Dear Editor,
Emerging studies focusing on severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) detection have aroused worldwide interest and attention. A recent study reported by Azzi et al. which demonstrated that saliva was a reliable tool for SARS-CoV-2 detection, and further validated separately by Iwasaki and Zhu, have provided promising evidence for dependable test source.1, 2, 3 To better validate the potential of saliva as an alternative specimen for coronavirus disease 2019 (COVID-19) diagnosis, especially in low viral load samples, we recruited 35 recovered COVID-19 inpatients who showed no symptoms and long-term positive nucleic acid in the respiratory tract and totally collected 183 specimens of saliva, sputum, nasopharyngeal swab (NPS), oropharyngeal swab (OPS), anal swab, and feces for reverse transcriptase digital polymerase chain reaction (RT-dPCR) evaluation. Our results confirmed that saliva is a self-collectable and reliable specimen source for accurate SARS-CoV-2 carrier screening.
SARS-CoV-2 is the novel coronavirus causing COVID-19 and has high transmissibility and distribution worldwide, making accurate and efficient SARS-CoV-2 detection for potential infected patients crucial in guiding effective treatment and public infection control. Sputum, NPS, OPS, anal swab, and feces have been collected for SARS-CoV-2 detection. For various drawbacks of these specimen collections, like discomfort caused to patients, risk to healthcare workers while accessing, and also being self-undoable, saliva is being considered as a promising alternative.1 And detection performances of saliva have been reported, such as high/low viral load,4 , 5 or less sensitive comparing to NPS.6 These differences exist because RT-qPCR and RT-dPCR are mostly accurate for high viral load specimens, whereas for low viral load specimens, RT-dPCR performs better than RT-qPCR.7 And ORF1ab/RdRp, E, S, or N genes detection using clinical specimens is currently the gold standard.8 Moreover, both Zhu and Iwasaki et al. reported that the viral load tested in saliva was identical at earlier stages but reduced to lower levels especially during recovering phase.2 , 3
Therefore, in our study, we aim to further validate the feasibility of saliva collection for COVID-19 diagnosis, especially in low viral load populations, which is a group of recovered COVID-19 inpatients who showed no symptoms and long-term positive nucleic acid in the respiratory tract. Thirty-five patients including 16 males and 19 females with a median age of 65 years old (IQR: 57–75.5) enrolled in this study, and 183 specimens (saliva: 36, sputum: 24, NPS: 28, OPS: 46, anal swab: 35, and feces: 14) were collected.
All 183 specimens have been processed for RT-dPCR detection by targeting ORF1ab, E, and N genes of SARS-CoV-2.9 The positive rates of SARS-CoV-2 detection in saliva, sputum, NPS, OPS, anal swab, and feces were 86%, 79%, 93%, 83%, 64%, and 36%, respectively. These results showed that NPS had the highest positive rate among the six types of specimens, followed by saliva detection that had slightly lower rate than NPS. This finding suggested that SARS-CoV-2 detection using NPS and saliva specimens had high screening efficiencies.
PPA and NPA analyses were conducted to further determine the detection performance of saliva in comparison with three most commonly used respiratory tract specimens, namely, OPS, NPS, and sputum. Among the 27 specimen pairs of saliva and OPS analyzed, 17 were positive, 2 were negative, 3 were positive in saliva but negative in OPS, and 5 were positive in OPS but negative in saliva. The PPA and NPA of saliva were 85.0% (95% CI 64.0%–94.8%) and 28.6% (95% CI 8.2%–64.1%), respectively (Table 1 ). Same comparisons were conducted between saliva and NPS (16 specimen pairs), saliva and sputum (20 specimen pairs) with result listed (Table 1). Of note, saliva had higher detection performance and less false negative than NPS, OPS, and sputum. Among the 21 individuals, the viral loads in the saliva for ORF1ab, E, and N gene detection were above the detection limit of RT-dPCR. These findings indicated that saliva has good detection performance and might avoid false negatives compared with other specimens for SARS-CoV-2 detection.
Table 1.
The comparison for the SARS-CoV-2 detection between saliva and OPS, NPS or sputum.
| OPS |
||||||
|---|---|---|---|---|---|---|
| Positive | Negative | Total | PPA | NPA | ||
| Saliva | Positive | 17 | 5 | 22 | 85.0% (95% CI 64.0%−94.8%) | 28.6% (95% CI 8.2%−64.1%) |
| Negative | 3 | 2 | 5 | |||
| Total | 20 | 7 | 27 | |||
| NPS | Total | PPA | NPA | |||
| Positive | Negative | |||||
| Saliva | Positive | 14 | 2 | 16 | 100.0% (95% CI 78.5%−100.0%) | 0.0% (95% CI 0.0%−65.8%) |
| Negative | 0 | 0 | 0 | |||
| Total | 14 | 2 | 16 | |||
| Sputum | Total | PPA | NPA | |||
| Positive | Negative | |||||
| Saliva | Positive | 12 | 4 | 16 | 80.0% (95% CI 54.8%−93.0%) | 20.0% (95% CI 3.6%−62.4%) |
| Negative | 3 | 1 | 4 | |||
| Total | 15 | 5 | 20 | |||
CI: Confidence interval.
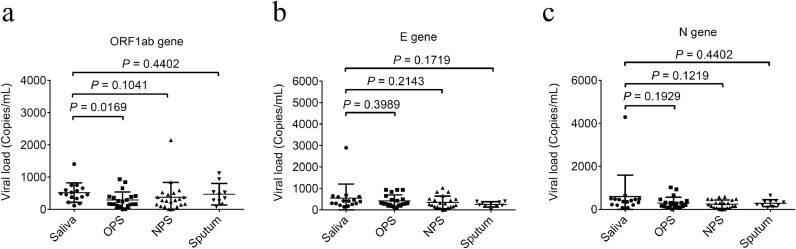
In Azzi's study, the use of the Ct values highlights a trend in viral load but does not allow a quantification of the viral copies/ml. Here, our result of viral load values of different specimens from 21 individuals showed that the viral loads of all saliva, sputum, OPS, and NPS specimens collected from recruited recovered COVID-19 inpatients were lower than 5 × 103 copies/mL (Fig. 1 ), which is below the qPCR limit.10 Although the viral load values of saliva were significantly higher than that of OPS (P = 0.0169) in the ORF1ab gene detection (Fig. 1). This finding suggested a tendency of higher viral load for saliva than for other types of specimens in SARS-CoV-2 detection, especially for OPS and NPS, which have been used as two recommended upper respiratory tract specimen types.
Fig. 1.
Viral load comparison in the specimens of saliva, sputum, OPS and NPS
Viral loads of SARS-CoV-2 in the specimens of saliva, sputum, OPS and NPS were detected by RT-dPCR targeting ORF1ab gene (a), N gene (b), and E gene (c) by RT-dPCR. The significance of difference between the sample types was analyzed by unpaired t-test using GraphPad Prism (version 6.01).
The main limitation of this study is small sample size of the recruited recovered COVID-19 inpatients, as well as low compliance of patients. And the viral load value of specimens collected from the recruited inpatients were mostly lower than 5 × 103 copies/mL, which was consistent with the detection in the specimens collected from asymptomatic SARS-CoV-2 carriers.7 However, from another perspective, this is also the highlight of our study that we recruited specific population with low viral load to prove the advantages of saliva test as an thoroughly screening of those SARS-CoV-2 carriers or recovering patients. Moreover, RT-dPCR assay used in our study revealed a high detection rate of SARS-CoV-2 in saliva. Finally, our results showed that the SARS-CoV-2 detection in saliva had the highest median viral load, which was significantly higher than that in OPS by ORF1ab genes.
In conclusion, our results implicated saliva as an alternative diagnostic specimen for SARS-CoV-2 screening, in light of its relatively high viral load, positive rate, clinical performance, reliability, stability, patient-acceptable, and healthcare worker safety.
Declaration of Competing Interest
The authors declare that they have no competing financial interests.
Acknowledgments
Ethics
This study was approved by the Medical Ethics Committee of Wuhan Infectious Disease Hospital (KY-2020-75.01) and was conducted from May 2020 to May 2021.
Acknowledgement
We thank all of the healthcare workers and patients involved in the study.
Funding
This work was funded by the Fundamental Research Funds for Central Public Welfare Scientific Research Institutes sponsored by National Institute of Metrology, PR China (No. 31-ZYZJ2001 and No. AKYYJ2009).
References
- 1.Azzi L., Carcano G., Gianfagna F., Grossi P., Gasperina D.D., Genoni A. Saliva is a reliable tool to detect SARS-CoV-2. J Infect. 2020;81:e45–e50. doi: 10.1016/j.jinf.2020.04.005. PubMed PMID: 32298676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu J., Guo J., Xu Y., Chen X. Viral dynamics of SARS-CoV-2 in saliva from infected patients. J Infect. 2020;81(3):e48–e50. doi: 10.1016/j.jinf.2020.06.059. PubMed PMID: 32593658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iwasaki S., Fujisawa S., Nakakubo S., Kamada K., Yamashita Y., Fukumoto T. Comparison of SARS-CoV-2 detection in nasopharyngeal swab and saliva. J Infect. 2020;81(2):e145–e1e7. doi: 10.1016/j.jinf.2020.05.071. PubMed PMID: 32504740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wyllie A.L., Fournier J., Casanovas-Massana A., Campbell M., Tokuyama M., Vijayakumar P. Saliva is more sensitive for SARS-CoV-2 detection in COVID-19 patients than nasopharyngeal swabs. medRxiv. 2020 2020.04.16.20067835. [Google Scholar]
- 5.Williams E., Bond K., Zhang B., Putland M., Williamson D.A. Saliva as a Noninvasive Specimen for Detection of SARS-CoV-2. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.00776-20. e00776-20 PubMed PMID: 32317257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becker D., Sandoval E., Amin A., De Hoff P., Diets A., Leonetti N. Saliva is less sensitive than nasopharyngeal swabs for COVID-19 detection in the community setting. medRxiv. 2020 2020.05.11.20092338. [Google Scholar]
- 7.Yu F., Yan L., Wang N., Yang S., Wang L., Tang Y. Quantitative Detection and Viral Load Analysis of SARS-CoV-2 in Infected Patients. Clin Infect Dis. 2020;71:793–798. doi: 10.1093/cid/ciaa345. PubMed PMID: 32221523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang B., Hu M., Ren Y., Xu X., Wang Z., Lyu X. Evaluation of seven commercial SARS-CoV-2 RNA detection kits based on real-time polymerase chain reaction (PCR) in China. Clin Chem Lab Med. 2020;58(9):e149–e153. doi: 10.1515/cclm-2020-0271. PubMed PMID: 32221523.32651975. [DOI] [PubMed] [Google Scholar]
- 9.Dong L., Zhou J., Niu C., Wang Q., Pan Y., Sheng S. Highly accurate and sensitive diagnostic detection of SARS-CoV-2 by digital PCR. medRxiv. 2020 doi: 10.1016/j.talanta.2020.121726. 2020.03.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wyllie A.L., Fournier J., Casanovas-Massana A., Campbell M., Tokuyama M., Vijayakumar P. Saliva or Nasopharyngeal Swab Specimens for Detection of SARS-CoV-2. N Engl J Med. 2020;383(13):1283–1286. doi: 10.1056/NEJMc2016359. PubMed PMID: 32857487. [DOI] [PMC free article] [PubMed] [Google Scholar]