Abstract
Background Although federal regulations mandate documentation of structured race data according to Office of Management and Budget (OMB) categories in electronic health record (EHR) systems, many institutions have reported gaps in EHR race data that hinder secondary use for population-level research focused on underserved populations. When evaluating race data available for research purposes, we found our institution's enterprise EHR contained structured race data for only 51% (1.6 million) of patients.
Objectives We seek to improve the availability and quality of structured race data available to researchers by integrating values from multiple local sources.
Methods To address the deficiency in race data availability, we implemented a method to supplement OMB race values from four local sources—inpatient EHR, inpatient billing, natural language processing, and coded clinical observations. We evaluated this method by measuring race data availability and data quality with respect to completeness, concordance, and plausibility.
Results The supplementation method improved race data availability in the enterprise EHR up to 10% for some minority groups and 4% overall. We identified structured OMB race values for more than 142,000 patients, nearly a third of whom were from racial minority groups. Our data quality evaluation indicated that the supplemented race values improved completeness in the enterprise EHR, originated from sources in agreement with the enterprise EHR, and were unbiased to the enterprise EHR.
Conclusion Implementation of this method can successfully increase OMB race data availability, potentially enhancing accrual of patients from underserved populations to research studies.
Keywords: electronic health records and systems, data quality, data completeness, clinical research informatics, data collection, recruitment
Background and Significance
To categorize race, the United States currently relies on federal Office of Management and Budget (OMB) standards. 1 2 However, critics of these standards deem them flawed and reductive, collapsing racial identity to the geographic region of a person's ancestry and ignoring social group identification factors such as religion, culture, and language. 3 While racial classifications may elide substantial distinctions within and between populations, they remain meaningful within the context of well-demonstrated differences in the prevalence, severity, and treatment of disease between racial groups, 4 5 as well as differences in how groups interact with the research enterprise, for example, less than 2% of clinical trials sponsored by the National Cancer Institute focus on any racial minority population as their primary emphasis. 6 Without accurate and comprehensive data on patient race, efforts to address these disparities are impossible to evaluate and address.
One particularly rich source of population-level demographic data are the electronic health record (EHR), which can serve as an important resource for cohort discovery and large-scale observational analyses. 7 However, despite efforts to improve the quality of data in EHR systems through the Meaningful Use incentive program, 8 quality of structured OMB race data remains poor, as many patients are missing data or have values discordant from patient self-report. 9 This has a concrete impact on the conduct of observational research or patient cohort discovery reliant on EHR data, given that patients missing structured race data may more likely be from underserved racial groups. 10
Objectives
At our institution, the enterprise EHR system contained structured OMB race data for only 51% of patients, limiting the ability of researchers to conduct population-level research on underserved populations. Accordingly, for patients currently lacking structured race data in the enterprise EHR, we developed and evaluated a method that makes OMB race values from other local structured and unstructured sources available to researchers as part of existing infrastructure for secondary use of EHR data. 7
Methods
Setting
Weill Cornell Medicine (WCM) is a multispecialty group practice with over 1,500 physicians serving more than 2 million patients at over 45 practice sites in the New York City area. WCM has an affiliation with New York-Presbyterian Hospital (NYPH), which serves as the primary inpatient and emergency setting for WCM patients.
For clinical documentation, care team members have used EpicCare Ambulatory in WCM outpatient practices since 2000 and Allscripts Sunrise Clinical Manager (SCM) in NYPH since 2007. For billing, WCM has used Epic while NYPH has used Eagle. Automated interfaces between clinical and billing systems have shared data on the basis of a common medical record number (MRN) for each patient. In October 2020, all WCM and NYPH workflows will migrate to a single enterprise EpicCare EHR system, henceforth defined as the “target EHR.”
Race Data Supplementation Method
For all patients existing in the target EHR system as of December 2019, we obtained available patient race databased on MRNs shared in four local sources. Three of these sources—inpatient EHR system, inpatient billing system, and Logical Observation Identifiers Names and Codes (LOINC)-coded clinical observations of race associated with genetic counseling panels 11 further described in Appendix A —contained structured patient registration data recorded by either the patient, registrar, or provider. The final source, the output of a natural language processing (NLP) pipeline on outpatient EHR notes, contained race information extracted from unstructured clinical text. Our NLP pipeline, built using the Apache Unstructured Information Management Architecture-based Leo system, was validated by a manual review of 400 notes and achieved precision of 0.885, recall of 0.939, and F score of 0.911 for classifying black patients. 10 We have described details of the rule-based approach to extract race and ethnicity entities 10 and made the code available at https://github.com/wcmc-research-informatics/CIREX .
Race values from data sources were standardized to OMB categories 2 : American Indian or Alaska native; Asian, black, or African American; “other combinations not described;” native Hawaiian or other Pacific Islander, and white as described in Appendix B . We defined a patient to be missing race information if the structured race field in the target EHR was “unknown” (i.e., no value was entered) or “declined” (i.e., patient chose to decline providing race and/or clinic staff documented “declined”). For each patient missing race information in the target EHR, we attempted to supplement an OMB race value into existing infrastructure for secondary use of EHR data from the four sources in a cascading order shown below in Fig. 1 . Race values in the target EHR system were not updated, respecting patient declinations even if patients provided data elsewhere.
Fig. 1.
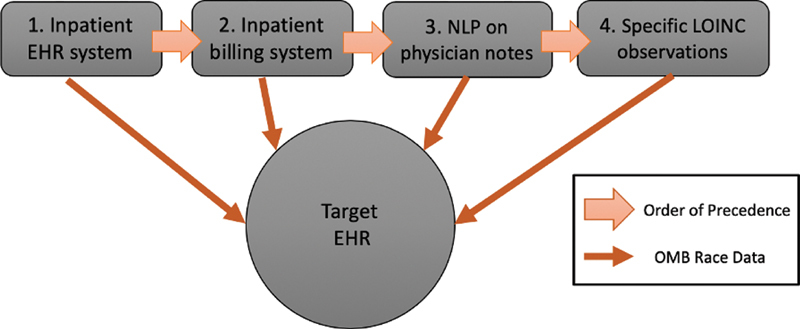
Order of precedence for race data supplementation method.
Evaluation
We evaluated our method with regard to data availability and data quality. To measure data availability, we calculated the percent change in OMB race databased on the number of patients in each race category before and after method implementation. Because the data quality of the backfill method is a direct result of the local sources' data quality, we measured each source on three of the five dimensions proposed by Weiskopf and Wang 12 —completeness, concordance, and plausibility—excluding two as discussed in Appendix C ( Fig. 2 ). Each of our data quality dimensions relied on the presence of a gold standard—which for the purposes of this study—was the target EHR's OMB race value, as we could not verify a patient's race value without access to patient self-reported data. To assess completeness, the presence of data, we quantified the number of patients with OMB race information in each source. To assess concordance, agreement between sources, we measured the agreement of OMB race values between the target EHR and the local source of interest. To assess plausibility or believability of the data, we conducted Chi-squared tests of independence between each local source's OMB race distribution and the target EHR's OMB race distribution, as well as a Chi-squared test of independence between the original target EHR's OMB race distribution and the distribution of race values yielded by the supplementation method. We conducted all analyses in Python 2.7 using pandas (0.25.3), NumPy (1.18.1), and SciPy (1.4.1) modules.
Fig. 2.
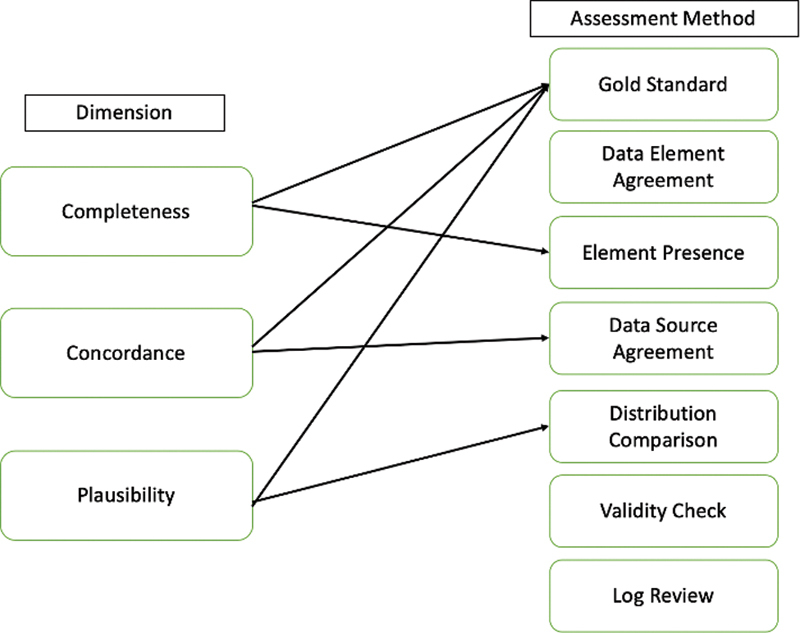
Weiskopf data quality framework applied to race data.
Results
As shown in Table 1 , the supplementation method increased the availability of structured OMB race values for patients from 51 to 55%. The three OMB race categories—black or African American, native Hawaiian/other Pacific Islander, and white—experienced the greatest change in data availability at 10%. In total, 28% (39,768) of patients with supplemented race values were from minority OMB race categories.
Table 1. Effect of race data supplementation method on office of management and budget race categories.
| Race | Pre-method | Post-method | Percent change |
|---|---|---|---|
| American Indian or Alaska native | 6,229 (<1%) | 6,570 (<1%) | 341 (6%) |
| Asian | 216,712 (13%) | 233,639 (13%) | 16,927 (8%) |
| black or African American | 220,536 (13%) | 242,165 (13%) | 21,629 (10%) |
| native Hawaiian/other Pacific Islander | 8,631 (1%) | 9,502 (1%) | 871 (10%) |
| “other combinations not described” | 409,760 (24%) | 429,501 (23%) | 19,741 (5%) |
| white | 819,288 (49%) | 901,792 (50%) | 82,504 (10%) |
| Total | 1,681,156 (51%) | 1,823,169 (55%) | 142,013 (9%) |
Regarding data quality, first we assessed completeness, determining that race data availability in the four local sources ranged from 7 to 78% in the following order: 7% in NLP on physician notes ( n = 1,885,390), 64% in the inpatient billing system ( n = 1,172,903), 66% in the inpatient EHR system ( n = 1,253,924), and 78% in LOINC observations ( n = 71,065).
Second, in measuring concordance, we observed near-perfect source agreement (98%) between the target EHR and inpatient EHR, as well as between the target EHR and inpatient billing system (97%). NLP on physician notes and LOINC observations had lower concordance at 81 and 77%, respectively. Among OMB race categories, White patients had the highest overall concordance across each of the local sources with the target EHR system, ranging from 90 to 99%. In comparison, patients recorded as American Indian or Alaska native, native Hawaiian/other Pacific Islander, and “other combinations not described” had much lower concordance between the target EHR and local systems.
Finally, we confirmed plausibility, or believability of the supplemented data, as the Chi-squared tests of independence revealed no statistically significant difference between the local source's OMB race distribution and target EHR's OMB race distribution. Similarly, the Chi-squared test of independence between the target EHR race distribution and the distribution of race values obtained via the supplementation method yielded a p- value of 0.99, indicating no statistically significant difference as demonstrated in Fig. 3 .
Fig. 3.
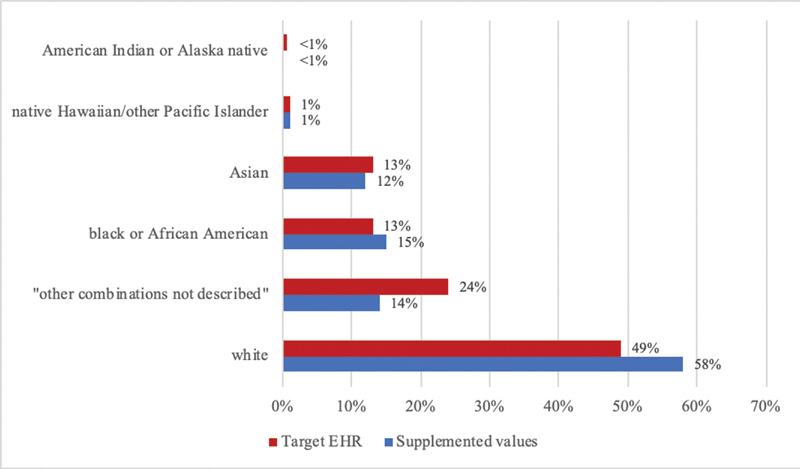
Race distributions of patients with a supplemented office of management and budget race value versus target electronic health record.
Discussion
Our supplementation method increased structured race data availability in the target EHR from 51 to 55%, populating OMB race data for 142,013 patients. The method made race data available for nearly 40,000 patients from minority OMB race categories and made the greatest impact on race data availability for black or African American (10%) and native Hawaiian/other Pacific Islander (10%) patients. Despite this improvement in data availability, race data are still only present for 55% (1.8 million) of patients in the target EHR system, suggesting that despite efforts to supplement race information from other sources, substantial gaps in data persist that may hinder clinical research.
Our data quality analysis demonstrated that race values gathered from local sources could supplement race information present in the target EHR system. The supplemented race values had a positive effect on completeness, came from sources concordant with the target EHR system, and were relatively unbiased from original race values in the target EHR system. This suggests that the implementation of this method can provide quality race data that can potentially bolster clinical trial accrual in underserved populations, improve the accuracy of population-scale disparities research, and help researchers further stratify patient cohorts for multiple scientific workflows.
The findings of our evaluation on this race data supplementation method elaborate on the findings of similar investigations on the quality of race and ethnicity data, which found that race data quality issues were prevalent in EHR systems. In one study, researchers reported that 49% of patients did not have an identified OMB race value, with evidence of improvement in race data collection after the implementation of Meaningful Use in 2011. 13
The method could potentially enhance the capture of other demographic variables captured in EHR systems such as ethnicity. 14 However, the method proved ineffective for ethnicity data at our institution, as ethnicity was not well captured within our source systems. Only 779 of the 1.2 million patients in the inpatient EHR system had an ethnicity value, all of which were Hispanic or Latino or Spanish Origin.
This study has limitations. First, because this is a single-site study, it is unknown whether results will generalize to other institutions. However, as many hospitals and health systems transition to a single enterprise EHR, 15 they may benefit from implementation of a similar method to bolster race data availability. Second, each local source maintained distinct and varying categories for mapping race data, making data standardization difficult. To solve this challenge, we focused specifically on OMB race categories, which resulted in loss of some meaningful race data due to lack of granularity of the categories. While other race categories— such as the CDC/HL7 race and ethnicity code set—can be more expressive, our analysis did not explore these race breakdowns per the current federal mandate. Third, our data quality evaluation assumed all OMB race values documented in the target EHR were correct. Without a source of patient self-report, we could not validate the OMB race value for each patient, and we assumed that the value in the target EHR was true. Fourth, we had no insight into the local sources' documentation workflow other than we know it is varied. Thus, it is unclear whether specific OMB race values were self-reported by the patient or documented by a registrar or provider, posing challenges on determining why sources were more concordant or plausible. Finally, in addressing the ethical implications of supplementing race for patients who declined in the enterprise EHR, we view the method to be democratizing access to race data already available in each local source.
Although the current standards for race categorization are problematic, these groupings remain meaningful in evaluating and addressing health disparities. As academic medical centers strive to provide researchers with tools to use EHR data for research, results from implementing this race data supplementation method may yield insight on how to make use of race data from local EHR sources. However, changes to front-end data entry workflows, such as a patient-facing self-report tool, may be necessary to more directly address race data quality issues. 16 Despite these challenges, our supplementation method enhanced the availability of race data for patients at our institution, especially for those within racial minority groups.
Conclusion
This study demonstrated a method that effectively made race data available from different local systems while maintaining quality data. Using this method, institutions can potentially make OMB race data more available for population-level research on racial minority groups.
Clinical Relevance Statement
This work is pertinent to our community as academic medical institutions are increasingly making use of EHR data for research. As care is moved to a single enterprise EHR, informaticians should also make data available from local legacy sources, particularly for data elements known for poor capture, such as race and ethnicity.
Multiple Choice Questions
1. What type of research benefits from improving Office of Management and Budget (OMB) race data availability and quality available to researchers as a part of existing infrastructure for secondary use of electronic health record data?
Health disparities research
Screening research
Clinical trials
All of the above
Correct Answer: The correct answer is option d (all of the above). Although critics argue that OMB race categories are flawed and reductive, many researchers have found that they do remain meaningful in the context of well-demonstrated differences in the prevalence, severity, and treatment of disease between racial groups, as well as differences in how groups interact with the research enterprise. Thus, not only does this benefit health disparities research, it will also benefit study of screening and clinical trials to ensure that minority groups receive equitable treatment.
2. What is a known issue limiting capture of race data in electronic health record (EHR) systems?
Federal mandate to capture race
Implementation of patient self-report tools
Standardized categories across EHR systems
Use of electronic capture
Correct Answer: The correct answer is option c (standardized categories across EHR systems). Despite the federal mandate to capture race according to the OMB race categories, many EHR systems do not adhere to these categorizations and report other race categories, such as “Middle Eastern” or “Asian Indian.” The federal mandate to capture race, implementation of patient self-report tools, and use of electronic capture should help to improve race data quality in EHR systems.
Appendix B: Race Data Standardization and Coding Errors.
Patients categorized as Asian Indian were included within the Asian category. Patients categorized as Middle Eastern or North African were included within the White category. Patients categorized as mixed were included within the “other combinations not described” category. Patients categorized as Jewish, Ashkenazi Jewish, and Sephardic Jewish are categorized as “unknown.” Patients with Office of Management and Budget (OMB) ethnicity values recorded in their structured race field (e.g., those categorized as Hispanic, Latino, and South American) were categorized as “other combinations not described.” Any patients with two different race categories listed (e.g., Asian and White) were categorized as “other combinations not described.” In addition to data standardization, several coding errors were encountered. Numerous genetic counseling panels encoded as LOINC observations intended to encode OMB ethnicity values contained OMB race values and vice versa. For this reason, both types of LOINC observations were included.
Funding Statement
Funding This study received support from New York-Presbyterian Hospital and Weill Cornell Medical College, including the Clinical and Translational Sciences Center (ULI TR000457) and Joint Clinical Trials Office.
Conflict of Interest None declared.
Protection of Human and Animal Subjects
The study was performed in compliance with the “Federal Policy for the Protection of Human Subjects” by the U.S. Department of Health and Human Services and was reviewed by the WCM Institutional Review Board.
Appendix A: Logical Observation Identifiers Names and Codes.
We used the following Logical Observation Identifiers Names and Codes (LOINC) identifiers: 21484–1, 32624–9, 42784–9, and 32624–9. These observations contain structured patient registration data from genetic counseling panels. They correspond to the following names in our enterprise electronic health record system: “race/ethnicity, mother,” race/ethnicity, patient,” and “ethnicity.” In many cases, we found that race data were incorrectly reported as ethnicity.
Appendix B: Race Data Standardization and Coding Errors.
Patients categorized as Asian Indian were included within the Asian category. Patients categorized as Middle Eastern or North African were included within the White category. Patients categorized as mixed were included within the “other combinations not described” category. Patients categorized as Jewish, Ashkenazi Jewish, and Sephardic Jewish are categorized as “unknown.” Patients with Office of Management and Budget (OMB) ethnicity values recorded in their structured race field (e.g., those categorized as Hispanic, Latino, and South American) were categorized as “other combinations not described.” Any patients with two different race categories listed (e.g., Asian and White) were categorized as “other combinations not described.” In addition to data standardization, several coding errors were encountered. Numerous genetic counseling panels encoded as LOINC observations intended to encode OMB ethnicity values contained OMB race values and vice versa. For this reason, both types of LOINC observations were included.
Appendix C: Exclusion of Two Dimensions from Weiskopf and Wang Data Quality Framework.
Two of the five data quality dimensions, correctness and currency, cannot be applied to race information. 12 The correctness of race information cannot be verified without patient self-report. In addition, the currency of race information was deemed not relevant, as we assumed a patient's race would be consistent throughout their lifetime.
References
- 1.Commonwealth Fund Who, when, and how: the current state of race, ethnicity, and primary language data collection in hospitals Commonwealth Fund. Available at: https://www.commonwealthfund.org/publications/fund-reports/2004/may/who-when-and-how-current-state-race-ethnicity-and-primary. Accessed July 15, 2020 [Google Scholar]
- 2.Office of Management and Budget . Recommendations from the interagency committee for the review of the racial and ethnic standards to the office of management and budget concerning changes to the standards for the classification of federal data on race and ethnicity. Fed Regist. 1997;62(131):36873–36945. [Google Scholar]
- 3.Burchard E G, Ziv E, Coyle N. The importance of race and ethnic background in biomedical research and clinical practice. N Engl J Med. 2003;348(12):1170–1175. doi: 10.1056/NEJMsb025007. [DOI] [PubMed] [Google Scholar]
- 4.Vyas D A, Eisenstein L G, Jones D S. Hidden in plain sight — reconsidering the use of race correction in clinical algorithms. N Engl J Med. 2020;383(09):874–882. doi: 10.1056/NEJMms2004740. [DOI] [PubMed] [Google Scholar]
- 5.Risch N, Burchard E, Ziv E, Tang H. Categorization of humans in biomedical research: genes, race and disease. Genome Biol. 2002;3(07):t2007. doi: 10.1186/gb-2002-3-7-comment2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen M S, Jr, Lara P N, Dang J HT, Paterniti D A, Kelly K. Twenty years post-NIH Revitalization Act: enhancing minority participation in clinical trials (EMPaCT): laying the groundwork for improving minority clinical trial accrual: renewing the case for enhancing minority participation in cancer clinical trials. Cancer. 2014;120 07:1091–1096. doi: 10.1002/cncr.28575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sholle E T, Kabariti J, Johnson S B. Secondary use of patients' electronic records (SUPER): an approach for meeting specific data needs of clinical and translational researchers. AMIA Annu Symp Proc. 2018;2017:1581–1588. [PMC free article] [PubMed] [Google Scholar]
- 8.Blumenthal D, Tavenner M. The “meaningful use” regulation for electronic health records. N Engl J Med. 2010;363(06):501–504. doi: 10.1056/NEJMp1006114. [DOI] [PubMed] [Google Scholar]
- 9.Polubriaginof F CG, Ryan P, Salmasian H.Challenges with quality of race and ethnicity data in observational databases J Am Med Inform Assoc 201926(8-9):730–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sholle E T, Pinheiro L C, Adekkanattu P.Underserved populations with missing race ethnicity data differ significantly from those with structured race/ethnicity documentation J Am Med Inform Assoc 201926(8-9):722–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McDonald C J, Huff S M, Suico J G. LOINC, a universal standard for identifying laboratory observations: a 5-year update. Clin Chem. 2003;49(04):624–633. doi: 10.1373/49.4.624. [DOI] [PubMed] [Google Scholar]
- 12.Weiskopf N G, Weng C. Methods and dimensions of electronic health record data quality assessment: enabling reuse for clinical research. J Am Med Inform Assoc. 2013;20(01):144–151. doi: 10.1136/amiajnl-2011-000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Polubriaginof F, Boland M R, Perotte A, Vawdrey D.Quality of race and ethnicity data in electronic health records. AMIAAvailable at: https://knowledge.amia.org/amia-59309-cri2016-1.3011827/t004-1.3012641/f004-1.3012642/a097-1.3012734/a099-1.3012729?qr=1. Accessed 2016
- 14.National Dental PBRN Collaborative Group . Thyvalikakath T P, Duncan W D, Siddiqui Z. Leveraging electronic dental record data for clinical research in the national dental PBRN practices. Appl Clin Inform. 2020;11(02):305–314. doi: 10.1055/s-0040-1709506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koppel R, Lehmann C U. Implications of an emerging EHR monoculture for hospitals and healthcare systems. J Am Med Inform Assoc. 2015;22(02):465–471. doi: 10.1136/amiajnl-2014-003023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polubriaginof F, Salmasian H, Shapiro A W.Patient-provided Data Improves Race and Ethnicity Data Quality in Electronic Health Records. AMIAAvailable at: https://knowledge.amia.org/amia-59309-cri2016-1.3011827/t004-1.3012641/f004-1.3012642/a097-1.3012734/a099-1.3012729?qr=1. Accessed 2016


