A fungal effector targets a heat shock–dynamin protein complex to modulate mitochondrial dynamics and reduce plant immunity.
Abstract
Mitochondria are essential for animal and plant immunity. Here, we report that the effector MoCDIP4 of the fungal pathogen Magnaporthe oryzae targets the mitochondria-associated OsDjA9-OsDRP1E protein complex to reduce rice immunity. The DnaJ protein OsDjA9 interacts with the dynamin-related protein OsDRP1E and promotes the degradation of OsDRP1E, which functions in mitochondrial fission. By contrast, MoCDIP4 binds OsDjA9 to compete with OsDRP1E, resulting in OsDRP1E accumulation. Knockout of OsDjA9 or overexpression of OsDRP1E or MoCDIP4 in transgenic rice results in shortened mitochondria and enhanced susceptibility to M. oryzae. Overexpression of OsDjA9 or knockout of OsDRP1E in transgenic rice, in contrast, leads to elongated mitochondria and enhanced resistance to M. oryzae. Our study therefore reveals a previously unidentified pathogen-infection strategy in which the pathogen delivers an effector into plant cells to target an HSP40-DRP complex; the targeting leads to the perturbation of mitochondrial dynamics, thereby inhibiting mitochondria-mediated plant immunity.
INTRODUCTION
Mitochondria are critical to the cellular metabolism and bioenergetics of eukaryotic cells (1). Besides generating reactive oxygen species (ROS) or being involved in programmed cell death, mitochondria also function as a signaling platform to activate downstream immune responses after perception of viral pathogen-associated molecular patterns (PAMPs) in mammals (2, 3). In response to various cellular stresses, mitochondria undergo coordinated cycles of fission and fusion, referred to as “mitochondrial dynamics,” to maintain cellular homeostasis. Accumulating evidence from mammals suggests that mitochondrial dynamics contribute directly to innate immune signaling against viral pathogens by mediating the mitochondrial antiviral signaling protein (MAVS), which is an adaptor of the retinoic acid–inducible gene 1–like receptors (RLRs) (4, 5). In mammalian cells, mitochondrial elongation enhances MAVS-mediated antiviral immunity, while mitochondrial fragmentation has the opposite effect (4–6). Similarly, when a pathogen attacks plant cells, mitochondria are thought to act as a signaling organelle to enhance defense responses by activating various signals such as nitric oxide, ROS, or salicylic acid (7). A functional link between mitochondrial dynamics and innate immunity, however, has not been documented in plants.
The mechanism of mitochondrial fusion and fission in eukaryotes is conserved and accurately controlled by a complex machinery composed of dynamin-related proteins (DRPs), which belong to the dynamin superfamily with the multidomain guanosine triphosphatase (GTPase) activity (8). In mammals, DRP1 regulates mitochondrial fission and is required for efficient antiviral response (4, 9). Mitochondrial elongation due to a mutation in DRP1 promotes endoplasmic reticulum (ER)–mitochondria interactions during viral infection, and this interaction enhances the association of MAVS with the stimulator of the interferon genes (STING) to augment the intracellular RLR signaling (4). In agreement with the findings in mammals, several studies reported the function of the following DRPs in plant mitochondrial fission: AtDRP1C, AtDRP1E, AtDRP3A, and AtDRP3B (10, 11). Although two DRPs have been reported to be involved in plant resistance as a negative regulator against hemibiotrophic or biotrophic pathogens, a direct link between mitochondrial dynamics and plant immunity has not been documented (12, 13).
Precise regulation of mitochondrial fission is crucial because its overactivation often reduces mitochondrial homeostasis (8). As a cochaperone of heat shock protein 70 (HSP70), HSP40 (also known as DnaJ protein) is essential for maintaining protein homeostasis under stress conditions and in many cellular processes (14). HSP40 contains a conserved DnaJ domain that is crucial for the binding of DnaJ to HSP70 and a C-terminal domain that is involved in substrate recognition (15). DnaJ proteins are usually involved in the maturation and stabilization of proteins (16, 17) and especially in the accumulation of immune receptors, such as RLRs in mammals (18) and EF-Tu receptors in Arabidopsis (19). However, many studies have shown that DnaJ proteins, in concert with the ubiquitin-proteasome system or autophagy, selectively degrade substrates to ensure cell homeostasis (20, 21). A previous study in our laboratory demonstrated that the rice DnaJ protein OsDjA6 is involved in rice PAMP-triggered immunity (PTI) regulation and interacts with an E3 ligase (22).
Because mitochondrial dynamics affect disease resistance, pathogens have evolved several strategies to interfere with the dynamics to suppress immune responses (23). Several pathogen effectors were recently found to directly target DRP1 and mitofusins, the key component of mitochondrial fission and fusion machinery to interfere with mitochondrial dynamics and block innate immune responses in mammalian cells (6, 24).
The current research concerns how the fungus Magnaporthe oryzae, the causal agent of rice blast disease, interferes with mitochondrial dynamics and blocks innate immune responses in rice plants. We found that MoCDIP4, an effector of M. oryzae, targets the rice DnaJ protein OsDjA9 and the dynamin-related GTPase OsDRP1E complex. MoCDIP4 causes OsDjA9 to remain in the ER, which therefore reduces the degradation of the mitochondrial fission factor OsDRP1E mediated by OsDjA9. The resulting increase in OsDRP1E levels accelerates mitochondrial fission and reduces mitochondrial size, ROS production, and defense-related gene expression. Mitochondrial size in OsDjA9 and OsDRP1E overexpression and knockout plants is well correlated with their resistance to M. oryzae, indicating that mitochondrial dynamics are involved in plant immunity. Our results reveal a previously unidentified fungal pathogen infection strategy that subverts immunity by interfering with mitochondrial dynamics in plant cells.
RESULTS
Glycosyl hydrolase protein MoCDIP4 contributes to M. oryzae pathogenicity
Our laboratory previously demonstrated that MoCDIP4, a cellulose-growth-specific protein, contains a predicted signal peptide (SP), a glycosyl hydrolase family 61, and a fungal-type cellulose-binding domain (fig. S1A) and can induce cell death in Nicotiana benthamiana and rice protoplasts (25). Using the yeast signal trap assay, we found that fusion of the signal peptide sequence of MoCDIP4 with the invertase gene resulted in the secretion of invertase in yeast, suggesting that MoCDIP4 is likely secreted into rice cells during M. oryzae infection (fig. S1B).
To determine the role of MoCDIP4 in pathogenicity, we generated MoCDIP4 knockout mutants (ΔMoCDIP4) and complementation strains (ΔMoCDIP4/MoCDIP4) in the M. oryzae strain Guy11 (fig. S1C) and used these strains for spray inoculation. Knockout of MoCDIP4 resulted in significantly reduced M. oryzae infection on rice plants, while complementation restored the full virulence of Guy11 (Fig. 1, A and B). MoCDIP4 overexpression in rice cultivar Nipponbare (NPB) (fig. S1D), in contrast, promoted susceptibility to M. oryzae, i.e., overexpression of MoCDIP4 in rice resulted in larger lesions and increased fungal biomass relative to NPB (fig. S1, E to H). In addition, the transcript levels of the pathogen-related (PR) genes PAL1 and WRKY45 were decreased in the transgenic plants (fig. S1I). Together, these data suggest that MoCDIP4 contributes to the virulence of M. oryzae and can suppress plant defense to the pathogen.
Fig. 1. MoCDIP4 contributes to the pathogenicity of M. oryzae and targets the DnaJ protein OsDjA9 in rice.
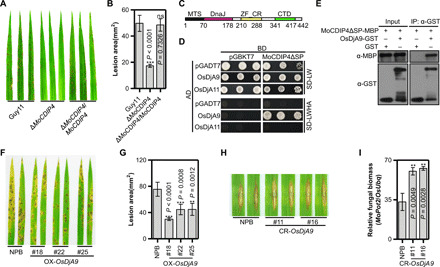
(A) Pathogenicity of the MoCDIP4 mutant to NPB seedlings. (B) Lesion area of spray-inoculated leaves in (A). Bars represent means ± SD, n = 5. Asterisks and ns (no significant difference) indicate statistical significance between the MoCDIP4 mutant and Guy11 according to Student’s t test. (C) The structure of OsDjA9. (D) Interaction between MoCDIP4 and OsDjA9 in yeast. OsDjA11 was used as the specific control. (E) Interaction between MoCDIP4 and OsDjA9 in glutathione S-transferase (GST) pulldown assay. (F) Spray inoculation of OsDjA9 overexpression plants with M. oryzae. (G) Lesion area of spray-inoculated leaves in (F). Bars represent means ± SD, n = 5. Asterisks indicate the significant differences between the transgenic line and NPB according to Student’s t test. (H) Punch inoculation of OsDjA9 CRISPR-Cas9–edited plants with M. oryzae. (I) Relative fungal biomass of punch-inoculated leaves in (H). Bars represent means ± SD, n = 3. Asterisks indicate the significant differences between the transgenic line and NPB according to Student’s t test.
MoCDIP4 targets the rice DnaJ protein OsDjA9
To investigate the role of MoCDIP4 in reducing host defense, we identified a partial cDNA clone (amino acids 109 to 396) of OsDjA9 (LOC_Os06g02620) (26) in a yeast two-hybrid (Y2H) screen of a rice cDNA library using MoCDIP4 as the bait (27). OsDjA9 encodes a rice DnaJ protein consisting of a mitochondrial targeting sequence (MTS), a DnaJ domain, a zinc finger CR (ZF_CR), and a C-terminal domain (CTD) (Fig. 1C). We confirmed that MoCDIP4 interacts with OsDjA9 in yeast and that the interaction is specific because MoCDIP4 did not interact with the OsDjA9 homolog OsDjA11 (LOC_Os12g07060) (Fig. 1D).
To further confirm the interaction between MoCDIP4 and OsDjA9, we performed an in vitro pulldown assay. The MoCDIP4 protein was effectively detected in the glutathione Sepharose beads with OsDjA9-GST (glutathione S-transferase; Fig. 1E). Furthermore, a luciferase complementation imaging (LCI) assay revealed strong luciferase activity in N. benthamiana leaves when MoCDIP4ΔSP-nLUC and OsDjA9-cLUC were coexpressed (fig. S2). Together, these results suggest that MoCDIP4 interacts with OsDjA9 in vitro and in vivo.
OsDjA9 positively regulates rice immunity against M. oryzae
To investigate the function of OsDjA9 in rice immunity, we generated OsDjA9 overexpression, CRISPR-Cas9–edited, and RNA interference (RNAi) transgenic plants of OsDjA9 in the NPB background (fig. S3, A to C) and tested their resistance to M. oryzae isolate RO1-1. Compared to NPB, the overexpression plants showed enhanced resistance to RO1-1 with smaller lesions and reduced fungal biomass (Fig. 1, F and G, and fig. S3D), while the gene-edited plants and the RNAi plants showed reduced resistance with increased fungal biomass (Fig. 1, H and I, and fig. S3, E and F). To investigate how OsDjA9 regulates rice immunity, we measured ROS accumulation in response to PAMP treatments. As shown in fig. S3G, the chitin-triggered ROS burst was greater in the overexpression plants than in the RNAi or NPB plants. The transcript levels of many PR genes, including PAL1, PBZ1, and PR10, were up-regulated in the overexpression plants (fig. S3H) and down-regulated in the RNAi plants (fig. S3I) compared to NPB. Together, these results demonstrate that OsDjA9 positively regulates PTI responses in rice.
OsDjA9 is colocalized with MoCDIP4 in the ER
To elucidate the relationship between MoCDIP4 and OsDjA9, we first examined the subcellular localization of MoCDIP4 and OsDjA9 in rice protoplasts and N. benthamiana. Confocal microscopy showed that MoCDIP4ΔSP signals extensively overlapped with those of the ER marker HDEL (Fig. 2A and fig. S4A) (28), indicating that MoCDIP4ΔSP is localized to the ER. Like DnaJ protein DnaJA3 in mammals (29), OsDjA9 was localized to both the cytoplasm and mitochondria in rice cells (Fig. 2B and fig. S4B, top). Further examination of the OsDjA9-GFP (green fluorescent protein) signals in rice protoplasts and N. benthamiana revealed that OsDjA9-GFP had a reticular structure of ER (Fig. 2B and fig. S4B, bottom). Cell fractionation and immunoblotting analyses in N. benthamiana further confirmed that MoCDIP4ΔSP was detected exclusively in the membrane fraction (Fig. 2C) and the major part of OsDjA9 was present in mitochondria and the membrane fraction, while a small amount was detected in the soluble fraction (Fig. 2D).
Fig. 2. MoCDIP4 is colocalized with OsDjA9 in the ER.
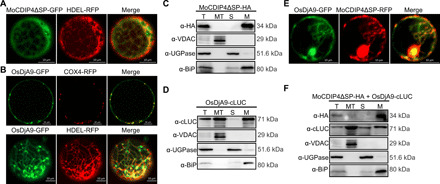
(A) Subcellular localization of MoCDIP4 in rice protoplasts. HDEL was used as the ER marker. (B) Subcellular localization of OsDjA9 in rice protoplasts. COX4 was used as a mitochondrial marker; HDEL was used as the ER marker. Detection of (C) MoCDIP4 and (D) OsDjA9 in different cell fractions extracted from infiltrated tobacco leaves. T, total extract; MT, mitochondria; S, soluble fraction; M, membrane fraction. (E) Colocalization of MoCDIP4 and OsDjA9 in rice protoplasts. (F) Detection of MoCDIP4 and OsDjA9 in different cell fractions extracted from tobacco leaves coinfiltrated with MoCDIP4 and OsDjA9. Scale bars (A, B, E), 10 μm.
Next, we coexpressed MoCDIP4 and OsDjA9 in rice protoplasts and N. benthamiana and found that the overlapped signals between MoCDIP4 and OsDjA9 were observed in the ER (Fig. 2E and fig. S4C). Cell fractionation and immunoblotting analyses further confirmed that both MoCDIP4ΔSP and OsDjA9 had accumulated in the membrane fraction (Fig. 2F). In addition, there were strong signals in the cells agro-infiltrated with MoCDIP4ΔSP-YN and OsDjA9-YC plasmids in a bimolecular fluorescence complementation (BiFC) assay (fig. S4D). Given the confocal images and subcellular fractionation data, it seems likely that MoCDIP4 interacts with OsDjA9 in the ER.
OsDjA9 interacts with the DRP OsDRP1E
To better understand the role of OsDjA9 in rice resistance, we search for rice proteins that interact with OsDjA9. Last, we focused on a DRP OsDRP1E, which negatively regulates programmed cell death (13), and its homologous protein DRP1 in mammals is related to morphological changes in mitochondria induced by DnaJA3 (30). In yeast, OsDjA9, but not the most homologous protein OsDjA11, interacted with OsDRP1E (Fig. 3A). The interaction between OsDjA9 and OsDRP1E was confirmed by both coimmunoprecipitation (Co-IP) and LCI assays in vivo (fig. S5, A and B).
Fig. 3. OsDjA9 interacts with the DRP OsDRP1E, which negatively regulates rice immunity.
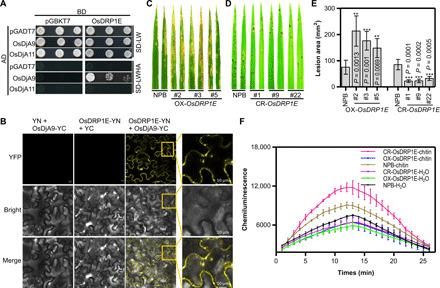
(A) Interaction between OsDjA9 and OsDRP1E in yeast. (B) BiFC analysis of the OsDjA9-OsDRP1E interaction in N. benthamiana. Yellow squares show the enlarged mitochondrial localization. Scale bars, 10 μm. Spray inoculation of OsDRP1E (C) overexpression and (D) CRISPR-Cas9–edited plants with M. oryzae. (E) Lesion area of spray-inoculated leaves in (C and D). Bars represent means ± SD, n = 5. Asterisks indicate the significant differences between the transgenic line and NPB according to Student’s t test. (F) ROS accumulation in OsDRP1E overexpression and CRISPR-Cas9–edited plants after chitin treatment. Bars represent means ± SD, n = 3.
OsDRP1E was previously reported to be localized to mitochondria (13). In this study, on the basis of confocal images and subcellular fractionation, we confirmed that OsDRP1E was localized to mitochondria, cytoplasm, and membrane fraction (fig. S5, C and D). To determine the colocalization of OsDjA9 and OsDRP1E, we coexpressed OsDjA9 and OsDRP1E in N. benthamiana and found that the OsDjA9 red fluorescence could overlap with the green signals of OsDRP1E, and both proteins were detected in mitochondria, cytoplasm, and membrane fraction (fig. S5, E and F). In addition, BiFC assay of OsDjA9 and OsDRP1E showed that stronger yellow fluorescent protein (YFP) signals were mainly detected in the cytoplasm and the punctate mitochondrial structure (Fig. 3B). These results indicate that OsDRP1E could interact with OsDjA9.
OsDRP1E negatively regulates rice immunity
Our laboratory previously demonstrated that the E409V mutation in OsDRP1E caused spontaneous cell death in rice and significantly increased resistance to fungal and bacterial pathogens. To confirm OsDRP1E function in rice immunity, we generated overexpression and CRISPR-Cas9–edited OsDRP1E plants (fig. S6, A to C). Inoculation results showed that the overexpression plants were more susceptible to M. oryzae and that the gene-edited plants were more resistant to M. oryzae than NPB plants (Fig. 3, C to E, and fig. S6, D to F). Unlike rice plants with the E409V mutation in OsDRP1E, most of the gene-edited plants did not display cell death lesions or a growth defect, but a few did at the late growth stage. To investigate OsDRP1E function in PTI responses, we measured ROS generation in response to PAMP treatments. The results revealed that ROS accumulation was lower in the overexpression lines and notably higher in the gene-edited plants (Fig. 3F) than in NPB in response to chitin treatment. In addition, the transcript levels of many PR genes, including PAL1, PR10, and WRKY45, were down-regulated in the overexpression plants (fig. S6G) and up-regulated in the gene-edited plants (fig. S6H) compared to NPB. Together, these results suggest that OsDRP1E negatively regulates rice PTI.
OsDjA9 destabilizes OsDRP1E via its DnaJ domain
To determine the relationship between OsDjA9 and OsDRP1E, we first assessed OsDRP1E transcript levels in OsDjA9 transgenic lines. Quantitative reverse transcription polymerase chain reaction (qRT-PCR) revealed that the expression of OsDRP1E was the same in OsDjA9 overexpression and RNAi transgenic plants and NPB plants (fig. S7A), suggesting that OsDjA9 does not regulate OsDRP1E at the transcript level. Immunoblotting, however, showed that OsDRP1E accumulated to a much lower level in rice protoplasts with OsDjA9 than with the control nLUC, MoCDIP4ΔSP, or the mutated OsDjA9QNA [substitution of HPD (histidine, proline, and aspartic acid) to QNA (glutamine, asparagine, and alanine) in the conserved DnaJ domain] (Fig. 4A). To determine which pathway regulates the degradation of OsDRP1E, we treated rice protoplasts with the 26S proteasome inhibitor MG132 and the autophagy inhibitors E64d, concanavalin A (Con A), and 3-methyladenine (3-MA) and found that OsDRP1E degradation was affected by the autophagy inhibitors but not by the 26S proteasome inhibitor (Fig. 4B). Similarly, OsDRP1E protein levels were lower in OsDjA9 overexpression protoplasts than in NPB and OsDjA9 knockout rice protoplasts, and this low abundance of OsDRP1E in OsDjA9 overexpression protoplasts could be increased by adding the three autophagy inhibitors (fig. S7C). In addition, a web-based analysis (https://ilir.warwick.ac.uk) identified a potential autophagy-related protein 8 (ATG8)–interacting motif in OsDRP1E. Our Y2H assay verified that OsDRP1E could interact with ATG8d (fig. S7D). In addition, the transcript levels of the autophagy-related ATG genes ATG8c, ATG8d, and ATG9 were higher in OsDjA9 overexpression rice protoplasts than those in NPB, MoCDIP4 overexpression, and OsDjA9 knockout rice protoplasts when transfected with the OsDRP1E-GFP plasmids (fig. S7E). Together, these results suggest that OsDjA9 may destabilize OsDRP1E via the autophagy pathway and that its DnaJ domain is important for the degradation.
Fig. 4. MoCDIP4 stabilizes OsDRP1E by decreasing the association of OsDjA9 with OsDRP1E in a competitive manner.
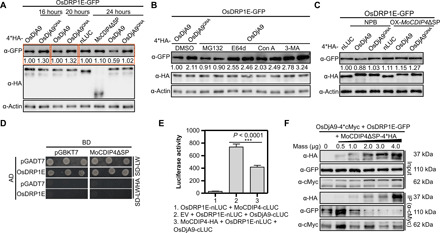
(A) OsDRP1E protein levels when coexpressed with nLUC, MoCDIP4ΔSP, OsDjA9, and OsDjA9QNA in rice protoplasts. (B) OsDRP1E protein levels when coexpressed with OsDjA9 after inhibitor treatment in rice protoplasts. MG132, 26S proteasome inhibitor; E64d, Con A, and 3-MA, autophagy inhibitors. (C) OsDRP1E protein levels when coexpressed with nLUC, OsDjA9, and OsDjA9QNA in NPB and MoCDIP4 overexpression rice protoplasts. (D) No interaction between MoCDIP4 and OsDRP1E in yeast. (E) Interaction between OsDjA9 and OsDRP1E when coexpressed with MoCDIP4 in LCI assay. Bars represent means ± SD, n = 3. Asterisks indicate the significant difference according to Student’s t test. (F) The dosage-dependent effect of MoCDIP4 on the OsDjA9-OsDRP1E interaction in Co-IP assay. OsDjA9-4*cMyc and OsDRP1E-GFP were coexpressed with increasing amounts of MoCDIP4ΔSP-4*HA in rice protoplasts. The expressed proteins were immunoprecipitated with anti-cMyc antibody, and crude and immunoprecipitated proteins were then analyzed with anti-HA (hemagglutinin), anti-cMyc, or anti-GFP antibodies. The actin protein (A to C) was used as the internal control. Band intensities determined by ImageJ were labeled below the bands. The sample labeled 1.00 was used as a control, and the band intensity of each sample was its ratio to the control. Each experiment was repeated at least three times.
MoCDIP4 competes with OsDRP1E for binding to OsDjA9
Because MoCDIP4 interacts with OsDjA9, we wondered whether MoCDIP4 affects OsDjA9-mediated degradation of OsDRP1E protein. Immunoblotting of rice protoplasts showed that OsDRP1E accumulated to a slightly higher level when coexpressed with MoCDIP4 than with the nLUC control [Fig. 4A, top (lane 6)]. In addition, in MoCDIP4 overexpression protoplasts, OsDRP1E protein levels [Fig. 4C, top (lane 5)] were higher than in NPB protoplasts [Fig. 4C, top (lane 2)] when transfected with OsDjA9, confirming that overexpression of MoCDIP4 stabilizes OsDRP1E protein. These data suggest that the interaction between MoCDIP4 and OsDjA9 affects OsDRP1E accumulation. In addition, qRT-PCR showed that OsDRP1E transcript levels did not significantly differ between MoCDIP4 overexpression lines and NPB (fig. S7B), indicating that MoCDIP4 does not regulate OsDRP1E transcription.
To determine how MoCDIP4 suppresses OsDjA9-mediated degradation of OsDRP1E, we first used Y2H and LCI assays to assess the interaction between MoCDIP4 and OsDRP1E. In both assays, the two proteins failed to interact with each other (Fig. 4, D and E, lane 1). Considering that both MoCDIP4 and OsDRP1E interact with OsDjA9, we then investigated whether MoCDIP4 interferes with the association of OsDjA9 with OsDRP1E in a competitive manner. LCI assay showed that the luciferase activity of the OsDjA9-OsDRP1E interaction was significantly decreased when MoCDIP4 was coexpressed with the two proteins (Fig. 4E, lane 3). A competitive Co-IP assay in vivo and GST pulldown assay in vitro confirmed that the protein levels of OsDRP1E coimmunoprecipitated by OsDjA9 were notably decreased when the amount of MoCDIP4 was increased (Fig. 4F, second panel from the bottom, and fig. S7F). However, immunoblotting showed that OsDjA9 had no effect on MoCDIP4 protein levels (fig. S8). Together, these results indicate that MoCDIP4 decreases the association between OsDjA9 and OsDRP1E in a competitive manner, resulting in OsDRP1E accumulation.
OsDRP1E and OsDjA9 are involved in rice mitochondrial dynamics
To gain insight into the possible function of OsDRP1E and OsDjA9 in mitochondrial dynamics, we used transmission electron microscopy (TEM) to observe mitochondrial morphology in the overexpression and knockout plants of the two genes (Fig. 5A). Relative to the mitochondria in NPB plants, the mitochondria were smaller in OsDRP1E overexpression lines (Fig. 5, B and C, lane 5) but were larger in OsDRP1E knockout plants (Fig. 5, B and C, lane 6). In contrast, the mitochondria were elongated in OsDjA9 overexpression plants (Fig. 5, B and C, lane 3) but shortened in OsDjA9 knockout plants (Fig. 5, B and C, lane 4) and MoCDIP4 overexpression plants (Fig. 5, B and C, lane 2) relative to those in NPB (Fig. 5, B and C, lane 1). The presence of larger mitochondria in OsDjA9 overexpression plants and OsDRP1E knockout plants was associated with enhanced resistance of these plants to M. oryzae. Similarly, the presence of smaller mitochondria in MoCDIP4 or OsDRP1E overexpression plants and OsDjA9 knockout plants was associated with enhanced susceptibility of these plants to M. oryzae. On the basis of these results, we conclude that MoCDIP4 and OsDjA9 modulate mitochondrial dynamics by affecting the abundance of OsDRP1E and mitochondrial size, which are correlated with resistance to M. oryzae.
Fig. 5. OsDRP1E and OsDjA9 affect rice mitochondrial dynamics.
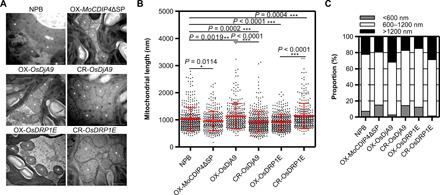
(A) TEM images of mitochondria from the transgenic rice (MoCDIP4, OsDjA9, and OsDRP1E) and NPB. *Mitochondria. Scale bars, 1 μm. (B) Length of mitochondria as measured with ImageJ based on multiple images acquired by TEM. Bars represent means ± SD, n ≥ 300. Asterisks indicate the significant difference according to Student’s t test. (C) Proportions of small, medium, and large mitochondria in the leaves of transgenic rice (MoCDIP4, OsDjA9, and OsDRP1E) and NPB.
DISCUSSION
In mammals, mitochondrial dynamics play an important role in innate immunity (4–6). In antiviral responses, mammalian cells with deletion of the mitochondrial fission factors DRP1 and FIS1 have an elongated mitochondrial network and increased RLR signaling (4). The current study indicated that elongation of mitochondrial network by depletion of OsDRP1E or overexpression of OsDjA9 enhances the rice resistance to the fungal pathogen M. oryzae, whereas MoCDIP4 or OsDRP1E overexpression lines and OsDjA9 knockout lines have short mitochondria and reduced resistance to M. oryzae. We have therefore found a previously unidentified infection strategy in which the pathogen subverts plant immunity by delivering an effector into host cells that reduces the stability of HSP40-DRP1 complex; the reduced stability alters plant mitochondrial dynamics and reduces defense responses.
Several studies have reported the function of the Arabidopsis DRP1E in mitochondrial fission (11, 31). Jin et al. (11) showed that mutation in AtDRP1E causes abnormal mitochondrial elongation; the authors concluded that AtDRP1E is required for mitochondrial fission, and similar results were previously reported for the human DRP1 (9). Three other studies, however, reported no notable differences in mitochondrial size between DRP1E mutant and the wild-type plants (12, 13, 31). To get an accurate estimate of mitochondrial size, we measured a large number of mitochondria (>300) in each transgenic line and NPB. In agreement with the results of Jin et al., we found that overexpression of OsDRP1E generates shortened mitochondria, whereas knockout of OsDRP1E causes abnormal elongation of mitochondria. Our data therefore confirm the role of DRP1E in plant mitochondrial fission.
When attacked by pathogens, plants must balance between protein maturation and degradation. To avoid proteotoxicity, DnaJ proteins function at the crossroad of protein stabilization and degradation by acting in concert with HSP70 system and protein degradation system via the ubiquitin-proteasome or autophagy pathway (21, 32). In this study, we found that the rice DnaJ protein OsDjA9 mediates OsDRP1E protein degradation via autophagy. Furthermore, the HPD-to-QNA mutation of OsDjA9 eliminates its ability to destabilize OsDRP1E, suggesting that DnaJ domain of OsDjA9 is essential for OsDRP1E degradation. Previous research revealed that HPD mutation of HSP40 could abolish the interaction with HSP70 (33), suggesting that HSP70 chaperone machinery may participate in OsDRP1E degradation. We therefore speculate that OsDjA9 cooperates with HSP70 system and autophagy to regulate mitochondrial dynamics and plant disease resistance by controlling OsDRP1E protein levels; the role of HSP70 in the OsDjA9-mediated regulation of OsDRP1E warrants additional investigation.
The ER is an intracellular organelle responsible for protein folding into native structure and posttranslational modification. Many ER proteins, such as BiP (binding immunoglobulin protein) and ERdj3B, are involved in ER quality control machinery for plasma membrane–associated pattern recognition receptor biogenesis in plants (19, 34, 35). In the current study, we found out that the ER-localized effector MoCDIP4 secreted by M. oryzae specifically targets rice DnaJ protein OsDjA9 to decrease the association between OsDjA9 and OsDRP1E in a competitive manner, resulting in OsDRP1E accumulation; the target therefore resembles that of the bacterial effector HopBF1, which mimics a host HSP90 client to subvert immunity (36). Whether MoCDIP4 is a substrate of OsDjA9 or not and how this effector disrupts the binding between OsDjA9 and OsDRP1E to release the latter remain to be elucidated.
On the basis of the results of this and previous studies, we propose a working model of how M. oryzae affects mitochondrial dynamics and plant immunity (Fig. 6). In the absence of M. oryzae infection, OsDRP1E functions as a fission factor that promotes mitochondrial division for plant growth, development, and responses to stresses. To maintain cellular homeostasis, OsDjA9 interacts with OsDRP1E and possibly cooperates with HSP70 and the autophagy pathway to negatively regulate OsDRP1E protein levels, thus balancing mitochondrial dynamics. When M. oryzae infects rice plants, the secreted MoCDIP4 interacts with OsDjA9, which decreases the association of OsDjA9 with OsDRP1E in a competitive manner, resulting in the stabilization of OsDRP1E. The resulting accumulation of OsDRP1E increases mitochondrial fission and reduces mitochondrial size, ROS burst, and rice immunity.
Fig. 6. A working model illustrates how M. oryzae affects mitochondrial dynamics and plant immunity.
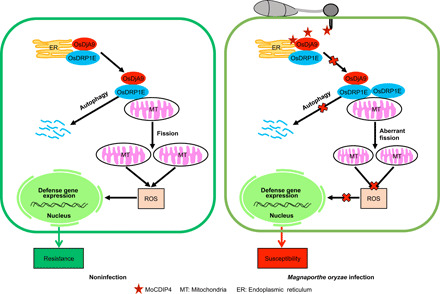
Without M. oryzae infection, the OsDjA9-OsDRP1E complex modulates mitochondrial dynamics for plant growth, development, and defense. During M. oryzae infection, secreted MoCDIP4 interacts with OsDjA9, which decreases the association of OsDjA9 with OsDRP1E in a competitive manner to stabilize OsDRP1E. The accumulation of OsDRP1E promotes mitochondrial fission, reduces mitochondrial size and ROS generation, and increases susceptibility to M. oryzae.
MATERIALS AND METHODS
Plant material and growth conditions
Rice plants were grown in a greenhouse for breeding and phenotype observations. Plants were grown in growth chambers (26°C/20°C day/night, 80% humidity, 12-hour photoperiod) for M. oryzae inoculation and ROS assay. Rice cultivar NPB was used in the study. The methods of rice transformation for the generation of transgenic lines were described previously (27). N. benthamiana was grown in a growth chamber at 22°C under a long-day photoperiod (16-hour light and 8-hour dark).
Rice blast inoculation and disease phenotype evaluation
M. oryzae was cultured as described previously (27). Briefly, the blast fungus isolates RO1-1 and Guy11, MoCDIP4 knockout mutant (ΔMoCDIP4), and complementation strain (ΔMoCDIP4/MoCDIP4) were cultured at room temperature on oatmeal medium [3% (w/v) oat and 1.5% (w/v) agar]. The fungal cultures were grown in the dark for 3 days and then under light for sporulation for 14 to 15 days. The spores were scraped from the surface of the agar and suspended in water [with 0.1% (v/v) Tween 20]. The suspension was passed through Miracloth before inoculation. For punch inoculation, fully expanded leaves from 1.5-month-old rice plants were invaded by spore suspension (5.0 × 105 spores/ml). Disease phenotypes were evaluated at 15 days after inoculation. The relative fungal biomass was calculated using DNA-based qPCR as reported previously (22). For spray inoculation, 21-day-old rice seedlings were sprayed with spore suspension (1.0 × 105 spores/ml); the inoculated plants were kept in a sealed container with high humidity for 24 hours and were then moved to a growth chamber for 6 days for disease development. The leaves were photographed at 7 days after inoculation, and the lesion area was measured using Photoshop.
Y2H assay
The Matchmaker gold Y2H system (Clontech) was used according to the manufacturer’s protocol. The yeast strain AH109 was cultured at 30°C to an optical density at 600 nm of 0.8, and the cells were collected by centrifugation at 2500 rpm for 5 min. The yeast cells were suspended in a polyethylene glycol/lithium acetate (PEG/LiOAc) solution (30% PEG3350 and 100 mM LiOAc). A 300-μl volume of yeast cells was mixed with 50 μg of carrier DNA, 200 ng of bait plasmids, and 200 ng of prey plasmids. After the yeast cells were subjected to heat shock at 42°C for 45 min, the cells were cooled to 30°C, and the pelleted cells by centrifuging were plated on the synthetic defined (SD) medium without Leu and Trp. Cotransformants were grown for 3 days at 30°C and were then cultured on selection medium (SD/-Leu-Trp-His-Ade) to detect the protein-protein interaction.
GST pulldown assay
The DNA fragment of MoCDIP4ΔSP was cloned into the vector pMAL-c2 to generate the MoCDIP4ΔSP-MBP fusion protein. OsDjA9 was constructed in pGEX-6p-1 vector to make the OsDjA9-GST fusion protein. MoCDIP4ΔSP-MBP, GST, and OsDjA9-GST plasmids were expressed in the BL21 strain of Escherichia coli and were then affinity purified with a maltose and glutathione matrix, respectively. After about 10 μg of GST or OsDjA9-GST proteins was mixed with 10 μg of MoCDIP4ΔSP-MBP proteins at 4°C for 2 hours with gentle shaking, 30 μl of prerinsed glutathione-Sepharose 4B beads (GE Healthcare) was added, and the preparation was kept at 4°C for another 2 hours. The beads were harvested and washed five times with cold GBB buffer containing 0.5 M NaCl [50 mM tris at pH 8.0, 120 mM NaCl, 0.5% NP-40, 1 mM phenylmethylsulfonyl fluoride, and 1 mM dithiothreitol (DTT)] and another two times with cold GBB buffer. Last, the beads were boiled with 1× SDS sample-loading buffer at 100°C for 5 min for immunoblot analysis.
For competitive GST pulldown assay, MoCDIP4ΔSP-MBP, OsDjA9-GST, and OsDRP1E-His plasmids were expressed in E. coli and were then affinity purified with a maltose, glutathione matrix, and Ni–nitrilotriacetic acid agarose, respectively. After about 10 μg of OsDjA9-GST and OsDRP1E-His proteins were mixed with increasing amounts of MoCDIP4ΔSP-MBP into glutathione-Sepharose 4B beads, the mixtures were incubated and washed, and then the beads were boiled for immunoblot analysis.
Luciferase complementation assay in N. benthamiana
The DNA fragments of the MoCDIP4ΔSP and OsDRP1E genes were fused to N terminus of luciferase (LUC) in the pCAMBIA-nLUC vector to produce MoCDIP4ΔSP-nLUC and OsDRP1E-nLUC constructs, respectively. Similarly, the MoCDIP4ΔSP and OsDjA9 genes were fused to C terminus of LUC in the pCAMBIA-cLUC vector to produce MoCDIP4ΔSP-cLUC and OsDjA9-cLUC. The Agrobacterium strain EHA105 carrying nLUC/cLUC fusion plasmids were equally mixed and coinfiltrated into N. benthamiana leaves. At 2 days after infiltration, leaf discs were taken and incubated with 200 μl of 100 mM d-luciferin potassium in a 96-well plate, and the luciferase activity was detected with a GloMax 96 microplate luminometer (Promega).
Rice protoplast isolation and Co-IP assays
The Co-IP assay in rice protoplasts was performed as previously described, with some modifications (37, 38). Briefly, 11-day-old etiolated seedlings were cut into 0.5-mm strips and incubated in the enzyme solution [1.0% Cellulase RS, 0.5% Macerozyme R-10, 0.6 M mannitol, 10 mM MES, 1 mM CaCl2, and 5 mM β-mercaptoethanol (pH 5.7)] for 6 hours in the dark with gentle shaking (80 rpm). After digestion, the strips were washed two times with W5 solution [154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 2 mM MES, and 5 mM glucose (pH 5.7)] and suspended in W5 solution. OsDjA9-4*HA plasmid (39) was cotransfected with OsDRP1E-GFP plasmid or empty vector (GFP) into 100 μl of rice protoplasts using the PEG4000-mediated transfection method (40). After 24 hours of incubation, the total proteins were extracted with native extraction buffer (50 mM tris-MES at pH 8.0, 0.5 M sucrose, 1 mM MgCl2, 10 mM EDTA, 5 mM DTT, and protease inhibitor cocktail) and incubated with 25 μl of prewashed GFP-Trap_A beads (ChromoTek) at 4°C for 2 hours. The beads were then washed three times with ice-cold dilution buffer. Last, the beads were boiled with 1× SDS sample-loading buffer at 100°C for 5 min for immunoblot analysis.
For competitive Co-IP assay, OsDjA9-4*cMyc and OsDRP1E-GFP plasmids were coexpressed with increasing amounts of MoCDIP4ΔSP-4*HA plasmids in NPB rice protoplasts. The expressed proteins were immunoprecipitated with anti-cMyc antibody, and crude and immunoprecipitated proteins were then analyzed with anti-HA (hemagglutinin), anti-cMyc, or anti-GFP antibodies.
Subcellular localization in rice protoplasts
The DNA fragments of the MoCDIP4ΔSP and OsDjA9 genes were fused to C terminus of GFP or RFP. HDEL was used as an ER marker (28), and COX4 (cytochrome c oxidase subunit 4) was used as the mitochondria marker (41). Transfection of the indicated GFP and RFP fusion plasmids into rice protoplasts was performed as described earlier, and the transfected protoplasts were observed with a confocal laser scanning microscope (Zeiss LSM Confocal, Oberkochen, Germany) at 24 hours after transfection.
Subcellular localization in N. benthamiana
The different GFP, RFP, or CFP fusion constructs were transformed into Agrobacterium strain EHA105 via electroporation and then infiltrated into N. benthamiana leaves. The transfected leaves were observed with a confocal microscope at 2 days after infiltration.
Cell fractionation assays
All cellular components were isolated as previously described (42). The solutions, tubes, and homogenizer were prechilled on ice. All centrifugation steps were at 4°C. The fresh leaves were ground in liquid nitrogen using a mortar and pestle and then suspended in 50 ml of homogenization buffer [0.3 M sucrose, 25 mM tetrasodium pyrophosphate, 1% polyvinylpyrrolidone-40, 2 mM EDTA, 10 mM KH2PO4, 1% bovine serum albumin, 20 mM sodium l-ascorbate, 1 mM DTT, and 5 mM cysteine (pH 7.5)]. The homogenized solution was passed through two layers of Miracloth and centrifuged two times at 1500g for 5 min; the resulting supernatant was then centrifuged at 20,000g for 15 min to separate the soluble fraction and crude mitochondria pellet. The soluble fraction, which contained cytosol and light membranes such as ER, was recentrifuged at 20,000g for 10 min and 100,000g for 60 min. The resultant supernatant was referred to as the soluble cytosol fraction, and the pellet was referred to as the membrane fraction. The crude mitochondria pellet was further resuspended in 2 ml of washing buffer [0.3 M sucrose and 10 mM N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid (pH 7.5)] and then gently layered over a discontinuous Percoll gradient composed of 1.4 ml of 18% Percoll, 7 ml of 27% Percoll, and 1.4 ml of 50% Percoll. The Percoll gradient was centrifuged at 40,000g for 45 min in a swinging-bucket rotor. The mitochondrial band was carefully collected from the interphase between 27 and 50% Percoll and was then diluted with washing buffer and centrifuged at 20,000g for 10 min. After this washing step was repeated two times, the final pellet was resuspended with washing buffer containing the protease inhibitor cocktail.
ROS detection
The ROS detection method was described previously (27). Leaves from 5-week-old rice plants were cut into 4-mm-diameter disks and incubated in distilled water for 12 hours in the dark. After the leaf disks were placed on a paper towel to remove excess water, they were submerged in 1.5-ml microcentrifuge tubes (three disks per tube) containing 100 μl of luminol; 1 μl of horseradish peroxidase; and 1 μl of distilled water (which served as the control), 10 μM flg22, or 0.8 μM chitin. The tube was immediately put in a GloMax 20/20 luminometer (Promega) to record the luminescence at 1-min intervals for 30 min.
Transcriptional analysis using qRT-PCR
Total RNA was extracted with the UNlQ-10 Column TRIzol Total RNA Isolation Kit (Sangon Biotech) according to the manufacturer’s protocol. Approximately 2 μg of RNA in a 20-μl reaction volume was treated with deoxyribonuclease I and used for cDNA synthesis using the TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix Kit (TransGen Biotech). With 1.0 μl of 1:5 diluted cDNA as the template, qRT-PCR was performed in a 20-μl reaction volume with SYBR Green I Supermix buffer in an ABI Prism 7500 PCR instrument. Gene expression levels were calculated on the basis of three technical replications.
Transmission electron microscopy
Ultrathin slices were prepared at the Center of Biomedical Analysis, Tsinghua University. Briefly, 6-week-old rice leaves (the second leaves from the top) were cut into 1-mm2 slices and fixed overnight with 2% paraformaldehyde and 2.5% glutaraldehyde in sodium phosphate buffer (pH 7.2) at 4°C. After three washes with 0.1 M phosphate buffer, samples were incubated with 1% osmium tetroxide in phosphate buffer for 1 hour, washed with phosphate buffer three times, and dehydrated with increasing concentrations of ethanol (50 to 100%) and acetone. Samples were progressively infiltrated in the different concentrations of acetone and resin, embedded in a polymerizing epon and resin, and lastly sectioned into ultrathin slices. The images were observed with a TEM (H-7650B, Hitachi Ltd.). The lengths of mitochondria were measured using ImageJ software.
Yeast signal sequence trap system
The yeast signal trap system was used for functional verification of predicted signal peptide of MoCDIP4 as previously described (43). The vector pSUC2T7M13ORI (pSUC2) contains a truncated invertase gene, SUC2, which lacks both the initiation Met and signal peptide. DNA encoding the predicted signal peptide of MoCDIP4 was synthesized by TsingKe (Beijing) and cloned into pSUC2 using EcoRI and Xho I enzyme sites; the derived construct was then transformed into the invertase negative yeast strain YTK12 by the lithium acetate method. Avr1b-pSUC2 and Mg87-pSUC2 were used as a positive and negative control, respectively (44). YTK12 and the transformants containing pSUC2-derived plasmids were grown on CMD-W plates (0.67% yeast N base without amino acids, 0.075% W dropout supplement, 2% sucrose, 0.1% glucose, and 2% agar) to ensure expression of the pSUC2-derived plasmids. The transformants were also plated on the YPRAA [1% yeast extract, 2% peptone, 2% raffinose, and antimycin A (2 mg/ml)] to detect invertase secretion.
Fungal transformation and pathogenesis analysis
MoCDIP4 knockout mutants in the strain Guy11 were generated by replacing the open reading frame of MoCDIP4 with an HPT gene (hygromycin-resistance gene) using a fusion PCR–based, split-marker deletion method (45). N terminus and C terminus of HPT were fused by PCR with the 2-kb upstream sequence and 2-kb downstream region of MoCDIP4 coding region, respectively. These two amplified fragments were cotransformed into the protoplasts of Guy11 strain by a PEG-mediated approach as described previously (46). Selection and confirmation of the MoCDIP4 knockout mutants were performed on the basis of resistance to hygromycin B and PCR screening.
The MoCDIP4 coding region driven under the control of the native promoter was cloned into the pKNGT vector and transformed into MoCDIP4 knockout mutant (45). Transformants were selected on the geneticin plates. MoCDIP4 transcript levels in the complementation strain were confirmed by RT-PCR. We used spray inoculation to evaluate the virulence of MoCDIP4 knockout mutant and complementation strain as described earlier.
Statistical analysis
Statistical parameters are reported in the figures and figure legends. qRT-PCR, fungal biomass, and ROS assay were usually calculated from three replicates, and bars represent means ± SD. Statistically significant differences were determined by two-tailed Student’s t test: *P < 0.05, **P < 0.01, and ***P < 0.001. Statistical analyses were done using Microsoft Excel 2016 and GraphPad Prism 6. Image analyses were generated from ImageJ version 1.49.
Supplementary Material
Acknowledgments
We thank Y. Liu from South China Agricultural University for providing CRISPR-Cas9 plasmids and technical assistance and J. Jin from the Institute of Botany, Chinese Academy of Sciences, for helpful suggestions in protein subcellular localization. Funding: This study was supported by the Natural Science Foundation of China (31671984, 31471737, and 31601591) and the National Transgenic Project (2012ZX08009001). Author contributions: G.X., G.-L.W., and X.W. designed the experiments. G.X., X.Z., Y.S., Z.L. N.J., J.L., B.D., Z.L., H.K., Y.N., W.L., and Z.G. performed the experiments. G.X., G.-L.W., and X.W. wrote the paper. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The nucleotide and protein sequences of MoCDIP4, OsDjA9, and OsDRP1E are downloaded from the databases called Magnaporthe oryzae 70-15–Ensembl Genomes 45 and Rice Genome Annotation Project. Correspondence and requests for materials should be addressed to G.-L.W.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/48/eabb7719/DC1
REFERENCES AND NOTES
- 1.O’Neill L. A. J., Pearce E. J., Immunometabolism governs dendritic cell and macrophage function. J. Exp. Med. 213, 15–23 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angajala A., Lim S., Phillips J. B., Kim J.-H., Yates C., You Z., Tan M., Diverse roles of mitochondria inimmune responses: Novel insights into immuno-metabolism. Front. Immunol. 9, 1605 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mohanty A., Tiwari-Pandey R., Pandey N. R., Mitochondria: The indispensable players in innate immunity and guardians of the inflammatory response. J. Cell Commun. Signal. 13, 303–318 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castanier C., Garcin D., Vazquez A., Arnoult D., Mitochondrial dynamics regulate the RIG-I-like receptor antiviral pathway. EMBO Rep. 11, 133–138 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Onoguchi K., Onomoto K., Takamatsu S., Jogi M., Takemura A., Morimoto S., Julkunen I., Namiki H., Yoneyama M., Fujita T., Virus-infection or 5′ppp-RNA activates antiviral signal through redistribution of IPS-1 mediated by MFN1. PLOS Pathog. 6, e1001012 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu C.-Y., Liang J.-J., Li J.-K., Lee Y.-L., Chang B.-L., Su C.-I., Huang W.-J., Lai M. M. C., Lin Y.-L., Dengue virus impairs mitochondrial fusion by cleaving mitofusins. PLOS Pathog. 11, e1005350 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colombatti F., Gonzalez D. H., Welchen E., Plant mitochondria under pathogen attack: A sigh of relief or a last breath? Mitochondrion 19 Pt B, 238–244 (2014). [DOI] [PubMed] [Google Scholar]
- 8.van der Bliek A. M., Shen Q., Kawajiri S., Mechanisms of mitochondrial fission and fusion. Cold Spring Harb. Perspect. Biol. 5, a011072 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smirnova E., Griparic L., Shurland D. L., van der Bliek A. M., Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell 12, 2245–2256 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujimoto M., Arimura S.-i., Mano S., Kondo M., Saito C., Ueda T., Nakazono M., Nakano A., Nishimura M., Tsutsumi N., Arabidopsis dynamin-related proteins DRP3A and DRP3B are functionally redundant in mitochondrial fission, but have distinct roles in peroxisomal fission. Plant J. 58, 388–400 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Jin J. B., Bae H., Kim S. J., Jin Y. H., Goh C.-H., Kim D. H., Lee Y. J., Tse Y. C., Jiang L., Hwang I., The Arabidopsis dynamin-like proteins ADL1C and ADL1E play a critical role in mitochondrial morphogenesis. Plant Cell 15, 2357–2369 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang D., Ade J., Frye C. A., Innes R. W., A mutation in the GTP hydrolysis site of Arabidopsis dynamin-related protein 1E confers enhanced cell death in response to powdery mildew infection. Plant J. 47, 75–84 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Z., Ding B., Zhou X., Wang G.-L., The rice dynamin-related protein OsDRP1E negatively regulates programmed cell death by controlling the release of cytochrome c from mitochondria. PLOS Pathog. 13, e1006157 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartl F. U., Bracher A., Hayer-Hartl M., Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Craig E. A., Marszalek J., How do J-proteins get Hsp70 to do so many different things? Trends Biochem. Sci. 42, 355–368 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du Y., Zhao J., Chen T., Liu Q., Zhang H., Wang Y., Hong Y., Xiao F., Zhang L., Shen Q., Liu Y., Type I J-domain NbMIP1 proteins are required for both Tobacco mosaic virus infection and plant innate immunity. PLOS Pathog. 9, e1003659 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jores T., Lawatscheck J., Beke V., Franz-Wachtel M., Yunoki K., Fitzgerald J. C., Macek B., Endo T., Kalbacher H., Buchner J., Rapaport D., Cytosolic Hsp70 and Hsp40 chaperones enable the biogenesis of mitochondrial β-barrel proteins. J. Cell Biol. 217, 3091–3108 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takashima K., Oshiumi H., Matsumoto M., Seya T., DNAJB1/HSP40 suppresses melanoma differentiation-associated gene 5-mitochondrial antiviral signaling protein function in conjunction with HSP70. J. Innate Immun. 10, 44–55 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nekrasov V., Li J., Batoux M., Roux M., Chu Z.-H., Lacombe S., Rougon A., Bittel P., Kiss-Papp M., Chinchilla D., van Esse H. P., Jorda L., Schwessinger B., Nicaise V., Thomma B. P. H. J., Molina A., Jones J. D. G., Zipfel C., Control of the pattern-recognition receptor EFR by an ER protein complex in plant immunity. EMBO J. 28, 3428–3438 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hafrén A., Hofius D., Rönnholm G., Sonnewald U., Mäkinen K., HSP70 and its cochaperone CPIP promote potyvirus infection in Nicotiana benthamiana by regulating viral coat protein functions. Plant Cell 22, 523–535 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen C.-Y., Jan C.-I., Lo J.-F., Yang S.-C., Chang Y.-L., Pan S.-H., Wang W.-L., Hong T.-M., Yang P.-C., Tid1-L inhibits EGFR signaling in lung adenocarcinoma by enhancing EGFR Ubiquitinylation and degradation. Cancer Res. 73, 4009–4019 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Zhong X., Yang J., Shi Y., Wang X., Wang G.-L., The DnaJ protein OsDjA6 negatively regulates rice innate immunity to the blast fungus Magnaporthe oryzae. Mol. Plant Pathol. 19, 607–614 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spier A., Stavru F., Cossart P., Interaction between intracellular bacterial pathogens and host cell mitochondria. Microbiol. Spectr. 7, BAI-0016-2019 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Suzuki M., Danilchanka O., Mekalanos J. J., Vibrio cholerae T3SS effector VopE modulates mitochondrial dynamics and innate immune signaling by targeting Miro GTPases. Cell Host Microbe 16, 581–591 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen S., Songkumarn P., Venu R. C., Gowda M., Bellizzi M., Hu J., Liu W., Ebbole D., Meyers B., Mitchell T., Wang G.-L., Identification and characterization of in planta-expressed secreted effector proteins from Magnaporthe oryzae that induce cell death in rice. Mol. Plant Microbe. In. 26, 191–202 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Sarkar N. K., Thapar U., Kundnani P., Panwar P., Grover A., Functional relevance of J-protein family of rice (Oryza sativa). Cell Stress Chaperones 18, 321–331 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park C.-H., Chen S., Shirsekar G., Zhou B., Khang C. H., Songkumarn P., Afzal A. J., Ning Y., Wang R., Bellizzi M., Valent B., Wang G.-L., The Magnaporthe oryzae effector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern-triggered immunity in rice. Plant Cell 24, 4748–4762 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Napier R. M., Fowke L. C., Hawes C., Lewis M., Pelham H. R., Immunological evidence that plants use both HDEL and KDEL for targeting proteins to the endoplasmic reticulum. J. Cell Sci. 102, 261–271 (1992). [DOI] [PubMed] [Google Scholar]
- 29.Lu B., Garrido N., Spelbrink J. N., Suzuki C. K., Tid1 isoforms are mitochondrial DnaJ-like chaperones with unique carboxyl termini that determine cytosolic fate. J. Biol. Chem. 281, 13150–13158 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Elwi A. N., Lee B., Meijndert H. C., Braun J. E., Kim S.-W., Mitochondrial chaperone DnaJA3 induces Drp1-dependent mitochondrial fragmentation. Int. J. Biochem. Cell Biol. 44, 1366–1376 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Kang B.-H., Busse J. S., Bednarek S. Y., Members of the Arabidopsis dynamin-like gene family, ADL1, are essential for plant cytokinesis and polarized cell growth. Plant Cell 15, 899–913 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Araki K., Nagata K., Protein folding and quality control in the ER. Cold Spring Harb. Perspect. Biol. 4, a015438 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsai J., Douglas M. G., A conserved HPD sequence of the J-domain is necessary for YDJ1 stimulation of Hsp70 ATPase activity at a site distinct from substrate binding. J. Biol. Chem. 271, 9347–9354 (1996). [DOI] [PubMed] [Google Scholar]
- 34.Li J., Zhao-Hui C., Batoux M., Nekrasov V., Roux M., Chinchilla D., Zipfel C., Jones J. D. G., Specific ER quality control components required for biogenesis of the plant innate immune receptor EFR. Proc. Natl. Acad. Sci. U.S.A. 106, 15973–15978 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu J.-X., Howell S. H., Endoplasmic reticulum protein quality control and its relationship to environmental stress responses in plants. Plant Cell 22, 2930–2942 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lopez V. A., Park B. C., Nowak D., Sreelatha A., Zembek P., Fernandez J., Servage K. A., Gradowski M., Hennig J., Tomchick D. R., Pawłowski K., Krzymowska M., Tagliabracci V. S., A bacterial effector mimics a host HSP90 client to undermine immunity. Cell 179, 205–218.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He F., Chen S., Ning Y., Wang G.-L., Rice (Oryza sativa) protoplast isolation and its application for transient expression analysis. Curr. Protoc. Plant Biol. 1, 373–383 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y., Su J., Duan S., Ao Y., Dai J., Liu J., Wang P., Li Y., Liu B., Feng D., Wang J., Wang H., A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7, 30 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He F., Zhang F., Sun W., Ning Y., Wang G.-L., A versatile vector toolkit for functional analysis of rice genes. Rice 11, 27 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoo S.-D., Cho Y.-H., Sheen J., Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Nelson B. K., Cai X., Nebenfuhr A., A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51, 1126–1136 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Zhao Y., Luo L., Xu J., Xin P., Guo H., Wu J., Bai L., Wang G., Chu J., Zuo J., Yu H., Huang X., Li J., Malate transported from chloroplast to mitochondrion triggers production of ROS and PCD in Arabidopsis thaliana. Cell Res. 28, 448–461 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jacobs K. A., Collins-Racie L. A., Colbert M., Duckett M., Golden-Fleet M., Kelleher K., Kriz R., La Vallie E. R., Merberg D., Spaulding V., Stover J., Williamson M. J., McCoy J. M., A genetic selection for isolating cDNAs encoding secreted proteins. Gene 198, 289–296 (1997). [DOI] [PubMed] [Google Scholar]
- 44.Gu B., Kale S. D., Wang Q., Wang D., Pan Q., Cao H., Meng Y., Kang Z., Tyler B. M., Shan W., Rust secreted protein Ps87 is conserved in diverse fungal pathogens and contains a RXLR-like motif sufficient for translocation into plant cells. PLOS ONE 6, e27217 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang J., Zhao X., Sun J., Kang Z., Ding S., Xu J.-R., Peng Y.-L., A novel protein Com1 is required for normal conidium morphology and full virulence in Magnaporthe oryzae. Mol. Plant Microbe Interact. 23, 112–123 (2010). [DOI] [PubMed] [Google Scholar]
- 46.Sweigard J. A., Carroll A. M., Farrall L., Chumley F. G., Valent B., Magnaporthe grisea pathogenicity genes obtained through insertional mutagenesis. Mol. Plant Microbe Interact. 11, 404–412 (1998). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/48/eabb7719/DC1


