Dear Editor,
In reference to the letter to the editor by Krupp et al.1 we have some data of interest obtained from our SARS-CoV2 culturing experience.
RT-PCR has become in the more sensitive method to detect SARS-CoV-2 infection. Amplification of genomic sequence is measured in cycle thresholds (Ct). Despite its high sensitivity and wider application, RT-PCR has an important limitation. It does not distinguish between infectious and non-infectious virus. Different studies regarding virus shedding in mild and severe COVID-19 patients during hospitalization showed that SARS-CoV-2 RNA may well be detected in the respiratory tract for up to 21, 32 and 34 days, respectively2, 3, 4.
Also, viral loads on different time courses of SARS-CoV-2 infection could show changes in disease stages. It was reported that viral load was high in the early and progressive phase of the disease and decrease gradually in the recovery phase, peaking at 4 to 6 days after onset of symptoms and gradually decline afterwards. However, initial viral load was not correlated with days after symptom onset. Nowadays the use of Ct values as direct measure of SARS-CoV-2 viral load should be taken with care because is not a standard measure of viral load in clinical samples, as it was demonstrated by Dahdouh et al.5.. Finally, the prolonged viral shedding is relevant for the control of infection in hospitals and discharge management, the correlation between detectable viral RNA and virus isolation in clinical samples remains unclear though4.
Viral transmission and infectivity are one of the most important determinants for prevention strategies in respiratory viral infections and can be substantially reduced by containment measures such as isolation and quarantine. Knowledge of SARS-CoV-2 viral shedding is of primary importance for the development of effective prevention and control measures. We investigated the infectivity of clinical samples obtained from patients with SARS-CoV-2, comparing the results obtained by RT-PCR with the growth capacity of the virus in vitro.
For all patients, 400 μL of nasopharyngeal swab fluid (NP) (UTMⓇ: Viral Transport, Copan Diagnostics, Inc., Murrieta, CA) were obtained. Samples were inactivated with 400 μL of AVL buffer (Qiagen) at a 1:1 ratio (400 μl AVL: 400 μl nasopharyngeal swab fluid) and incubated for 10 min at room temperature before nucleic acid extraction. Non-inactivated fractions were conserved at −80 °C for viral growth assays.
MagMax Express 96 (Applied Biosystems™, MA, USA) was used for nucleic acid extraction according to manufacturer's instructions. TaqMan2019-nCoV Assay Kit v1 (ThermoFisher, MA, USA) was used for detection of viral RNA.
Those samples conserved at −80 °C were re-evaluated using the same methods extraction and RT-PCR kit after a freeze-thaw cycle before viral growth assays in cell culture.
All procedures for viral culture followed the laboratory biosafety guidelines and were conducted in a biosafety level 3 facility. 250 µl of nasopharyngeal swab fluid (NP), freshly obtained or conserved at −80 °C, were inoculated in confluent Vero E6 cells monolayers (ATCC CRL-1586) in 250 µl of Minimum Essential Medium culture medium with 4% fetal calf serum and 1% glutamine. Vero E6 cells were growth in Dulbecco's Minimal Essential Medium (DMEM) supplemented with 10% fetal bovine serum and Antibiotic-Antimycotic Gibco™ solution and in a humidified 37 °C incubator with 5% CO2.
After 4 days infected cells were fixed with 10% formaldehyde in phosphate-buffered saline (PBS) for 30 min at room temperature and then stained with crystal violet and observed to confirm cytopathic effect (CPE). Culture supernatants were collected from each well, for RNA extraction and SARS-CoV-2 RT-PCR.
A cell culture is suspected of hosting virus replication based on the presence of CPE including damage to the monolayer, cell clearing and morphological changes, and if the RT-PCR was positive and the Ct was at least 2 cycles lower than that of the original sample.
In all the studied samples, the presence of other microorganism that could produce a cytopathic effect was ruled out after performing the multiplex PCR FTDTM Respiratory pathogens 21 (Fast Track Diagnostic. Siemens, Madrid, Spain).
A total of 72 specimens of NP from 66 patients were analysed in this study. Medical records for these patients regarding epidemiological and demographic characteristics, symptom history and relevant exposure data on admission were retrospectively reviewed. Sixty-six samples were positive and six were negative for SARS-CoV-2 genome by PCR. A total of five (7.57%) asymptomatic, forty-six (69.69%) mildly symptomatic and fifteen patients (22.72%) with severe pneumonia due to SARS-CoV-2 were identified. Seven patients died due to severe pneumonia by SARS-CoV-2. The average age of the patients was 52.64 years (range, 25 to 89), and 42 patients were male (63.63%).
Fever was present in 31 out of the forty-six mildly symptomatic patients (31/46, 67.39%), and cough was present in 29 of them (29/46, 63.04%).
Five patients had neither clinical symptom (5/66, 7.57%), two of them were close contacts of positive confirmed cases, two were previous positive cases of SARS-CoV-2 PCR and the fifth case was detected in a routine screening prior to hospital admission due to digestive bleeding.
Specimens collected between February and April (44 out of 72) were stored at −80 °C until being analysed for SARS-CoV-2 viral growth. Samples collected between May and June (28 out of 72) were conserved for no more of 48 h at 6 °C until virus culturing. Samples collected during the first period had lower SARS-CoV-2 targets Ct than those obtained during the second period (p = 0.0002). We obtained 17 viral isolates of SARS-CoV-2 (25.75%) from inoculated Vero E6 cells showing CPE, 14/44 (31.81%) from NP that underwent a single freeze-thaw cycle (first period) and 3/28 (10.71%) of samples without a freeze-thaw cycle (second period). The positive culture´s rate was higher in samples with a freezing-thawing cycle (P value= 0.0225). Infection by other respiratory microorganism was not detected.
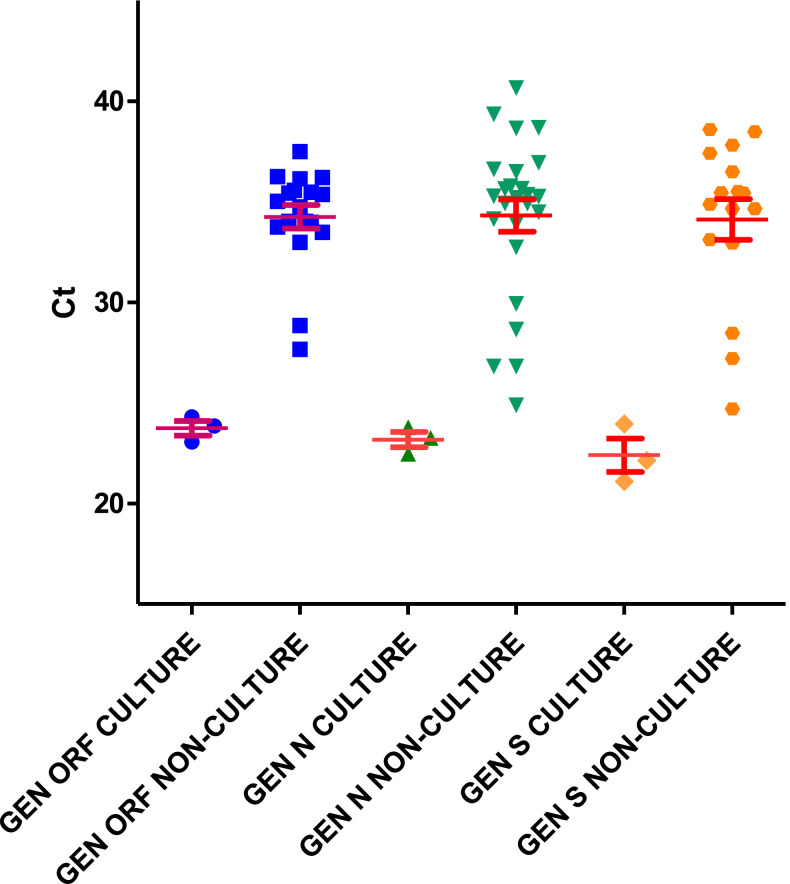
We also compared the RT-PCR Ct of the culturable and nonculturable samples. We found a significative difference between Ct of culturable and nonculturable samples (p< 0.0001). The average Ct values in the specimens with culturable virus were 25.97±4.59 for ORF1ab, 25.43±4.58 for gene N and 25.54±5.1 for gene S in samples previously freezed and thawed, and 23.69±0.88, 23.27±0.66 and 23.06±1.28, respectively, in fresh samples (Fig. 1, Fig. 2 ).
Fig. 1.
Samples without freeze-thaw.
Fig. 2.
Samples freeze-thaw.
The highest Ct value in samples with positive cultures was found to be 36.08, 37.73 and 37.41 for the ORF1ab, N and S genes, respectively in a specimen previously freeze-thawed from a woman with cough and fever. This sample had been taken one day after symptoms onset.
The average time elapsed since the onset of symptoms to sample collection was 7.05 days in patients with positive viral growth (range:0–12 days) and 13.51 days (range: 1–106 days) in patients without viral growth (p< 0.0001).A positive or negative RT-PCR result might not be enough for an accurate identification of the infectious potential of the patients. The best indicator of infectiousness is the viral culture of samples from patients with SARS-CoV-26 , 7. Different studies show that there is an inverse relationship between Ct, viral culturability and date of symptom onset8 , 9.
The duration of viral shedding in NP evaluated by RT-PCR for SARS-CoV-2 as a determining factor to recover the virus in culture. The range reported was between 7 and 35 days after symptom onset4. In our study, the maximum time after the onset of symptoms in which the virus was isolated was 12 days.
Positive cultures were associated with low Ct values. Kujawski et al. obtained viral growth from NP samples with PCR Ct values between 17.0 and 39.0. In the work of Singanayagam et al. the RT-PCR cycle threshold median values was 31.15, and the estimated probability of recovery of virus from samples with Ct>35 was 8.3%. In our series, we obtained viral growth in samples with Ct between 21.54 and 37.73 in gene N, which agrees with previously published data4 , 10 .
On the other hand, we studied the effect of storing the samples at −80 °C and the subsequent thawing on the viability of SARS-CoV-2. The results suggest that a freeze-thaw cycle did not significantly affect the positive culture rate as previously described6
In conclusion, Cts values cannot be used as unique tool to identify those patients who can be infective despite a positive SARS-CoV-2 PCR. Different studies demonstrated that viral growth could be obtained with high Cts. Our results also suggest that the moment of sample collection after onset of symptoms is an important variable that has to be taken into account.
Footnotes
SARS-CoV-2 Working Group: A. Gutiérrez-Arroyo, I. Bloise, F. Lázaro-Perona, M.I Quiles-Melero, G. Ruiz-Carrascoso, I. Falces-Romero, M. Ruiz-Bastián, C. Toro-Rueda, S. García-Bujalance, B. Gómez-Arroyo, C. García-Sánchez, V. Guedez-López, M. Gracia Liras-Hernández, M. Sánchez-Castellano, P. García-Clemente, P. González- Donapetry.
References
- 1.Krupp K., Madhivanan P., Perez-Velez C.M. Should qualitative RT-PCR be used to determine release from isolation of COVID-19 patients? J Infect. 2020;81:452–482. doi: 10.1016/j.jinf.2020.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zou L., Ruan F., Huang M. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basile K., McPhie K., Carter I. Cell-based culture of SARS-CoV-2 informs infectivity and safe de-isolation assessments during COVID-19. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singanayagam A., Patel M., Charlett A. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveill. 2020;25:465. doi: 10.2807/1560-7917.ES.2020.25.32.2001483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dahdouh E., Lázaro-Perona F., Romero Gomez M.P., Mingorance J., Garcia-Rodriguez J. Ct values from SARS-CoV-2 diagnostic PCR assays should not be used as direct estimates of viral load. J Infect. 2020 doi: 10.1016/j.jinf.2020.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang C.-.G., Lee K.-.M., Hsiao M.-.J. Culture-based virus isolation to evaluate potential infectivity of clinical specimens tested for COVID-19. J Clin Microbiol. 2020;58:270. doi: 10.1128/JCM.01068-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.La Scola B., Le Bideau M., Andreani J., et al. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. 2020;:1–3. [DOI] [PMC free article] [PubMed]
- 8.Zhou F., Fan G., Liu Z., Cao B. SARS-CoV-2 shedding and infectivity - Authors' reply. Lancet. 2020;395:1340. doi: 10.1016/S0140-6736(20)30869-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou F., Yu T., Du R. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.COVID-19 Investigation Team Clinical and virologic characteristics of the first 12 patients with coronavirus disease 2019 (COVID-19) in the United States. Nat Med. 2020;26:861–868. doi: 10.1038/s41591-020-0877-5. [DOI] [PMC free article] [PubMed] [Google Scholar]