biochemical characterization of a reductive release mechanism from nonribosomal peptide synthetases. Proc Natl Acad Sci U S A 98:11136–11141. doi: 10.1073/pnas.201167098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weinert BT, Scholz C, Wagner SA, Iesmantavicius V, Su D, Daniel JA, Choudhary C. 2013. Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep 4:842–851. doi: 10.1016/j.celrep.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Z, Tan M, Xie Z, Dai L, Chen Y, Zhao Y. 2011. Identification of lysine succinylation as a new post-translational modification. Nat Chem Biol 7:58–63. doi: 10.1038/nchembio.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alleyn M, Breitzig M, Lockey R, Kolliputi N. 2018. The dawn of succinylation: a posttranslational modification. Am J Physiol Cell Physiol 314:C228–C232. doi: 10.1152/ajpcell.00148.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bochmann SM, Spieß T, Kötter P, Entian K-D. 2015. Synthesis and succinylation of subtilin-like lantibiotics are strongly influenced by glucose and transition state regulator AbrB. Appl Environ Microbiol 81:614–622. doi: 10.1128/AEM.02579-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cochrane SA, Surgenor RR, Khey KM, Vederas JC. 2015. Total synthesis and stereochemical assignment of the antimicrobial lipopeptide cerexin A1. Org Lett 17:5428–5431. doi: 10.1021/acs.orglett.5b02779. [DOI] [PubMed] [Google Scholar]
- 50.Winn M, Fyans JK, Zhuo Y, Micklefield J. 2016. Recent advances in engineering nonribosomal peptide assembly lines. Nat Prod Rep 33:317–347. doi: 10.1039/c5np00099h. [DOI] [PubMed] [Google Scholar]
- 51.Yang X, Huang E, Yousef AE. 2017. Brevibacillin, a cationic lipopeptide that binds to lipoteichoic acid and subsequently disrupts cytoplasmic membrane of Staphylococcus aureus. Microbiol Res 195:18–23. doi: 10.1016/j.micres.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 52.Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, Medema MH, Weber T. 2019. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res 47:W81–W87. doi: 10.1093/nar/gkz310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Altenbuchner J. 2016. Editing of the Bacillus subtilis genome by the CRISPR-Cas9 system. Appl Environ Microbiol 82:5421–5427. doi: 10.1128/AEM.01453-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rink R, Kuipers A, de Boef E, Leenhouts KJ, Driessen AJM, Moll GN, Kuipers OP. 2005. Lantibiotic structures as guidelines for the design of peptides that can be modified by lantibiotic enzymes. Biochemistry 44:8873–8882. doi: 10.1021/bi050081h. [DOI] [PubMed] [Google Scholar]
- 55.Wiegand I, Hilpert K, Hancock RE. 2008. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 56.Doern CD. 2014. When does 2 plus 2 equal 5? A review of antimicrobial synergy testing. J Clin Microbiol 52:4124–4128. doi: 10.1128/JCM.01121-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.