Genome-wide and source attribution studies based on Campylobacter species have shown their importance for the understanding of foodborne infections. Although the use of multilocus sequence typing based on 7 genes from C. jejuni is a powerful method to structure populations, when applied to C. coli, results have not clearly demonstrated its robustness. Therefore, we aim to provide more accurate data based on the identification of single-nucleotide polymorphisms. Results from this study reveal an important number of host-segregating SNPs, found in proteins involved in motility, membrane functions, or DNA repair systems. These findings offer new, interesting opportunities for further study of C. coli adaptation to its environment. Additionally, the results demonstrate that poultry is potentially the main reservoir of C. coli in France.
KEYWORDS: Campylobacter coli, SNP, source attribution, genomics, genotyping
ABSTRACT
Campylobacter is among the most common causes of gastroenteritis worldwide. Campylobacter jejuni and Campylobacter coli are the most common species causing human disease. DNA sequence-based methods for strain characterization have focused largely on C. jejuni, responsible for 80 to 90% of infections, meaning that C. coli epidemiology has lagged behind. Here, we have analyzed the genome of 450 C. coli isolates to determine genetic markers that can discriminate isolates sampled from 3 major reservoir hosts (chickens, cattle, and pigs). These markers then were applied to identify the source of infection of 147 C. coli strains from French clinical cases. Using STRUCTURE software, 259 potential host-segregating markers were revealed by probabilistic characterization of single-nucleotide polymorphism (SNP) frequency variation in strain collections from three different hosts. These SNPs were found in 41 genes or intergenic regions, mostly coding for proteins involved in motility and membrane functions. Source attribution of clinical isolates based on the differential presence of these markers confirmed chickens as the most common source of C. coli infection in France.
IMPORTANCE Genome-wide and source attribution studies based on Campylobacter species have shown their importance for the understanding of foodborne infections. Although the use of multilocus sequence typing based on 7 genes from C. jejuni is a powerful method to structure populations, when applied to C. coli, results have not clearly demonstrated its robustness. Therefore, we aim to provide more accurate data based on the identification of single-nucleotide polymorphisms. Results from this study reveal an important number of host-segregating SNPs, found in proteins involved in motility, membrane functions, or DNA repair systems. These findings offer new, interesting opportunities for further study of C. coli adaptation to its environment. Additionally, the results demonstrate that poultry is potentially the main reservoir of C. coli in France.
INTRODUCTION
Campylobacter is the leading cause of bacterial gastroenteritis worldwide (1), with around 800,000 campylobacteriosis cases in the United States (2) and 200,000 in the European Union (3) each year. Demographic, dietary, and surveillance program variations have made it difficult to generalize the understanding of Campylobacter epidemiology to all countries. For example, while there are an estimated 68,000 foodborne infections every year in France (4), the number attributable to Campylobacter is not clearly defined, and there are questions about the relative importance of different Campylobacter species (5–7).
C. jejuni and C. coli are part of the commensal microbiota of many bird and animal species (8). Human infection typically occurs via consumption of contaminated water or meat, especially chicken (9–11), or direct contact with animals (livestock farming). Infection is usually self-limiting with mild symptoms, including abdominal cramps, diarrhea, and fever. However, more severe symptoms, such as bloodstream infections and vascular disease, can occur, particularly at extreme ages, in immunosuppressed, diabetic, or cancer patients, and, in rare cases, postinfectious complications include Guillain-Barré syndrome (12) and irritable bowel syndrome (13). Prolonged or severe campylobacteriosis can require the administration of macrolide (azithromycin) or quinolone (ciprofloxacin) (14, 15) antibiotics, but increasing resistance, particularly among C. coli isolates (16), is reducing treatment options.
C. coli is responsible for an increasing number of infections, accounting for approximately 15% of all campylobacteriosis cases (6). While much research focuses on C. jejuni, accounting for about 85% of cases, there are proportional differences between countries, potentially reflecting variations in diet (17) and host source (18, 19). European studies typically have associated C. coli with pigs and sheep (5, 20, 21). However, intensive agricultural practices in recent decades have dramatically changed the distribution of livestock species on earth, creating opportunities for host transitions (22). This has likely driven changes to the natural host associations of both C. jejuni and C. coli, which are regularly isolated from cattle and chickens (9). This host melting pot has also dramatically affected the evolution of livestock-associated C. coli, leading to the emergence of a dominant disease-causing C. coli lineage, the ST-828 clonal complex (CC-828) (23), which has a mosaic genome, with over 10% of the genes having been acquired from C. jejuni by horizontal gene transfer (24–26). This genome plasticity is particularly of concern for C. coli, which acquires antimicrobial resistance genes more easily than C. jejuni (14, 16).
Genotyping methods, such as multilocus sequence typing (MLST) (27, 28), have improved our understanding of Campylobacter population structure, revealing host-specialist and host-generalist lineages (29). This host association has underpinned the development of methods that quantitatively attribute the source of human infections (9, 11). However, rapid host switching by host generalist Campylobacter, including C. coli CC-828, often can confound these methods, because, for some lineages, strains associated with one host source can be found in another (22, 30). The adoption of whole-genome sequencing techniques and the availability of curated genome databases (31) have allowed the incorporation of a broader number of host-segregating epidemiological markers in source attribution methods (32, 33). This additional genome information has increased the resolution, allowing the attribution of invasive/noninvasive strains from poultry (34) as well as geographical attribution of UK/U.S. isolates (19). However, almost all studies focused exclusively on C. jejuni (35), and no study aimed to specifically identify host-segregating markers in C. coli genomes.
In this study, we analyzed 450 C. coli genomes from public databases with defined sampling sources, including chickens, cattle, and pigs. Using comparative genomics approaches, we (i) tested the ability of traditional MLST-based methods to determine the source of C. coli with isolates from known source reservoirs; (ii) identified host-segregating SNPs in C. coli genomes; and (iii) determined the relative contribution of different C. coli infection sources in France. MLST was found to be a good proxy for more complex whole-genome SNP-based analysis, showing similar power for segregating isolates from the cattle host. However, additional discrimination of isolates from chicken and pig hosts was achieved by identifying genome-wide host-segregating SNPs. In the final probabilistic model, using 259 host-segregating SNPs, chicken was found to be the most common source of C. coli infection in France.
RESULTS
CC-828 isolates segregate by host.
From all 3 data sets, Data Sets S1, S2, and S3 in the supplemental material (see also Materials and Methods), nearly all isolates belonged to clonal complex 828 (CC-828; 780 isolates out of 900). The second most common clonal complex identified was ST-1150 (26), with four isolates, sampled from chicken. From the allelic profile minimum spanning tree, 3 clusters can be identified corresponding to the source of isolation (Fig. 1). Cattle isolates clustered together, with 162 isolates (64.8% of all cattle isolates) assigned to ST-1068 (36). Chicken and pig isolates belonged to 78 and 83 sequence types, respectively (contrary to cattle, with 27 different sequence types), with 24.2% of isolates belonging to ST-828, ST-829, ST-825, ST-854, and ST-1119. Furthermore, 40.1% of all clinical isolates belonged to ST-825, ST-827, ST-832, and ST-860. Initial evidence for a role of chicken as a reservoir for human infection was provided by the clustering of clinical isolates together with isolates from chicken on the phylogenetic tree. The second tree, constructed using the maximum-likelihood approach from concatenated SNP sequences, revealed distinctive partitioning of isolates according to source (Fig. 2). C. coli strains isolated from cattle constitute a very distinct cluster; 168 isolates (67.2% of all cattle isolates) are located at the bottom of the tree and belonged to ST-1068. Distances were also shorter within cattle populations than chicken and pig isolates, where more variability was observed in both clades. While many clinical isolates clustered among chicken isolates, six clinical isolates were found along a long branch of the chicken’s clade; interestingly, these isolates were attributed to pig using STRUCTURE (described below).
FIG 1.
Phylogenic tree based on MLST analysis. The minimum spanning tree was generated using GrapeTree from the sequence types of all 896 C. coli isolates, based on 7 MLST genes (aspA, glnA, gltA, glyA, pgm, tkt, and uncA) extracted using the PubMLST platform. Orange represents isolates isolated from chickens, green from cattle, and magenta from pigs. Red is for clinical isolates. Circle sizes are proportional to the number of isolates, and the scale bar represents a genetic distance of 1.
FIG 2.

Phylogenic tree built from concatenated selected SNPs. The tree was designed using maximum-likelihood phylogeny between 896 isolate sequences built from the concatenation of all genotypes of the selected SNPs (n = 259). Orange nodes are the chicken population isolates, green nodes are cattle isolates, pink nodes are pig isolates, and red nodes are clinical isolates. The orange circle shows an estimation of the chicken cluster, the green circle shows the cattle cluster, and the pink circle shows the pig cluster. The scale bar represents a genetic distance of 0.24. Clinical isolates are located mostly within the chicken cluster, which is consistent with the probabilistic attribution model.
Host-segregating SNPs differentiate C. coli strains isolated from different hosts.
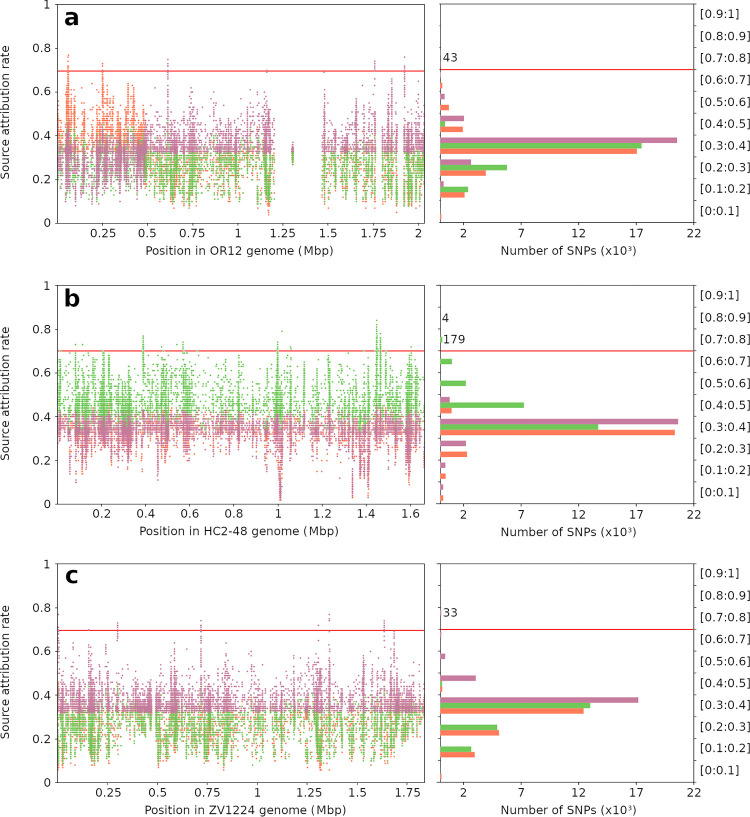
Putative host-segregating SNPs were identified by aligning all 450 isolates selected for marker determination against three C. coli reference genomes. The alignment of isolates against the OR12 C. coli reference strain identified 283,320 variant sites. To remove weakly discriminating polymorphisms, SNP versions represented in more than two-thirds of all isolates were filtered, leaving 26,131 variant sites. Similar alignment and filtering performed against the HC2-48 strain resulted in 202,111 variants, filtered to 24,395, and alignment against the ZV1224 reference identified 242,574 SNPs, which were filtered to 20,827. Host-segregating SNPs were identified by performing source attribution tests using each variant individually and all 450 isolates. SNPs with at least 70% accuracy for at least one source in the self-attribution test included 43, 183, and 33 from each alignment with the OR12, HC2-48, and ZV1224 reference strains, respectively (Table 1). Most of the self-attribution tests showed rates fluctuating between 30% and 40% (51.2%, 50.5%, and 48% of all variants for the chicken, cattle, and pig variants, respectively); 33% indicates a complete inability to differentiate 3 individuals (Fig. 3). In total, 259 host-segregating SNPs from 41 nucleotide sequences were carried forward for further analyses.
TABLE 1.
Variant-calling comparison between three references of Campylobacter coli
| Reference | Variant callinga (raw) | Filtrationb | Selected SNPsc |
|---|---|---|---|
| OR12 (chicken) | 283,320 | 26,131 | 43 |
| HC2-48 (cattle) | 202,111 | 24,395 | 183 |
| ZV1224 (pig) | 242,574 | 20,827 | 33 |
Number of SNPs determined after aligning all isolates from the marker determination (n = 450) data set to 3 different C. coli references: OR12 isolated from chicken, HC2-48 from cattle, and ZV1224 from pig.
Number of SNPs after the filtration of genotypes that represent more than two-thirds of all isolates.
Selected SNPs with 70% or greater total correct self-attributions.
FIG 3.
Host-segregating rate of all variants obtained from the alignment of 450 marker determination isolates against 3 references. Source attribution rates (y axes) were obtained by testing 26,131, 24,395, and 20,827 SNPs from OR12 (a), HC2-48 (b), and ZV1224 (c) references, respectively, and are shown here according to their genome position (left, x axis) and variant proportions (right, x axis). STRUCTURE software was run 3 times for each SNP (average attribution rates are shown here), using 390 randomly selected C. coli isolates as the training data set and 60 randomly selected isolates as the test data set. Orange represents attribution rates and the number of SNPs for chicken source, green for cattle source, and magenta for pig source. A total of 259 SNPs showed attribution rates greater than 70% (red line) for one or more sources and were carried forward for further analyses: 43, 183, and 33 SNPs from chicken, cattle, and pig references, respectively. Scores fluctuated between 30% and 40%, and the highest attribution rates for each host reservoir were found in the corresponding source reference. However, the OR12 reference showed two distinct regions of the genome, one part containing variants discriminating the chicken source and another part the pig source. Two low-variability regions (blanks), where no SNPs from the variant calling step were selected, are also visible.
To contextualize host-segregating SNPs within genes, BLAST-x annotation identified 32 coding regions for known proteins, 5 hypothetical proteins, and 4 intergenic regions (Table S1). Several SNPs (n = 27) were found in proteins involved in motility, which plays an important role in bacterial host adaptation: 12 and 4 SNPs in flagellar proteins FliK (with 2 SNPs in its basal body rod modification protein FlgD) and FliD, respectively, known to modulate flagellar hook length (37) and to act as immunodominant proteins (38); 5 SNPs from methyl-accepting chemotaxis proteins (TLP-like protein [39]) or intergenic regions before methyl-accepting chemotaxis proteins; and 4 SNPs in one aerotaxis receptor belonging to CetC, a protein involved in regulating energy taxis (40). Another protein involved in bacterial adaptation to its environment has also been identified from the OR12 chicken reference (3 SNPs), SbmA (41), a peptide antibiotic transporter described in many Gram-negative bacteria. SNPs were also found in proteins involved in metabolism and membrane functions: 3 SNPs from a histidine kinase, 5 SNPs from a single-domain globin protein, known to play a role against NO and nitrosative stress (42), and a LamB/YcsF family protein with 5 SNPs. Two phosphate-binding proteins showed the presence of one SNP from the OR12 chicken reference variant calling as well as one SNP from the ZV1224 pig reference. Proteins involved in DNA activities have also been identified, with a total of 56 SNPs: DNA recombination/repair protein RecA, excinuclease ABC subunit C (UvrC) (43), two restriction endonucleases from HC2-48 and ZV1224 references, and one transcriptional regulator. Two hypothetical proteins from OR12 and ZV1224 with 11 and 8 host-segregating SNPs, respectively, have been found to be the same protein; its domains and amino acid sequence, depending on the source, should be further investigated. Finally, a total of 110 SNPs were within 2 hypothetical proteins (from the HC2-48 cattle reference), which reflected highly variable and isolate-specific regions and should not be taken into account.
Genome-wide host-segregating SNPs provide more accurate source attribution than MLST alleles.
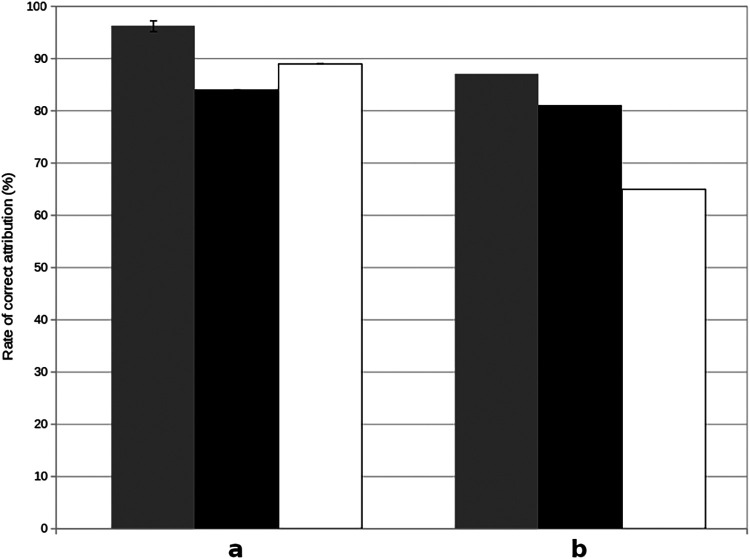
The degree of SNP segregation among isolates from different hosts and, hence, the potential as a marker for source attribution using STRUCTURE, was quantified. Self-attributions of chicken and pig isolates within the marker determination data set were consistently correct (Table 2). Using 43 SNPs detected from OR12 alignment as host-segregating markers allowed an average (± standard deviation [SD]) correct self-attribution of 88.35% (±6.2%), 63.75% (±9.2%), and 96.2% (±4.1%) for chickens, cattle, and pigs, respectively. Using 183 SNPs from the HC2-48 alignment, correct self-attribution was achieved for chicken, cattle, and pig isolates with 91.05% (±5.7%), 75% (±9.7%), and 42.45% (±18.7%) accuracy, respectively, and 74.95% (±13.9%), 19.65% (±10.1%), and 94.65% (±5.2%) for the 33 SNPs from ZV1224 alignment. A low self-attribution rate of cattle isolates using SNPs from the pig reference was observed. These isolates were not correctly attributed and were considered 50% chicken and 50% pig. When using all the SNPs simultaneously (n = 259), correct self-attribution showed average scores of 91.95% (±5.86%), 77% (±8.65%), and 95.25% (±4.4%) for chickens, cattle, and pigs, respectively. This is a considerable improvement of self-attribution using the 7 MLST genes, which returned average scores of 73.6% (±9.1%), 76.8% (±9.4%), and 74.4% (±9.5%) for chickens, cattle, and pigs, respectively. Source attribution of cattle C. coli isolates of the marker determination data set was similar between the two types of markers (genotype or allele), whereas SNPs performed significantly better for chicken and pig populations than the 7 MLST genes. Finally, the discriminatory power of host-segregating SNPs and MLST genes was evaluated by performing source reattribution of 299 C. coli isolates from the validation data set. SNPs showed correct reattribution proportions of 96.2% (±1.03%), 84% (±0%), and 89% (±0%) and MLST gene scores of 87% (±0%), 81% (±0%), and 65% (±0%) for chicken, cattle, and pig populations, respectively (Fig. 4). Overall, SNPs were able to better reattribute C. coli marker determination and validation isolates to their source than MLST genes, especially for chicken and pig populations.
TABLE 2.
Rates of correct self-attributions of marker determination isolates using 5 different sets of markersa
| Set of markers | Value (%) for self-attributed isolates of: |
|||||
|---|---|---|---|---|---|---|
| Chicken (n = 150) |
Cattle (n = 150) |
Pig (n = 150) |
||||
| Rate of correct attribution | SD | Rate of correct attribution | SD | Rate of correct attribution | SD | |
| 43 SNPs (OR12) | 88.4 | ±6.24 | 63.8 | ±9.22 | 96.2 | ±4.09 |
| 183 SNPs (HC2-48) | 91.1 | ±5.74 | 75.0 | ±9.69 | 42.5 | ±18.74 |
| 33 SNPs (ZV1224) | 75.0 | ±13.9 | 19.7 | ±10.08 | 94.7 | ±5.23 |
| 259 SNPs (all) | 92.0 | ±5.86 | 77.0 | ±8.65 | 95.3 | ±4.4 |
| 7 genes (MLST) | 73.6 | ±9.06 | 76.8 | ±9.42 | 74.4 | ±9.50 |
Discriminating strengths of selected SNPs and MLST genes were estimated using marker determination isolates. From 450 initial isolates, random selections of 390 and 60 isolates were used for training and self-attribution (sources set to “unknown”), respectively. Self-attributions were performed 100 times using selected SNPs from chicken alignment (n = 43), cattle alignment (n = 183), pig alignment (n = 33), and all alignments (n = 259) and 50 times using MLST genes (n = 7). Since multiple tests were performed for each set of markers using 60 randomly selected isolates, standard deviations were calculated.
FIG 4.
Correct reattribution proportions of 299 validation isolates using determined SNPs and MLST genes. (a and b) Source attribution strength of selected SNPs (a) and MLST genes (b) estimated using STRUCTURE software. A total of 299 isolates were tested (from the validation data set) using marker determination isolates for training (n = 450). Source attributions were performed 10 times using all selected SNPs (n = 259) and MLST genes (n = 7). Gray bars represent the rate of correct source attribution for chicken population isolates, black bars for cattle isolates, and white bars for pig isolates. An isolate was considered correctly source reattributed with a STRUCTURE score greater than 70%.
Chickens are a major source of C. coli infection in France.
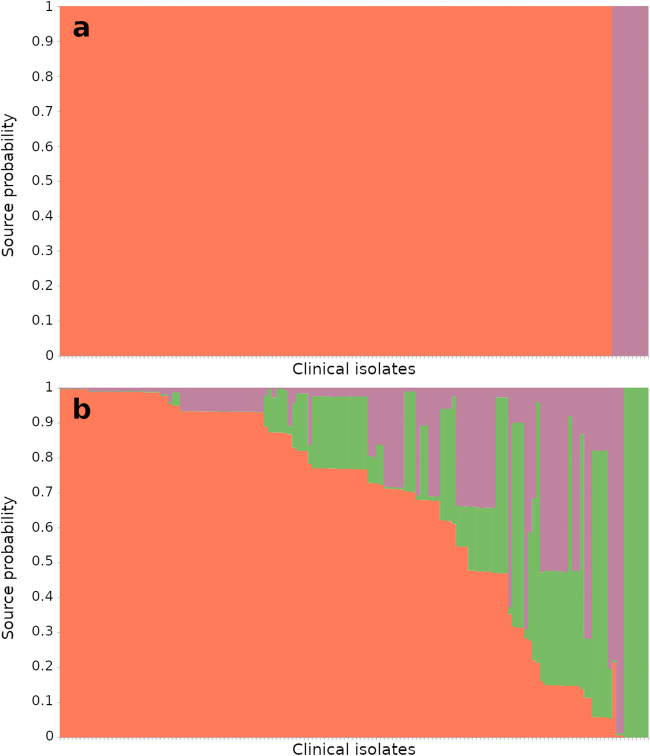
Source attribution of clinical isolates was performed using MLST alleles and all host-segregating SNPs with correct self-attribution >70% (n = 259) in the marker determination and training data set using STRUCTURE (Fig. 5). Using MLST genes, 89 clinical isolates (60.5%) were attributed to chickens, 13 to cattle (9%), and 6 to pigs (4%), and 39 clinical isolates (26.5%) showed attribution scores lower than 70% and, therefore, were considered inconclusive attributions. Inconclusive attributions specifically concern 3 commonly found sequence types, ST-827, ST-1055, and ST-1595, representing 48.7% of inconclusive attributions (n = 19). In contrast, using the 259 SNPs, 138 isolates (94%) were attributed to chickens, 9 to pigs (6%) (with an average source probability equal to 100%), and none to the cattle population. Therefore, whatever the approach (MLST or SNPs), a large proportion of C. coli clinical isolates were attributed to chickens. However, the attribution scores were more variable with MLST (on average, around 80%), whereas for the genome-wide host-segregating SNPs, the clinical isolates were more efficiently attributed to their infection source (Table 3).
FIG 5.
Population proportions of clinical isolates from source attribution. (a and b) Source attribution of the clinical data set using selected SNPs (n = 259) (a) and MLST genes (n = 7) (b). Clinical isolates (n = 147) are represented on the x axis and their attribution probabilities on the y axis in orange for chicken source, green for cattle source, and pink for pig source. The poultry reservoir was estimated as the main source of C. coli contamination in France, with 138 isolates (94%) attributed using host-segregating SNPs and 89 isolates (61%) using MLST (isolates selected with source probabilities of greater than 70%).
TABLE 3.
Source attribution scores of French clinical isolatesa
| Set of markers | % (avg score) of clinical isolates from: |
|||
|---|---|---|---|---|
| Chicken | Cattle | Pig | Inconclusive | |
| 259 SNPs | 93.88 (100.0) | 0.0 (0.0) | 6.12 (100.0) | 0.0 (0.0) |
| 7 MLST genes | 60.54 (88.35) | 8.84 (86.91) | 4.08 (83.22) | 26.53 (50.59) |
Data for source attribution of French clinical isolate data set using selected SNPs (n = 259) and MLST genes (n = 7). Shown are the distributions of estimated sources among clinical isolates, with average score as the average individual attribution rate. Using determined SNPs, source attribution rates for clinical isolates were constant, whereas using MLST genes, source attribution showed variable results.
DISCUSSION
The increasing availability of bacterial isolate genome collections and bioinformatics tools for large-scale analysis provides significant opportunities for understanding the genetic basis of phenotype variation in bacteria. Host adaptation is a key feature in the epidemiology of zoonotic pathogens (44), such as Campylobacter, and there has been considerable effort to identify host-associated genetic variation that can improve our understanding of the evolution and origin of infecting strains. Comparative genomic analyses have revealed core and accessory genome variation within C. jejuni that is associated with a given host/environment (45, 46), and this has been used to identify genome-wide host-segregating markers for source attribution (32). However, little comparable work has focused on C. coli.
Genetic variation in bacterial genomes reflects not only adaptation to different hosts/sources but also temporal and geographic variation among sample collections (19). Some studies avoid the potential confounding effect of phylogeographic variation by using national isolate collections, for example, Campylobacter attribution studies performed in Scotland (24, 47), Switzerland (48), New Zealand (49), and Germany (17). This has been informative for understanding the source of human infection; however, because of the strong segregation of genetic variation by host (18), it remains possible that collections from multiple countries could be combined to create international isolate collections. This would consolidate research effort and provide the large genome collections necessary for probabilistic attribution models and potential to identify universal host-segregating markers.
Here, we analyzed C. coli isolates from Europe and the United States using the conventional MLST method established by Dingle et al. in 2001 (27) and specific host-segregating SNPs. A single clonal complex (CC-828) dominated among the isolates independently of source and geographical location, representing 780 isolates out of 900. The predominance of CC-828 isolates in C. coli (66% to 81% of all isolates [17, 24, 36]), with the ST-1150 complex accounting for most of the remaining isolates (26), confounds efforts to identify host association at the clonal complex level that is possible for C. jejuni (18). However, within CC-828 there was evidence for sequence types that were more commonly isolated from particular hosts. For example, ST-829, ST-832, ST-825, and ST-860 predominated among chicken isolates, ST-827 was more common in pigs, and ST-1068 was nearly exclusive to cattle, consistent with previous studies (36, 50). Similar low diversity in cattle C. coli isolates was previously described among ruminant isolates from Scotland (47). A weaker host association signal, based upon MLST alleles, compared to that of C. jejuni has made it difficult to distinctively partition C. coli by source (49). However, genotype segregation in C. coli provided initial evidence that the genomes of these isolates would contain host-segregating genetic signatures.
Estimating the discriminating power of genetic markers can be performed by determining the probability that a given genetic element, such as a single mutation, will be found among isolates from a given host (self-attribution). As in previous studies (32, 33), we used STRUCTURE software and self-attribution to determine the predictive power of putative host-segregating markers. Moreover, a recent review (35) mentioned that MLST genes were used for self-attribution tests in 6 studies for both C. coli and C. jejuni (11, 24, 32, 33, 48, 51). However, correct attribution rates for C. coli showed inconsistent results for chickens (63 to 95%), cattle (26 to 89%), and pigs (70 to 94%), suggesting that an SNP-based approach is advantageous for source attribution of C. coli. In fact, we showed here that SNPs as host-segregating markers provided more accurate results for chickens, cattle, and pigs, with 92% (±5.9%), 77% (±8.7%), and 95.3% (±4.4%) correct attribution rates, respectively. While the difficulty in precise self-attribution using MLST genes is undoubtedly linked to reduced resolution, as CC-828 isolates dominate among C. coli populations (23), the transmission of C. coli between different host species would also reduce the discriminatory power of source-specific markers, potentially leading to incorrect source attribution (22). Thus, adjusting for single mutation determination provided promising candidates for accurate source attribution of human C. coli isolates. Of 669,019 SNPs from the alignment of 450 genomes against 3 references, 259 SNPs in genes associated with cell membrane (transporters and binding proteins), chemotaxis (FliK, FliD, and TLP-like protein), DNA activities (RecA, UvrC), or energy (CetC) functions were chosen for attributing 147 clinical C. coli isolates to sources.
It is known that poultry are a major reservoir for human C. jejuni infection (8), with a ratio of 9:1 for C. jejuni and C. coli, respectively (36). Previous studies focusing on the source of C. coli infection have come to contrasting conclusions. In France, Sweden, the United Kingdom, and the United States, the high prevalence of C. coli in pigs led to the assumption of the role of this reservoir in human infection (5, 20, 21), up to a ratio of 9:1 in favor of C. coli (36). However, in New Zealand, where human C. coli infection is also common, there is a low prevalence in pigs (49). Estimates of the relative contribution of different host sources to human infection varies among studies (11, 17, 24, 47–49, 52), with attribution to poultry (38 to 86%), ruminants (0 to 55%), and pigs (1 to 32%) all being implicated. With the exception of two studies, examining rural populations in Scotland and New Zealand, that largely attribute human C. coli infections to sheep (47) and ruminants (49), source attribution studies typically assign a principal role for poultry in human infection.
It is likely that there are differences in the major reservoirs of C. coli infection in different countries, but quantifying this requires accurate estimation. Estimates based on MLST loci provided source probabilities with some uncertainty. Specifically, although approximately 40% of the 147 French clinical isolates sampled in this study were clearly attributed (>90% probability) (Fig. 5), the remaining isolates showed variable scores, with many attributed <60% probability. Overall, MLST allele-based analyses did assign chicken as a major reservoir for C. coli, with 89 isolates (61%) attributed with a score equal to or greater than 70%. However, this proportion was greatly increased with more accurate attribution scores when using host-segregating SNPs in the attribution model. Specifically, chicken was predicted to be the source of C. coli infection for 138 isolates, constituting 94% of the clinical samples. In comparison, two recent studies showed that sources of infection of C. jejuni are more evenly shared between chicken and cattle populations in France, with approximately 50% for chicken and 40% for cattle, respectively (33, 34). To draw source attribution comparisons between North America and France, additional analyses were performed using 265 clinical isolates exclusively from the United States (see Fig. S2 and Table S2 in the supplemental material). The chicken source again was estimated as the main source of C. coli contamination in the United States, as well as in France, but in a lower proportion (67.9% against 94%), followed by cattle (11.7%) and pig (20.4%). It would be interesting, in a complementary study, to compare the eating habits of animals in these two countries.
In conclusion, the added resolution provided by genome-wide host-segregating markers not only improves source attribution for C. coli but also provides important information about the major infection reservoirs that had been missed in some previous studies (21). By combining whole-genome analysis with national surveillance programs and source attribution modeling, it was possible to identify the chicken reservoir as a major source of C. coli infection in France and abroad. These findings will support ongoing surveillance and the development of targeted interventions aimed at reducing the burden of human campylobacteriosis.
MATERIALS AND METHODS
Campylobacter coli isolate data sets.
A total of 450 C. coli isolate genomes from two major regions where campylobacters are a leading cause of foodborne infections, North America and Europe, were selected for the determination of host-segregating markers (see Data Set S1 in the supplemental material). To reduce the detection of region-specific markers, these genomes were randomly selected from multiple countries within these two regions. This included even numbers (n = 150) of chicken, cattle, and pig C. coli genomes to avoid bias in the identification of host-specific markers. This first data set was comprised of 151 isolates from PubMLST databases (31) and 299 isolates from the U.S. National Antimicrobial Resistance Monitoring System (NARMS) project (53). PubMLST genomes comprised 34% of that first data set and included 47%, 7%, and 47% of all chicken, cattle, and pig marker determination isolates, respectively. NARMS genomes comprised 66% of the data set and included 53%, 93%, and 53% of all chicken, cattle, and pig marker determination isolates. These data sets were entirely composed of European and North American genomes. European isolates represented 29% of the data set, including 41%, 1%, and 45% of all chicken, cattle, and pig isolates, respectively, while North American isolates comprised 71% of the data set, including 59%, 99%, and 55% of all chicken, cattle, and pigs isolates. North American isolates were mostly selected from the United States (n = 315). The remaining isolates (n = 4) were selected from Canada. A total of 424 isolates (94%) were obtained from 2005 to 2019.
A second data set (validation data set), comprised of 300 supplementary C. coli isolates of known source reservoirs, was used to test the discriminatory strength of the host-segregating SNPs previously obtained (Data Set S2). This data set comprised North American C. coli isolates from the NARMS project, 100 for each source. Finally, 150 French clinical isolates comprised a last set of genomes (clinical data set) and were used to attribute the putative source reservoir of clinical isolates (Data Set S3). This comprised 150 clinical isolates from French laboratories and a hospital surveillance network, sampled from stools between 2015 and 2017. Clinical isolates were chosen to represent patients from diverse geographic regions in France, with a sex ratio of 1.03 and a mean age of 39.4 ± 2.8 years.
Clinical isolate genomes had an average genome length of 1.7 Mbp (±69.7 kbp) and an average number of contigs of 43. C. coli marker determination isolates were, on average, 1.76 Mbp (±81.2 kbp) in length and comprised 83 contigs, and C. coli validation isolates were, on average, 1.78 Mbp (±74.7 kbp) in length over 78 contigs (Fig. S1). This is consistent with other published C. coli genomes, estimated to ∼1.7 Mbp in length (54). Furthermore, no significant difference in C. coli genome sizes from different hosts has been observed. C. coli strains isolated from chickens were, on average, 1.78 Mbp in length (±106 kbp), 1.77 Mbp (±61.6 kbp) for cattle isolates and 1.77 Mbp (±61 kbp) for pig isolates.
DNA extraction, genome sequencing, and assembly.
DNA from clinical isolates was extracted using the MagNA Pure 6 DNA and viral NA SV kit, and DNA purification was performed from bacterial lysis on a MagNA Pure 96 system (Roche Applied Science, Manheim, Germany). Quantification and purity checks (260/280 and 260/230 ratios) were determined by spectrophotometry (NanoDrop Technologies, Wilmington, DE) before sequencing. Paired-end next-generation sequencing was performed on DNA samples using Illumina HiSeq 4000 technology (Integragen, Evry, France). Additionally, FastQC v0.11.8 (55) was used to run data quality tests. Genomic data were cleaned and genomes were assembled using Sickle v1.33 (56) and SPAdes v3.10.1 (57), respectively. Genomes then were filtered to remove poor-quality contigs: sequences with a length smaller than 160 nucleotides and a k-mer coverage of less than 20× were removed. One isolate (2015_0475) showed an abnormal genome size of 2.5 Mbp after filtration and was excluded from subsequent analyses.
Characterization of genomic variation.
In silico, MLST was performed for a comparative analysis with host-segregating SNPs. Profiles were obtained for all 900 isolates using 7 housekeeping genes (aspA, glnA, gltA, glyA, pgm, tkt, and uncA) determined for Campylobacter species (27). Sequence types (STs) and clonal complexes (CC; groups of isolates with a sequence type that share four or more loci [27]) were defined using the sequence tag tool of PubMLST (58). Using this method, two clinical isolates (2016_1990 and 2017_2288) and one validation isolate (FSIS11705596) were misidentified as C. coli and were actually C. jejuni and, thus, were removed from the data set. The updated validation and clinical data sets then were comprised of 299 and 147 isolates, respectively. A phylogenetic tree was constructed according to all sequence types using GrapeTree (59). A second tree was built based on every host-segregating marker determined in this study to make a direct comparison with the MLST tree. A multi-fasta file containing sequences from concatenated SNPs of all isolates (n = 896) was created. Sequences were aligned using Muscle v3.8.1551 (60), and a Newick format tree from the maximum-likelihood method was generated using Fasttree v2.1.11 (61). The Microreact online platform was used to visualize the tree (62).
To identify candidate SNPs, genome-wide variant calling was performed primarily by aligning all isolates from the marker determination data set (n = 450) to C. coli reference genomes. Three references from each source were chosen to target source-specific genomic regions and capture all potential markers, including the OR12 strain isolated from a chicken (NZ_CP019977.1) (63), HC2-48 strain isolated from a cow (NZ_CP013034.1) (64), and ZV1224 strain isolated from a pig (NZ_CP017875.1) (65). The bwa v0.7.17 (66) tool, developed for mapping sequences against given genomes, was used here to align each isolate to OR12, HC2-48, and ZV1224 references. Alignment files were sorted using SAMtools v1.9 (67). Genotypes were determined with bcftools v1.9 “mpileup” variant-calling tool (67), and 3 variant-calling files (vcf) were generated (one for each reference). A script was written in Python (see “Data availability,” below) to filter all SNP variations found in more than 2 out of 3 isolates. Since a source represents 33% of the total data set (150 isolates out of 450), a proportion greater than 66% means that the same SNP variation is likely to be found in each of the 3 selected sources. Therefore, this step enabled the removal of weakly discriminating polymorphisms and reduced the computational time of subsequent analyses.
Identification of host-segregating markers.
To identify host-segregating markers, source attribution tests of marker determination isolates (of known sources) were performed using all previously selected SNPs individually to identify host-segregating markers. A matrix was constructed of all genotypes in the 450-marker determination isolate data set (nucleotides were translated into numbers: 1 for A, 2 for T, etc.). Source attribution tests were performed in triplicate for each SNP using STRUCTURE (68), with the no admixture model, 3 putative populations (K = 3), 10,000 iterations, and a burn-in period of 10,000 iterations. For each STRUCTURE test, 60 different random isolates (20 from each population) were set to “unknown source” (POPFLAG = 0) to estimate the probability of correct self-attribution and then to evaluate the SNP host-segregating strength. Each SNP with 70% or more total correct self-attributions for at least one source was selected; a minimum source attribution rate of 66% (here rounded up to 70%) indicates that a variant is discriminating between at least 1 out of 3 sources. Additionally, genomic sequences containing the selected SNPs were extracted from the corresponding reference (OR12, HC2-48, or ZV1224) and annotated using the blast-x online tool (69).
Validation of the discriminatory power of host-segregating markers.
To validate the capability of the selected SNPs to discriminate isolates from different populations, STRUCTURE tests were run again using the marker determination data set and different sets of markers: SNPs contained in the same coding DNA sequence, all SNPs determined from OR12, HC2-48, and ZV1224 alignments, and all SNPs from all alignments. One hundred tests were then performed using each set of SNPs and 60 random isolates per test for self-attribution (POPFLAG = 0) (“no admixture model,” K = 3, 10,000 iterations, and a burn-in period of 10,000 iterations). Additionally, source attribution of 299 validation isolates of known source reservoirs, which were not used for SNP determination, was performed. Specifically, each SNP was obtained using the SAMtools mpileup option. STRUCTURE was run 10 times using marker determination isolates as the training data set (n = 450) and validation data set as unknown source isolates (POPFLAG = 0). STRUCTURE model parameters remained unchanged. Each validation isolate was attributed to its source based on the average attribution rate of all 10 tests. An isolate was considered correctly source reattributed with a STRUCTURE score greater than 70%. In each case, the same method was performed simultaneously with MLST alleles to compare the discriminating strength of both types of marker (SNP or allele).
Source attribution of clinical isolates.
Similar to validation analysis, source attribution of C. coli clinical isolates was performed using determined host-segregating markers to identify the main source of infection in France. For each SNP (n = 259), every genotype was extracted from all clinical isolates using the SAMtools mpileup option. STRUCTURE was run 10 times using marker determination isolates as the training data set (n = 450) and clinical data set (n = 147) as unknown source isolates (POPFLAG = 0) (K = 3, 10,000 iterations, and a burn-in period of 10,000 iterations). Each clinical isolate was attributed to a source based on the average attribution rate of all 10 tests. Source attribution of clinical isolates was performed simultaneously with MLST alleles to compare proportions of each source between both types of markers (SNP or allele).
Data availability.
All 900 C. coli genomes are available using identifiers (IDs) listed in Data Sets S1, S2, and S3, which provide BioSample and PubMLST IDs for NCBI and PubMLST databases, respectively.
The personal vcf filter Python script is available on GitHub under QuentinJehanne (2020, April 8), QuentinJehanne/ccoli_2020: v1 of a personal vcf filter (v1.0.0) (https://doi.org/10.5281/zenodo.3744758).
Supplementary Material
ACKNOWLEDGMENTS
We thank all of the laboratories that sent Campylobacter isolates to our reference center.
This study was financed by internal funding of the French National Reference Center for Campylobacters and Helicobacters (Bordeaux, France) (www.cnrch.fr).
The material is original research and has not been previously published or submitted for publication elsewhere. We have no conflict of interest to declare.
Authors made the following contributions: conceptualization, Quentin Jehanne and Philippe Lehours; funding acquisition, Philippe Lehours; investigation, Quentin Jehanne, Ben Pascoe, and Samuel K. Sheppard; project administration, Quentin Jehanne, Samuel K. Sheppard, and Philippe Lehours; resources, Lucie Bénéjat, Astrid Ducournau, Alice Buissonnière, Evangelos Mourkas, Ben Pascoe, and Samuel K. Sheppard; supervision, Philippe Lehours and Samuel K. Sheppard; validation, Quentin Jehanne, Samuel K. Sheppard, and Philippe Lehours; visualization, Quentin Jehanne, Ben Pascoe, Samuel K. Sheppard, and Philippe Lehours; writing–original draft, Quentin Jehanne and Philippe Lehours; writing–review and editing, Quentin Jehanne, Samuel K. Sheppard, Ben Pascoe, Francis Mégraud, Emilie Bessède, and Philippe Lehours. All authors have read and agreed to the published version of the manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Blaser MJ. 1997. Epidemiologic and clinical features of Campylobacter jejuni infections. J Infect Dis 176(Suppl 2):S103–S105. doi: 10.1086/513780. [DOI] [PubMed] [Google Scholar]
- 2.Scallan E, Griffin PM, Angulo FJ, Tauxe RV, Hoekstra RM. 2011. Foodborne illness acquired in the United States–unspecified agents. Emerg Infect Dis 17:16–22. doi: 10.3201/eid1701.p21101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.European Food Safety Authority and European Centre for Disease Prevention and Control. 2018. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSA J 16:e05500. doi: 10.2903/j.efsa.2018.5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Cauteren D, Le Strat Y, Sommen C, Bruyand M, Tourdjman M, Da Silva NJ, Couturier E, Fournet N, de Valk H, Desenclos J-C. 2017. Estimated annual numbers of foodborne pathogen-associated illnesses, hospitalizations, and deaths, France, 2008–2013. Emerg Infect Dis 23:1486–1492. doi: 10.3201/eid2309.170081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horrocks SM, Anderson RC, Nisbet DJ, Ricke SC. 2009. Incidence and ecology of Campylobacter jejuni and coli in animals. Anaerobe 15:18–25. doi: 10.1016/j.anaerobe.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 6.French National Reference Center for Campylobacters & Helicobacters (Bordeaux Hospital University Center). 2019. 2018 Campylobacters surveillance report. French National Reference Center for Campylobacters & Helicobacters, Bordeaux, France. [Google Scholar]
- 7.Fitzgerald C. 2015. Campylobacter. Clin Lab Med 35:289–298. doi: 10.1016/j.cll.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 8.Kapperud G, Rosef O. 1983. Avian wildlife reservoir of Campylobacter fetus subsp. jejuni, Yersinia spp., and Salmonella spp. in Norway. Appl Environ Microbiol 45:375–380. doi: 10.1128/AEM.45.2.375-380.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheppard SK, Dallas JF, MacRae M, McCarthy ND, Sproston EL, Gormley FJ, Strachan NJC, Ogden ID, Maiden MCJ, Forbes KJ. 2009. Campylobacter genotypes from food animals, environmental sources and clinical disease in Scotland 2005/6. Int J Food Microbiol 134:96–103. doi: 10.1016/j.ijfoodmicro.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson DJ, Gabriel E, Leatherbarrow AJH, Cheesbrough J, Gee S, Bolton E, Fox A, Fearnhead P, Hart CA, Diggle PJ. 2008. Tracing the source of campylobacteriosis. PLoS Genet 4:e1000203. doi: 10.1371/journal.pgen.1000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sheppard SK, Dallas JF, Strachan NJC, MacRae M, McCarthy ND, Wilson DJ, Gormley FJ, Falush D, Ogden ID, Maiden MCJ, Forbes KJ. 2009. Campylobacter genotyping to determine the source of human infection. Clin Infect Dis 48:1072–1078. doi: 10.1086/597402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nachamkin I, Allos BM, Ho T. 1998. Campylobacter species and Guillain-Barré syndrome. Clin Microbiol Rev 11:555–567. doi: 10.1128/CMR.11.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grover M. 2014. Role of gut pathogens in development of irritable bowel syndrome. Indian J Med Res 139:11–18. [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Y, Feye KM, Shi Z, Pavlidis HO, Kogut M, J Ashworth A, Ricke SC. 2019. A historical review on antibiotic resistance of foodborne Campylobacter. Front Microbiol 10:1509. doi: 10.3389/fmicb.2019.01509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salazar-Lindo E, Sack RB, Chea-Woo E, Kay BA, Piscoya ZA, Leon-Barua R, Yi A. 1986. Early treatment with erythromycin of Campylobacter jejuni-associated dysentery in children. J Pediatr 109:355–360. doi: 10.1016/S0022-3476(86)80404-8. [DOI] [PubMed] [Google Scholar]
- 16.Mourkas E, Florez‐Cuadrado D, Pascoe B, Calland JK, Bayliss SC, Mageiros L, Méric G, Hitchings MD, Quesada A, Porrero C, Ugarte‐Ruiz M, Gutiérrez‐Fernández J, Domínguez L, Sheppard SK. 2019. Gene pool transmission of multidrug resistance among Campylobacter from livestock, sewage and human disease. Environ Microbiol 21:4597–4613. doi: 10.1111/1462-2920.14760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosner BM, Schielke A, Didelot X, Kops F, Breidenbach J, Willrich N, Gölz G, Alter T, Stingl K, Josenhans C, Suerbaum S, Stark K. 2017. A combined case-control and molecular source attribution study of human Campylobacter infections in Germany, 2011–2014. Sci Rep 7:1–12. doi: 10.1038/s41598-017-05227-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sheppard SK, Colles F, Richardson J, Cody AJ, Elson R, Lawson A, Brick G, Meldrum R, Little CL, Owen RJ, Maiden MCJ, McCarthy ND. 2010. Host association of Campylobacter genotypes transcends geographic variation. Appl Environ Microbiol 76:5269–5277. doi: 10.1128/AEM.00124-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pascoe B, Méric G, Yahara K, Wimalarathna H, Murray S, Hitchings MD, Sproston EL, Carrillo CD, Taboada EN, Cooper KK, Huynh S, Cody AJ, Jolley KA, Maiden MCJ, McCarthy ND, Didelot X, Parker CT, Sheppard SK. 2017. Local genes for local bacteria: evidence of allopatry in the genomes of transatlantic Campylobacter populations. Mol Ecol 26:4497–4508. doi: 10.1111/mec.14176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ogden ID, Dallas JF, MacRae M, Rotariu O, Reay KW, Leitch M, Thomson AP, Sheppard SK, Maiden M, Forbes KJ, Strachan NJC. 2009. Campylobacter excreted into the environment by animal sources: prevalence, concentration shed, and host association. Foodborne Pathog Dis 6:1161–1170. doi: 10.1089/fpd.2009.0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kempf I, Kerouanton A, Bougeard S, Nagard B, Rose V, Mourand G, Osterberg J, Denis M, Bengtsson BO. 2017. Campylobacter coli in organic and conventional pig production in France and Sweden: prevalence and antimicrobial resistance. Front Microbiol 8:955. doi: 10.3389/fmicb.2017.00955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dearlove BL, Cody AJ, Pascoe B, Méric G, Wilson DJ, Sheppard SK. 2016. Rapid host switching in generalist Campylobacter strains erodes the signal for tracing human infections. ISME J 10:721–729. doi: 10.1038/ismej.2015.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thakur S, Morrow WEM, Funk JA, Bahnson PB, Gebreyes WA. 2006. Molecular epidemiologic investigation of Campylobacter coli in swine production systems, using multilocus sequence typing. Appl Environ Microbiol 72:5666–5669. doi: 10.1128/AEM.00658-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheppard SK, Dallas JF, Wilson DJ, Strachan NJC, McCarthy ND, Jolley KA, Colles FM, Rotariu O, Ogden ID, Forbes KJ, Maiden MCJ. 2010. Evolution of an agriculture-associated disease causing Campylobacter coli clade: evidence from national surveillance data in Scotland. PLoS One 5:e15708. doi: 10.1371/journal.pone.0015708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheppard SK, McCarthy ND, Falush D, Maiden MCJ. 2008. Convergence of Campylobacter species: implications for bacterial evolution. Science 320:237–239. doi: 10.1126/science.1155532. [DOI] [PubMed] [Google Scholar]
- 26.Sheppard SK, Didelot X, Jolley KA, Darling AE, Pascoe B, Meric G, Kelly DJ, Cody A, Colles FM, Strachan NJC, Ogden ID, Forbes K, French NP, Carter P, Miller WG, McCarthy ND, Owen R, Litrup E, Egholm M, Affourtit JP, Bentley SD, Parkhill J, Maiden MCJ, Falush D. 2013. Progressive genome-wide introgression in agricultural Campylobacter coli. Mol Ecol 22:1051–1064. doi: 10.1111/mec.12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dingle KE, Colles FM, Wareing DRA, Ure R, Fox AJ, Bolton FE, Bootsma HJ, Willems RJL, Urwin R, Maiden MCJ. 2001. Multilocus sequence typing system for Campylobacter jejuni. J Clin Microbiol 39:14–23. doi: 10.1128/JCM.39.1.14-23.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clark CG, Bryden L, Cuff WR, Johnson PL, Jamieson F, Ciebin B, Wang G. 2005. Use of the Oxford multilocus sequence typing protocol and sequencing of the flagellin short variable region to characterize isolates from a large outbreak of waterborne Campylobacter sp. strains in Walkerton, Ontario, Canada. J Clin Microbiol 43:2080–2091. doi: 10.1128/JCM.43.5.2080-2091.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheppard SK, Cheng L, Méric G, Haan CD, Llarena A-K, Marttinen P, Vidal A, Ridley A, Clifton-Hadley F, Connor TR, Strachan NJC, Forbes K, Colles FM, Jolley KA, Bentley SD, Maiden MCJ, Hänninen M-L, Parkhill J, Hanage WP, Corander J. 2014. Cryptic ecology among host generalist Campylobacter jejuni in domestic animals. Mol Ecol 23:2442–2451. doi: 10.1111/mec.12742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gripp E, Hlahla D, Didelot X, Kops F, Maurischat S, Tedin K, Alter T, Ellerbroek L, Schreiber K, Schomburg D, Janssen T, Bartholomäus P, Hofreuter D, Woltemate S, Uhr M, Brenneke B, Grüning P, Gerlach G, Wieler L, Suerbaum S, Josenhans C. 2011. Closely related Campylobacter jejuni strains from different sources reveal a generalist rather than a specialist lifestyle. BMC Genomics 12:584. doi: 10.1186/1471-2164-12-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jolley KA, Bray JE, Maiden MCJ. 2018. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res 3:124. doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thépault A, Méric G, Rivoal K, Pascoe B, Mageiros L, Touzain F, Rose V, Béven V, Chemaly M, Sheppard SK. 2017. Genome-wide identification of host-segregating epidemiological markers for source attribution in Campylobacter jejuni. Appl Environ Microbiol 83:e03085-16. doi: 10.1128/AEM.03085-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thépault A, Rose V, Quesne S, Poezevara T, Béven V, Hirchaud E, Touzain F, Lucas P, Méric G, Mageiros L, Sheppard SK, Chemaly M, Rivoal K. 2018. Ruminant and chicken: important sources of campylobacteriosis in France despite a variation of source attribution in 2009 and 2015. Sci Rep 8:1–10. doi: 10.1038/s41598-018-27558-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berthenet E, Thépault A, Chemaly M, Rivoal K, Ducournau A, Buissonnière A, Bénéjat L, Bessède E, Mégraud F, Sheppard SK, Lehours P. 2019. Source attribution of Campylobacter jejuni shows variable importance of chicken and ruminants reservoirs in noninvasive and invasive French clinical isolates. Sci Rep 9:8098. doi: 10.1038/s41598-019-44454-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cody AJ, Maiden MC, Strachan NJ, McCarthy ND. 2019. A systematic review of source attribution of human campylobacteriosis using multilocus sequence typing. Euro Surveill 24:1800696. doi: 10.2807/1560-7917.ES.2019.24.43.1800696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller WG, Englen MD, Kathariou S, Wesley IV, Wang G, Pittenger-Alley L, Siletz RM, Muraoka W, Fedorka-Cray PJ, Mandrell RE. 2006. Identification of host-associated alleles by multilocus sequence typing of Campylobacter coli strains from food animals. Microbiology 152:245–255. doi: 10.1099/mic.0.28348-0. [DOI] [PubMed] [Google Scholar]
- 37.Kamal N, Dorrell N, Jagannathan A, Turner SM, Constantinidou C, Studholme DJ, Marsden G, Hinds J, Laing KG, Wren BW, Penn CW. 2007. Deletion of a previously uncharacterized flagellar-hook-length control gene fliK modulates the sigma54-dependent regulon in Campylobacter jejuni. Microbiology 153:3099–3111. doi: 10.1099/mic.0.2007/007401-0. [DOI] [PubMed] [Google Scholar]
- 38.Yeh H-Y, Hiett KL, Line JE, Seal BS. 2014. Characterization and antigenicity of recombinant Campylobacter jejuni flagellar capping protein FliD. J Med Microbiol 63:602–609. doi: 10.1099/jmm.0.060095-0. [DOI] [PubMed] [Google Scholar]
- 39.Clark C, Berry C, Demczuk W. 2019. Diversity of transducer-like proteins (Tlps) in Campylobacter. PLoS One 14:e0214228. doi: 10.1371/journal.pone.0214228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reuter M, van Vliet AHM. 2013. Signal balancing by the CetABC and CetZ chemoreceptors controls energy taxis in Campylobacter jejuni. PLoS One 8:e54390. doi: 10.1371/journal.pone.0054390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Runti G, Ruiz M del CL, Stoilova T, Hussain R, Jennions M, Choudhury HG, Benincasa M, Gennaro R, Beis K, Scocchi M. 2013. Functional characterization of SbmA, a bacterial inner membrane transporter required for importing the antimicrobial peptide Bac7(1–35). J Bacteriol 195:5343–5351. doi: 10.1128/JB.00818-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elvers KT, Wu G, Gilberthorpe NJ, Poole RK, Park SF. 2004. Role of an inducible single-domain hemoglobin in mediating resistance to nitric oxide and nitrosative stress in Campylobacter jejuni and Campylobacter coli. J Bacteriol 186:5332–5341. doi: 10.1128/JB.186.16.5332-5341.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bertrand-Burggraf E, Selby CP, Hearst JE, Sancar A. 1991. Identification of the different intermediates in the interaction of (A)BC excinuclease with its substrates by DNase I footprinting on two uniquely modified oligonucleotides. J Mol Biol 219:27–36. doi: 10.1016/0022-2836(91)90854-Y. [DOI] [PubMed] [Google Scholar]
- 44.Sheppard SK, Guttman DS, Fitzgerald JR. 2018. Population genomics of bacterial host adaptation. Nat Rev Genet 19:549–565. doi: 10.1038/s41576-018-0032-z. [DOI] [PubMed] [Google Scholar]
- 45.Sheppard SK, Didelot X, Meric G, Torralbo A, Jolley KA, Kelly DJ, Bentley SD, Maiden MCJ, Parkhill J, Falush D. 2013. Genome-wide association study identifies vitamin B5 biosynthesis as a host specificity factor in Campylobacter. Proc Natl Acad Sci U S A 110:11923–11927. doi: 10.1073/pnas.1305559110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yahara K, Méric G, Taylor AJ, Vries SD, Murray S, Pascoe B, Mageiros L, Torralbo A, Vidal A, Ridley A, Komukai S, Wimalarathna H, Cody AJ, Colles FM, McCarthy N, Harris D, Bray JE, Jolley KA, Maiden MCJ, Bentley SD, Parkhill J, Bayliss CD, Grant A, Maskell D, Didelot X, Kelly DJ, Sheppard SK. 2017. Genome-wide association of functional traits linked with Campylobacter jejuni survival from farm to fork. Environ Microbiol 19:361–380. doi: 10.1111/1462-2920.13628. [DOI] [PubMed] [Google Scholar]
- 47.Roux F, Sproston E, Rotariu O, Macrae M, Sheppard SK, Bessell P, Smith-Palmer A, Cowden J, Maiden MCJ, Forbes KJ, Strachan NJC. 2013. Elucidating the etiology of human Campylobacter coli infections. PLoS One 8:e64504. doi: 10.1371/journal.pone.0064504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kittl S, Heckel G, Korczak BM, Kuhnert P. 2013. Source attribution of human Campylobacter isolates by MLST and Fla-typing and association of genotypes with quinolone resistance. PLoS One 8:e81796. doi: 10.1371/journal.pone.0081796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nohra A, Grinberg A, Midwinter AC, Marshall JC, Collins-Emerson JM, French NP. 2016. Molecular epidemiology of Campylobacter coli strains isolated from different sources in New Zealand between 2005 and 2014. Appl Environ Microbiol 82:4363–4370. doi: 10.1128/AEM.00934-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rotariu O, Dallas JF, Ogden ID, MacRae M, Sheppard SK, Maiden MCJ, Gormley FJ, Forbes KJ, Strachan NJC. 2009. Spatiotemporal homogeneity of Campylobacter subtypes from cattle and sheep across northeastern and southwestern Scotland. Appl Environ Microbiol 75:6275–6281. doi: 10.1128/AEM.00499-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smid JH, Gras LM, Boer A. d, French NP, Havelaar AH, Wagenaar JA, van Pelt W. 2013. Practicalities of using non-local or non-recent multilocus sequence typing data for source attribution in space and time of human campylobacteriosis. PLoS One 8:e55029. doi: 10.1371/journal.pone.0055029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mossong J, Mughini-Gras L, Penny C, Devaux A, Olinger C, Losch S, Cauchie H-M, van Pelt W, Ragimbeau C. 2016. Human campylobacteriosis in Luxembourg, 2010–2013: a case-control study combined with multilocus sequence typing for source attribution and risk factor. Sci Rep 6:20939. doi: 10.1038/srep20939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karp BE, Tate H, Plumblee JR, Dessai U, Whichard JM, Thacker EL, Hale KR, Wilson W, Friedman CR, Griffin PM, McDermott PF. 2017. National antimicrobial resistance monitoring system: two decades of advancing public health through integrated surveillance of antimicrobial resistance. Foodborne Pathog Dis 14:545–557. doi: 10.1089/fpd.2017.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pearson BM, Rokney A, Crossman LC, Miller WG, Wain J, van Vliet AHM. 2013. Complete genome sequence of the Campylobacter coli clinical isolate 15–537360. Genome Announc 1:e01056-13. doi: 10.1128/genomeA.01056-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wingett SW, Andrews S. 2018. FastQ Screen: a tool for multi-genome mapping and quality control. F1000Res 7:1338. doi: 10.12688/f1000research.15931.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Joshi NA, Fass JN. 2011. Sickle: a sliding-window, adaptive, quality-based trimming tool for FastQ files (version 1.33). https://github.com/najoshi/sickle. Accessed 1 December 2018.
- 57.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jolley KA, Maiden MCJ. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou Z, Alikhan N-F, Sergeant MJ, Luhmann N, Vaz C, Francisco AP, Carriço JA, Achtman M. 2018. GrapeTree: visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res 28:1395–1404. doi: 10.1101/gr.232397.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Price MN, Dehal PS, Arkin AP. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Argimón S, Abudahab K, Goater RJE, Fedosejev A, Bhai J, Glasner C, Feil EJ, Holden MTG, Yeats CA, Grundmann H, Spratt BG, Aanensen DM. 2016. Microreact: visualizing and sharing data for genomic epidemiology and phylogeography. Microb Genom 2:e000093. doi: 10.1099/mgen.0.000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.O’Kane PM, Connerton IF. 2017. Characterization of aerotolerant forms of a robust chicken colonizing Campylobacter coli. Front Microbiol 8:513. doi: 10.3389/fmicb.2017.00513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marasini D, Fakhr MK. 2016. Complete genome sequences of the plasmid-bearing Campylobacter coli strains HC2-48, CF2-75, and CO2-160 isolated from retail beef liver. Genome Announc 4:e01004-16. doi: 10.1128/genomeA.01004-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marasini D, Fakhr MK. 2017. Complete genome sequences of plasmid-bearing multidrug-resistant Campylobacter jejuni and Campylobacter coli strains with type VI secretion systems, isolated from retail turkey and pork. Genome Announc 5:e01360-17. doi: 10.1128/genomeA.01360-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 1303.3997 [q-bio.GN]. https://arxiv.org/abs/1303.3997.
- 67.Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All 900 C. coli genomes are available using identifiers (IDs) listed in Data Sets S1, S2, and S3, which provide BioSample and PubMLST IDs for NCBI and PubMLST databases, respectively.
The personal vcf filter Python script is available on GitHub under QuentinJehanne (2020, April 8), QuentinJehanne/ccoli_2020: v1 of a personal vcf filter (v1.0.0) (https://doi.org/10.5281/zenodo.3744758).