Abstract
Somatic mutations affecting CREBBP and EP300 are a hallmark of Diffuse Large B Cell Lymphoma (DLBCL). These mutations are frequently monoallelic, within the histone acetyltransferase (HAT) domain and usually mutually exclusive, suggesting that they might affect a common pathway and their residual WT expression is required for cell survival. Using in vitro and in vivo models, we found that inhibition of CARM1 activity (CARM1i) slows DLBCL growth and that the levels of sensitivity are positively correlated with the CREBBP/EP300 mutation load. Conversely, treatment of DLBCLs that do not have CREBBP/EP300 mutations with CARM1i and a CBP/p300 inhibitor revealed a strong synergistic effect. Our mechanistic data show that CARM1i further reduces the HAT activity of CBP genome wide and downregulates CBP target genes in DLBCL cells, resulting in a synthetic lethality that leverages the mutational status of CREBBP/EP300 as a biomarker for the use of small molecule inhibitors of CARM1 in DLBCL and other cancers.
Introduction
Non-Hodgkin lymphoma (NHL) comprises a group of malignant neoplasms derived from B or T cell progenitors or mature B or T cells. The two most common NHL subtypes are Follicular lymphoma (FL) and Diffuse Large B Cell Lymphoma (DLBCL). FL initially presents with a slow progression but eventually 40–50% of these cancers transform into aggressive forms of DLBCL (1). Although with chemo- and immuno- therapy DLBCL cures can be achieved, many patients are still refractory and succumb to progressive or relapsed disease (2). Both FL and DLBCL derive from B cells that undergo the germinal center (GC) reaction, where B cells are generated and selected to produce high affinity antibodies (3–5).
Mutations of histone-modifying enzymes are a genetic hallmark of GC-BCLs. Somatic mutations in the genes that encode the histone acetyltransferases (HATs) CBP and p300 are among the highest recurrent disease alleles in FL and DLBCL. Reports show that up to 22% of DLBCLs and 68% of FLs have mutations in CREBBP (6–11). Most of these mutations are monoallelic and in the HAT domain, leading to an inability to acetylate target proteins, due to reduced binding of acetyl-CoA (9). On the other hand, up to 10% of DLBCLs and 23% of FLs have mutations in EP300 (6, 8–10). Interestingly, mutations in these two HATs are often mutually exclusive, suggesting that they might affect a common pathway that is essential for the survival of the cancer cells.
CBP/p300 co-activator activity can be modulated by numerous factors including methylation by Co-activator-Associated Methyltransferase 1 (CARM1) (12–15). CARM1 is one of several sequence-related protein arginine methyltransferases, termed PRMTs, responsible for methylating arginine residues in mammalian cells. These methylated arginine residues are thought to generate or prevent docking of effector molecules to adjacent sites (16, 17). The initial identification of CARM1 (also called PRMT4) as a co-activator of the steroid receptor provided the first evidence for the involvement of arginine methylation in transcriptional regulation (18). CARM1 is recruited to gene promoters and asymmetrically dimethylates histone H3 (H3R17me2a and H3R26me2a) and chromatin bound proteins and splicing factors (19). CARM1 activity has also been mapped to enhancer regions in studies involving a ChIP-chip approach in MCF7 breast cancer cells that showed CARM1 activity in distinct classes of ERα binding sites termed enhancer-rich clusters (20). A comprehensive study on estrogen receptor α transcriptional regulation identified five CBP arginine residues that are methylated in vivo by CARM1. CARM1-dependent CBP methylation resulted in gene-selective association of estrogen-recruited meCBP species with different HAT activities and specified distinct target gene hubs (12).
CARM1 null mice die shortly after birth (21). Further studies on these animals revealed roles for CARM1 in T cell development (22), adipocyte differentiation (23), chondrocyte proliferation (24) and proliferation and differentiation of pulmonary epithelial cells (25). For all of these in vivo functions, the methyltransferase activity of CARM1 is required (26). Emerging evidence suggests that CARM1 functions as an oncogene in human cancers (27). Overexpression of CARM1 has been reported in multiple cancer types including prostate, liver, colon and breast (28–31). CARM1 stimulates cell growth in breast cancer (32, 33) and a recent study described that CARM1 promotes EZH2-mediated silencing of tumor suppressor genes in ovarian cancer cells (34). Furthermore, the ectopic overexpression of CARM1 in mammary gland epithelial cells cause hyperproliferation of these cells and increased branching of the mammary gland, and also spontaneous mammary tumors after about 20 months (35). These data suggest that CARM1 small molecule inhibitors may have therapeutic value for the treatment of a number of different cancers.
Indeed, this has been an active area of research, and TP-064 was identified as the first small potent molecule inhibitor of CARM1 arginine methylation activity, which exhibited in vitro efficacy against Multiple Myeloma cell lines (36). Recently, another small molecule inhibitor of CARM1 methylation activity that is bioavailable and with potent in vivo efficacy in preclinical models of Multiple Myeloma was also developed: EZM2302, also called GSK3359088 (37). Another recent report shows that this inhibitor is efficient against acute myeloid leukemia (38). These small molecule inhibitors of CARM1 arginine methylation activity provide the opportunity for testing the therapeutic potential of CARM1 inhibition in a number of additional cancer settings, as well as in other diseases.
Based on the analysis of large-scale RNAi screens using cancer cell lines showing that DLBCLs are dependent on CARM1, we hypothesized that small molecule inhibition of CARM1 activity may represent a potential therapy for these cancers. We found that inhibition of CARM1 activity has potent growth arrest effects both in vitro and in vivo models of DLBCL and that the levels of sensitivity to CARM1 inhibition are positively correlated with the CREBBP/EP300 mutation load, with DLBCLs that harbor lesions in both CREBBP/EP300 showing the highest sensitivity. In keeping with this finding, treatment of DLBCLs that do not have mutations on CREBBP/EP300 with a CARM1 inhibitor and a specific CBP/p300 inhibitor revealed a strong synergistic effect. Our functional and mechanistic studies reveal that targeting CARM1 activity in CREBBP/EP300 mutated DLBCLs causes synthetic lethality. Our findings leverage the mutational status of CREBBP/EP300 as a biomarker for the use of CARM1 inhibitors in DLBCL and possibly other cancers. In addition, they also suggest that combining CARM1 inhibitors with CBP/p300 inhibitors may have a promising therapeutic effect in CREBBP/EP300 WT DLBCLs.
Materials and Methods
Animal Xenograft Studies
NOD SCID mice from Jackson Laboratory were subcutaneously injected with 107 U2932 or Toledo cells in the right flank. Tumor formation was monitored every day until they reached palpable size (75–100mm3), then mice were randomly separated into Vehicle and CARM1i groups. Mice were then treated with 150 mg/kg EZM2302 dissolved in 0.5% methylcellulose (CARM1iEZM), or 0.5% methylcellulose alone (Vehicle) by oral gavage twice daily for 9 days. Tumor size was monitored every other day using digital calipers in two dimensions. Upon completion of treatment, mice were sacrificed, and final tumor size and weight was determined. Tumor and organs were taken for western blot, and blood was analyzed for abnormalities using the HEMAVET HV950FS instrument (Drew Scientific Inc. FL, USA). All animal experiments were approved by the IACUC of the University of Texas MD Anderson Cancer Center.
Cell Culture and Inhibitor Experiments
CARM1fl/fl ER-Cre mouse embryonic fibroblasts (MEFs) were kindly provided by Dr. Bedford. Cells were treated with 2uM 4-OHT for 6 days or 1uM TP-064 (CARM1iTP) for 4 days and harvested for western blot and RNA-seq. RL and HT isogenic DLBCL lines with CRISPR-cas9 deletion of CREBBP were kindly provided by Dr. Michael Green. All cell lines were externally verified by IMPACT II PCR profiling at IDEXX BioResearch Labs to be pathogen and mycoplasma negative. All cell lines were cultured in RPMI-1640 (ATCC 30–2001) supplemented with 10% FBS and 5% penicillin/streptomycin. CARM1iTP was dissolved in DMSO, and cells were treated with a final concentration of 5uM for 8 days in the growth curve assay, 6 days in the WB, RNA and ChIP assays. For inhibitor experiments please see Supplemental Methods.
For RNA-seq, ChIP-seq, Flow Cytometry, In Vitro Overexpression, Immunoprecipitation, Western blot, qPCR, and statistical analysis, please see Supplemental Methods.
Results
DLBCL cells are dependent on CARM1 arginine methylation activity
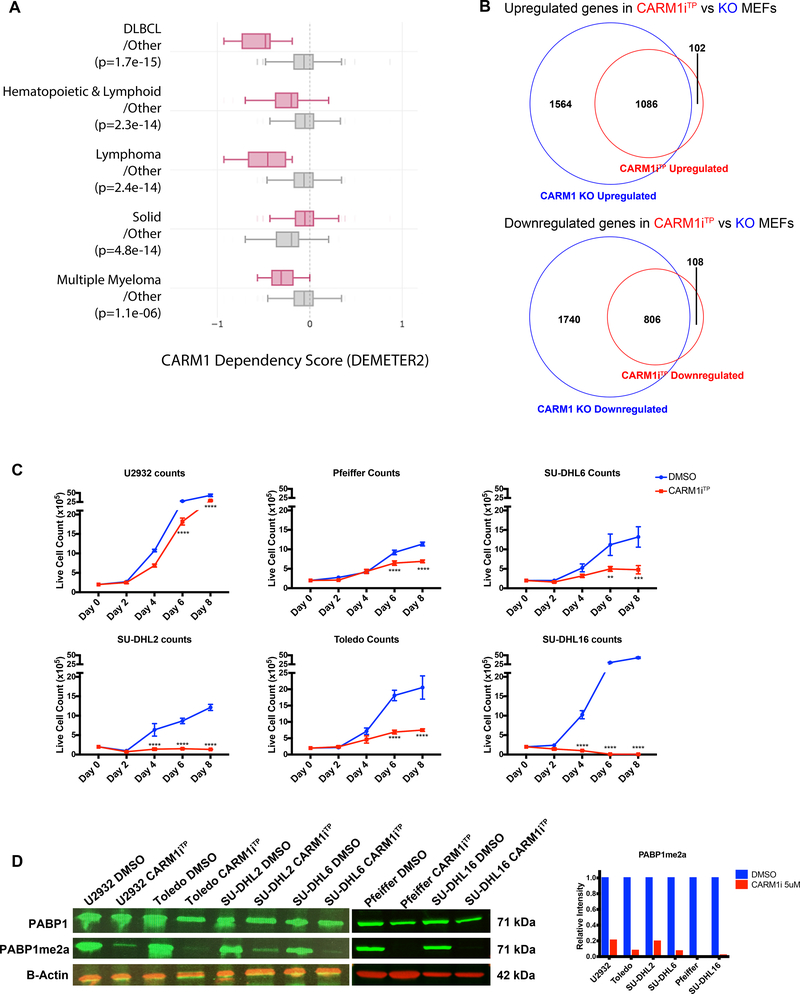
CARM1 is overexpressed in multiple cancer cell lines and patient samples (28–31). To identify cancer types potentially dependent on CARM1, we analyzed the dependency scores obtained from large-scale RNAi screens using cancer cell lines. As shown in Figure 1A, hematopoietic and lymphoid lineages appear to be dependent on CARM1, with DLBCL lines showing the most dependency. TP-064 is a potent small molecule inhibitor of CARM1 arginine methylation activity with potent in vitro efficacy in Multiple Myeloma cell lines (36). To understand the dependency of DLBCL cells on CARM1 activity, we profiled the sensitivity of 6 DLBCL lines to this small molecule inhibitor (CARM1iTP). Before proceeding with using TP-064 in the DLBCL lines, we wanted to compare the transcriptional effects of inhibition of CARM1 methyltransferase activity using TP-064 with the actual deletion of CARM1. We chose to utilize a mouse embryonic fibroblast (MEF) line that harbors Carm1flox/flox alleles expressing cre-recombinase fused to the hormone-binding domain of the estrogen receptor (Carm1f/f ER-Cre MEFs) as a tool to compare transcriptional alterations between genetic deletion of CARM1 and inhibition of CARM1 activity. After confirming that hydroxytamoxifen (OHT) treatment resulted in deletion of Carm1 (Supplementary Figure 1A upper panel), we performed gene expression analysis by RNA-sequencing (RNA-seq) in Carm1f/f ER-Cre MEFs either untreated or treated with OHT and on WT MEFs treated with DMSO or 1uM of CARM1iTP. In both cases we observed a significant reduction in CARM1 methylation activity, as shown using the anti-H3R17me2a antibody, which recognizes many CARM1 substrates(39) (Supplementary Figure 1A middle panel). As shown in the Venn diagrams in Figure 1B, around 90% of the genes upregulated in WT MEFs upon CARM1iTP treatment were also upregulated upon deletion of CARM1; the same was observed for downregulated genes. These data show that CARM1iTP treatment, that inhibits the arginine methylation activity of CARM1, has mostly ‘on target’ effects. We then proceeded to treat six independent DLBCL lines with CARM1iTP. The treatment concentration of 5μM was chosen as our standard treatment based on similar in vitro growth experiments in multiple myeloma cell lines (36) where micromolar concentrations of TP-064 showed great efficiency for inhibition of CARM1 methylation activity as well as for inhibition of cell growth. Although the levels of sensitivity were variable, we observed a significant growth inhibitory effect in the lines tested (Figure 1C). This sensivity was observed in as low as 1uM CARM1iTP dose (Supplementary Figure 1B). PABP1 (polyadenylate-binding protein 1) was shown to be a specific target for CARM1 methylation activity and antibodies to detect this CARM1-mediated arginine methylation in PABP1 have been identified and are commercially available (40). We therefore determined the methylation status of PABP1 in the cell lines upon treatment with CARM1iTP. As shown in Figure 1D, the levels of asymmetrically dimethylated PABP1 were greatly reduced upon treatment with 5μM CARM1iTP. Lower doses of CARM1i decreased the activity of CARM1, but to a lesser extent than the 5uM dose (Supplementary Figure 1C). Finally, there was no correlation between levels of CARM1 protein and sensitivity to CARM1 inhibition in these cells (Supplementary Figure 1D).
Figure 1. Human DLBCL cells are dependent on CARM1 arginine methyltransferase activity.
A) Cancer cell line Dependency Score (DEMETER2) of parallel two-group comparisons across genes obtained from http://depmap.org. Enriched lineages have a p-value of < 0.0005, and include DLBCL, Hematopoietic and Lymphoid, Lymphoma, Solid, and Multiple Myeloma cell lines. Box plots show the upper and lower quartile and the median. Whiskers indicate 95% confidence intervals. Each labeled group contains lines within the labeled class (pink) versus lines in other classes (grey). B) Venn diagram representation of RNA-seq performed on WT and CARM1flox/flox ER-Cre MEFs treated with hydroxytamoxifen (OHT) or WT MEFs treated with DMSO or 1uM TP-064 (CARM1iTP). The upper diagram shows the overlap of upregulated genes in CARM1iTP vs DMSO compared to those downregulated in KO vs WT MEFs. The lower diagram shows the overlap of downregulated genes in CARM1iTP vs DMSO compared to those downregulated in KO vs WT MEFs. RNA-seq experiments were performed on two independent biological replicates. C) Growth curves of 6 human DLBCL cell lines treated with DMSO or 5uM CARM1iTP for 8 days. Cells were counted every other day, and treatment was replaced. Error bars represent SEM, n=3. Statistical significance was calculated each day using unpaired t-tests. D) Western blot analysis and quantification of PABP1 methylation in 6 human DLBCL lines treated with DMSO or CARM1iTP. B-Actin was used as a loading control. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Transcriptional networks regulated by CARM1 activity in DLBCL cells
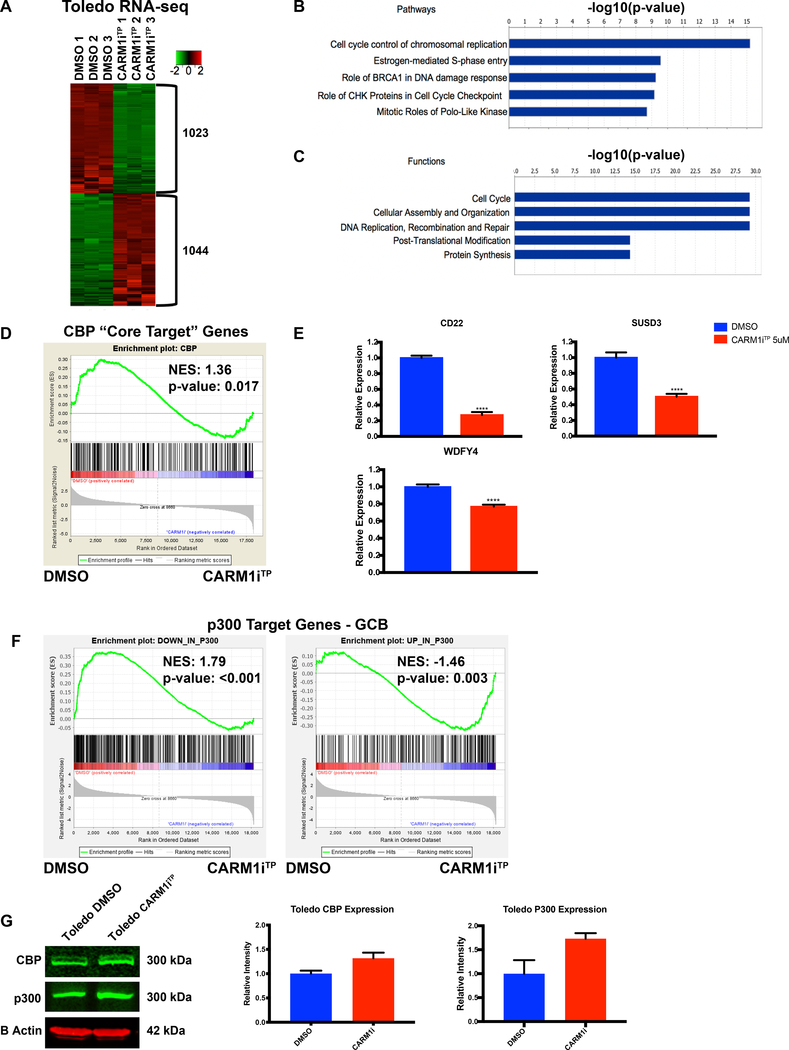
To define what transcriptional networks are regulated by CARM1 activity in DLBCL, we performed genome-wide transcriptome analysis by RNA sequencing (RNA-seq) in Toledo cells treated with CARM1iTP. We chose this line because it presented with a strong sensitivity to CARM1 inhibition, at the same time allowing us to collect enough live cells to proceed with downstream studies (Figure 1C). We found approximately 2,000 genes that were significantly modulated (around half up-regulated and half down-regulated) by inhibition of CARM1 activity in these cells (Figure 2A, Fold change, FC≥2 and False Discovery Rate, FDR≤0.05). The list of these up and down-regulated genes is shown in Supplementary Tables 1&2, respectively. Analysis of the RNA-seq dataset using IPA (Ingenuity Pathway Analysis ®) revealed high enrichment of pathways involved in cell cycle control and DNA repair (Figure 2B). Consistent with this, among the most significant cellular functions were also the Cell Cycle and DNA Replication, Recombination and Repair (Figure 2C). This agrees with the growth arrest we observed in cells treated with CARM1iTP (Figure 1C). We also performed gene analysis in Toledo cells treated with TP-064 to reassure that the effects on cell cycle genes are on-target, and not due to broad toxicity caused by higher dose treatments. As shown in Supplementary Figure 2A, at 5uM TP-064 we observed a deregulation of genes involved in the cell cycle. We specifically tested some of the genes in the E2F pathway because these have been shown to be downregulated upon CARM1 knock down in AML cell lines (38). Consistently with this, we observed a downregulation of the genes tested that include E2F1 and E2F targets CDC25A, and TOP2A. These data further validate on target effects of TP-064 treatment.
Figure 2. Transcriptional networks regulated by CARM1 activity in human DLBCL cells.
A) Heat map of genome-wide transcriptional changes in DMSO vs CARM1iTP-treated Toledo cells. Diagram shows three biological replicates. FC >/=2, FDR </= 0.05. B) IPA analysis of enriched pathways in the differential expression analysis. C) IPA analysis of enriched cellular functions in the differential expression analysis. D) GSEA plot evaluating transcriptional changes in CBP “core target” genes between DMSO and CARM1iTP-treated cells. CBP “core targets” are defined as genes bound by CBP in human germinal center B cells, and downregulated in CBP-deficient mouse BCLs. E) RT-qPCR confirmation of transcriptional changes in three CBP target genes shown to be significantly downregulated in the RNA-seq experiments. Statistical significance was calculated using unpaired t-tests. Error bars represent SEM, n=3. F) GSEA plot evaluating transcriptional changes in p300 target genes between DMSO and CARM1iTP-treated cells. Left panel shows downregulated p300 target genes, right panel shows upregulated p300 target genes. G) Western blot analysis and quantification of CBP and p300 protein expression levels in Toledo cells treated with DMSO or CARM1iTP. B-Actin was used as a loading control. Error bars indicate SD. N=2. All experiments were performed on the Toledo cell line, treated for 6 days with DMSO or 5uM of CARM1iTP. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
We also performed Gene Set Enrichment Analysis (GSEA) of this RNA-seq data using gene sets known to be deregulated in DLBCL. We found that CARM1iTP treatment resulted in a global reduction in expression of a well-established CBP-target gene signature in B cells (Figure 2D, FDR= 0.02). This gene signature is a list of CBP “core target” genes (bound by CBP in human GC B cells and downregulated in mouse CBP-deficient GC B cells defined in (41)). We independently validated three of these target genes (CD22, SUSD3 and WDFY4) and found that they were significantly downregulated upon CARM1iTP treatment (Figure 2E). A recent study describes the RNA-seq signatures of p300 and CBP in mouse germinal center B cells, and the cell cycle and DNA repair and replication pathways were found to be enriched in the p300 signature (genes in these pathways appear downregulated in p300 deficient GCB cells when compared with WT counterparts). These pathways are reminiscent of the pathways enriched in our RNAseq (Figure 2B&C). Thus we wondered whether this p300 signature was also enriched in our dataset. GSEA analysis showed that genes downregulated by p300 deletion in mouse GCB cells were significantly downregulated in Toledo cells treated with CARM1i vs DMSO-treated cells (Figure 2F, left panel); the same was observed for upregulated genes (Figure 2F, right panel). The set of downregulated genes in CBP-deficient vs WT mouse GC B cells was also enriched in our Toledo CARM1i vs DMSO-treated cells, although the p value is slightly higher than what we use as cut-off for significance (Supplementary Figure 2B). Finally, we confirmed that CARM1iTP did not reduce the CBP nor p300 protein levels, thus showing that downregulation of CBP and p300 target genes is not the result of lower levels of CBP and/or p300 protein (Figure 2G).
In summary, transcriptional analysis of CARM1iTP -treated DLBCL cells shows that CARM1 regulates the expression of genes involved in cell cycle and DNA repair in DLBCL cells and sustains the expression of a subset of CBP- and p300-target genes.
CARM1 inhibition in DLBCL leads to loss of H3K27ac at CBP/p300 chromatin bound regions
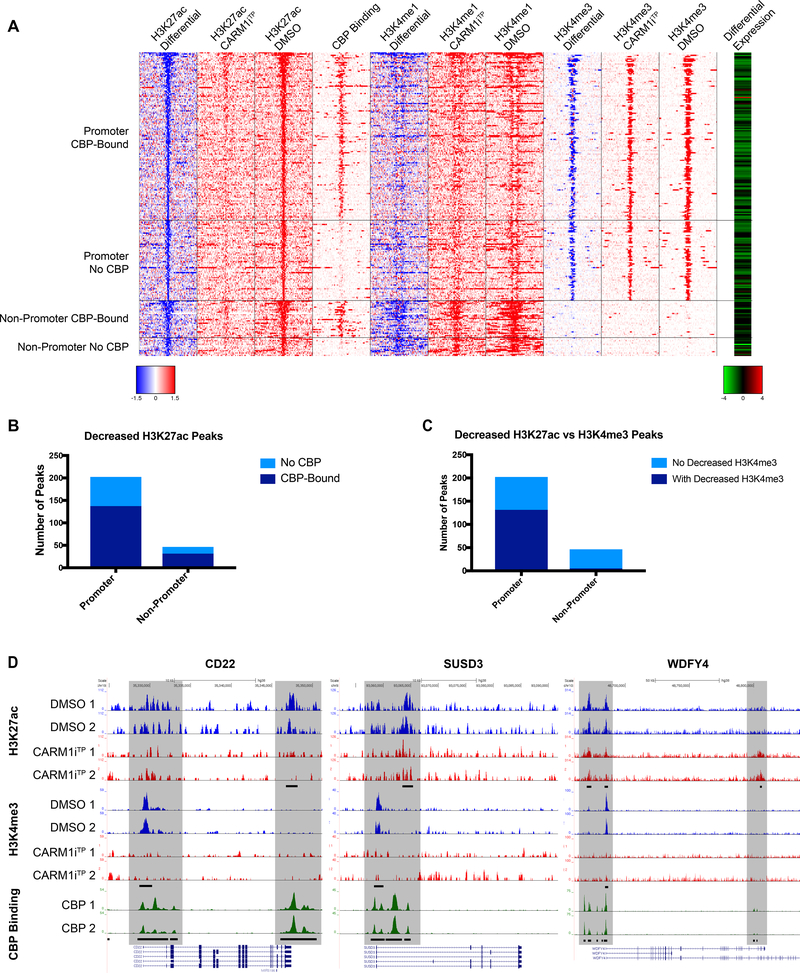
To determine the impact of inhibition of CARM1 activity on chromatin regulatory landscapes, we performed chromatin immunoprecipitation sequencing (ChIP-seq) in Toledo cells with antibodies directed against specific histone modifications defining well-characterized active chromatin states (H3K4me3, H3K4me1 and H3K27ac). Both CBP and p300 are well-known substrates for CARM1 methylation (12–15). CARM1 methylates CBP arginine residues that were shown to play a crucial role in ERα transcriptional regulation (12). Based on this and our RNA-seq analysis that showed downregulation of CBP “core target” genes in CARM1i treated cells (Figure 2D), we sought to determine whether inhibition of CARM1 activity in DLBCL cells preferentially affects CBP-bound chromatin regions. Previous studies have identified the genomic regions bound by CBP in human GC B cells (41). We thus compared our ChIP-seq datasets with these CBP bound regions. As shown in the heat map in Figure 3A, CARM1 inhibition led to a loss of H3K27ac peaks in promoter regions as well as outside promoter regions and the majority of these were bound by CBP (Figure 3A&B, pvalues of association with CBP binding by Fisher’s exact test were 3.3e-10 and 2.7e-10, respectively).
Figure 3. Deregulation of promoter and enhancer regions bound by CBP/p300 upon CARM1 inhibition.
A) Heatmap of Toledo ChIP-Seq signal around −10kb to 10kb of decreased H3K27ac peaks in CARM1iTP-treated cells. The decreased H3K27ac peaks were grouped by in-promoter and non-promoter, and also bound by CBP or not bound by CBP. CBP Binding column denotes whether CREBBP is bound at the locus in human GC B cells(41). The most right column in red-green color scale is the log2 expression ratio (log2 CARM1iTP-treated / DMSO) from RNA-Seq (Figure 2) for the genes that have the closest TSS to the H3K27ac peaks. For differential ChIP signal, blue color and red color indicate decreased signal and increased signal in CARM1iTP-treated cells, respectively. For log2 expression ratio, green color and red color indicate downregulated expression and upregulated expression in CARM1iTP-treated cells, respectively. B) Decreased H3K27ac peaks shown within promoter and outside promoter regions. Light blue bars represent no CBP binding in the region. Dark blue bars represent CBP binding within the region. P values of association with CBP binding by Fisher’s exact test were 3.3e-10 and 2.7e-10, respectively. C) Decreased H3K27ac peaks shown within promoter and outside promoter regions. Light blue bars represent no decrease in H3K4me3 in the region. Dark blue bars represent a concurrent decrease in both H3K4me3 and H3K27ac within the region. P values of association with CBP binding by Fisher’s exact test were 1.81e-4 and 9.22e-22, respectively. D) Representative ChIP-seq tracks of three CBP-target genes verified to have a significant decrease in expression in Figure 2E. Shaded areas indicate CBP-bound regions where peaks are significantly changed (FDR of 0.1 or lower) in CARM1iTP treated cells. Differential peaks are denoted by black bars in H3K27ac and H3K4me3 tracks (FDR of 0.1 or lower), CBP-bound peaks denoted by black bars in CBP track. Tracks represented include Input-subtracted H3K27ac peaks and H3K4me3 peaks measured in the Toledo B cell lymphoma line, and CBP binding measured in human germinal center B cells(41).
H3K27ac regions “outside promoter” were also marked by H3K4me1 in the DMSO samples but had reduced H3K4me1 upon CARM1 inhibition. The co-existence of these two marks at these regions suggests that these are active enhancer regions (42). Furthermore, the majority of these H3K27ac peaks lost at the “outside promoter” regions were not marked by H3K4me3, further supporting that these are enhancer regions (Figure 3A&C). Finally, Figure 3A (far right column) shows that predicted genes that lost H3K27ac peaks were also downregulated in the RNAseq dataset of Figure 2A. Figure 3D shows representative ChIP-seq read density tracks for the three CBP target genes validated in Figure 2E that exemplify the downregulation of H3K27ac and H3K4me3 at these CBP bound genes. Thus, we conclude that CARM1 inhibition in DLBCL cells leads to a loss of H3K27ac at CBP chromatin-bound regions. As seen in Figure 3A, there is a substantial number of genomic regions with loss of H3K27ac and H3K4me3 peaks that are not bound by CBP. These regions may be altered by other CARM1 coactivator functions. CARM1 interacts with a large number of proteins, modifies histones, and functions as a transcriptional coactivator within the nucleus (27), therefore inhibition of CARM1 activity may indirectly result in loss of chromatin marks associated with gene activation independent of its interaction with CBP. In summary, not all CARM1-associated chromatin changes may involve CBP, even though the majority of decreased H3K27ac peaks observed in our ChIP-seq dataset are bound by CBP.
To directly show that CARM1 methylates CBP, we transiently overexpressed a Flag-tagged CBP protein in cells treated with DMSO or CARM1i for 72h after transfection and performed immunoprecipitation to pull down CBP-Flag. We were able to detect CARM1 CBP methylation using a pan CARM1 substrate antibody (CARM1sub) with the potential to recognize many CARM1 substrates including CBP, to detect the methylation of CBP (please see methods for generation of this antibody). The levels of CARM1-specific CBP methylation were reduced in CARM1i treated cells when compared to DMSO-treated controls (Supplementary Figure 1E). These data show that CARM1 is able to methylate CBP and modulate its HAT activity at specific regions within the genome where CBP is bound.
CREBBP/EP300 mutation load in DLBCLs positively correlates with sensitivity to CARM1 inhibition
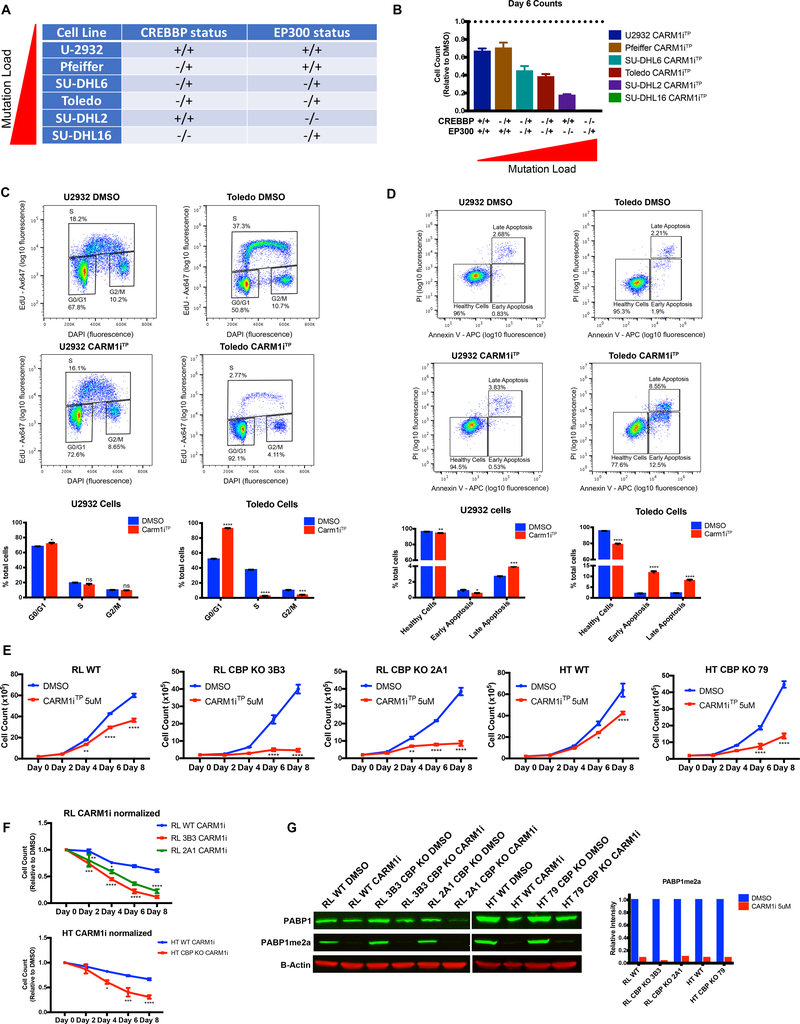
Somatic mutations in CREBBP and EP300 are frequent events in FL and DLBCL (6–11). Most of these mutations are monoallelic, in the HAT domain and are often mutually exclusive, suggesting that they might affect a common pathway that is essential for the survival of the cancer cells (6, 8–10). Our genome wide data suggesting that CARM1 activity is required for the activation of CBP target genes, prompted us to investigate whether the mutation status of CREBBP and EP300 influences DLBCL sensitivity to CARM1iTP. Previous studies using the DLBCL lines tested in Figure 1C showed that SUDHL16 and Toledo cells harbor mutations in both CREBBP and EP300 (43, 44), SUDHL2 carries a homozygous mutation in EP300 (9, 44), SUDHL6 carries heterozygous mutations in CREBBP and EP300 (9, 44), Pfeiffer carries a heterozygous mutation in CREBBP (11, 45), and U2932 is WT for both CREBBP and EP300 (9, 44, 45) (Figure 4A and Supplementray Table 3). As shown in Figure 4B we observed a positive correlation between CREBBP/EP300 mutation load and sensitivity to CARM1 inhibition. This genetic sensitivity to CARM1i does not necessarily correlate with protein expression, which suggests that cellular sensitivity is due to decreased activity of CBP and p300 at specific genomic regions rather than change in expression of the proteins (Supplementary Figures 3A&B). To further explore this finding, we chose one of the CREBBP/EP300 WT lines, U2932, and one line with mutations on both CREBBP/EP300, Toledo, to perform cell cycle and apoptosis analysis. As shown in Figure 4C, the impact of CARM1iTP treatment on the cell cycle, namely arresting the cells in G1, was far greater in Toledo cells than in U2932. The same was observed for cell death by Annexin V detection (Figure 4D). This experiment strongly suggests that CREBBP mutations are causing higher sensitivity to CARM1i in Toledo cells when compared to U2932, which are WT for CREBBP. However, there is still the possibility that differences in the background of the two distinct cell lines could be playing a role in CARM1i sensitivity. To directly confirm that mutations which reduce CBP activity are sufficient to increase sensitivity to CARM1i, we treated two pairs of isogenic lines: parental RL cells and RL cells with CRISPR-cas9 deletion of CREBBP, and HT parental cells and HT cells with CRISPR-cas9 deletion of CREBBP (Figure 4E–G, Supplementary Figure 3D). As shown in Figure 4E and F, CBP-deficient cells were more sensitive to CARM1 inhibition compared with the parental counterparts. Taken together, our data show that DLBCL cells harboring genetic lesions in CREBBP/P300 are highly sensitive to inhibition of CARM1 activity, suggesting a synthetic lethality to be further explored.
Figure 4. CREBBP/EP300 mutation load correlates with sensitivity to CARM1 inhibition.
A) Table denoting the CREBBP and EP300 mutation status of 6 human DLBCL lines, +/+ indicates no mutations in either allele, −/+ indicates a monoallelic mutation, and −/− indicates a biallelic mutation (see main text and Supplementary Table 3 for references). B) Graph representing cell numbers relative to the DMSO control at Day 6 of 5uM CARM1iTP treatment from Figure 1C. Mutation status of CREBBP and EP300 for each line is denoted below. Error bars represent SEM. C) Flow cytometry for cell cycle analysis showing the percentage of U2932 and Toledo cells in S phase (EdU+) after treatment with DMSO or 5uM CARM1iTP for 6 days. One representative plot per line and treatment for three biological replicates is shown. Bar graphs represent the frequency of cells in G0/G1, S, or G2 phase in DMSO vs CARM1iTP cells. D) Flow cytometry for cell death analysis showing the percentage of U2932 and Toledo cells in early apoptosis (Annexin V+, PI-), and late apoptosis (Annexin V+, PI+). One representative plot per line and treatment for three biological replicates is shown. Bar graphs represent the frequency of early and late apoptotic cells in DMSO vs CARM1iTP cells. Statistical significance was calculated using unpaired t-tests. Error bars represent SEM. All experiments were performed on cells treated with DMSO or 5uM CARM1iTP for 6 days. E) Growth curves of 2 human DLBCL isogenic cell lines (RL wild-type vs CBP KO clones 3B3 and 2A1, HT wild-type vs HT CBP KO clone 79) treated with DMSO or 5uM CARM1iTP for 8 days. Cells were counted every other day, and treatment was replaced. Error bars represent SEM, n=3. Statistical significance was calculated each day using unpaired t-tests. F) DMSO-normalized growth curves of 2 human DLBCL isogenic cell lines (RL wild-type vs CBP KO clones 3B3 and 2A1, HT wild-type vs HT CBP KO clone 79) treated 5uM CARM1iTP for 8 days. Cells were counted every other day, and treatment was replaced. Graph represents growth curves for each line treated with CARM1iTP normalized to the respective values for DMSO in the same line on each day. Error bars represent SEM, n=3. Statistical significance was calculated using one-way ANOVA. G) Western blot analysis and quantification of PABP1 methylation in 2 isogenic DLBCL lines treated with DMSO or CARM1iTP. B-Actin was used as a loading control. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Synergy between CARM1 inhibition and CBP/p300 activity inhibition in DLBCL cells
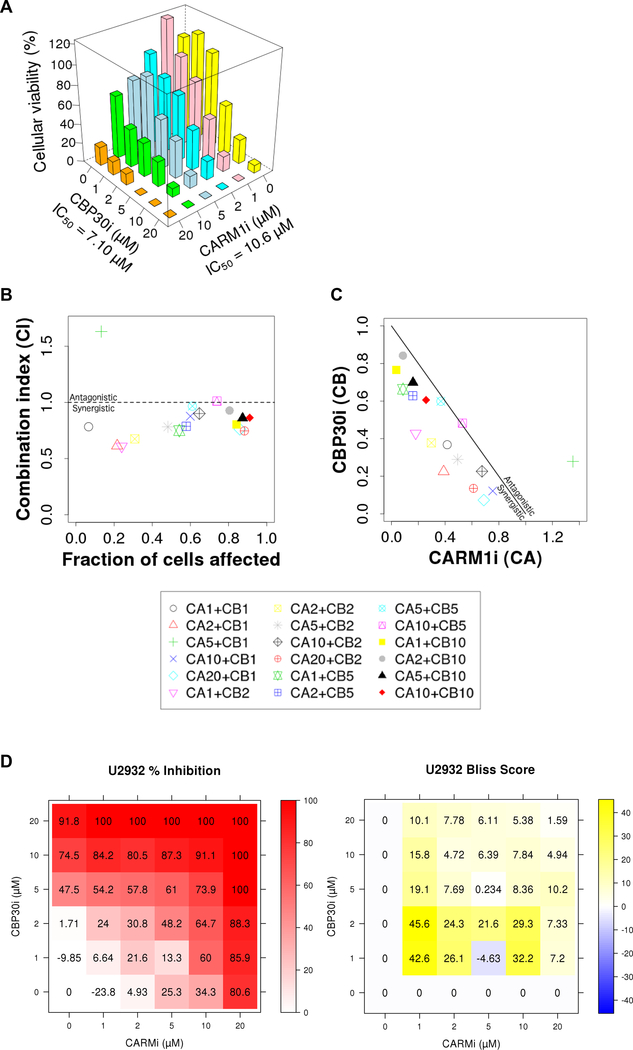
To further investigate the potential synergistic effect of loss of CARM1 and CBP/p300 activity on DLBCL growth, we combined CARM1iTP with CBP30, a selective and potent CBP/p300 bromodomain inhibitor (46), at different concentrations on U2932 cells. As individual agents, 5–20uM of either drug decreased the cell growth. In a combinatorial setting, lower concentrations of CARM1iTP and CBP30 (1–5uM) inhibited the growth of U2932 cells, suggesting a synergistic effect (Figure 5A). We used the combination index (CI) and the Bliss independence model as alternative methods to evaluate synergism between CARM1iTP and CBP30 in a quantitative manner (47). As shown in Figure 5B&C, the CI analysis with both the “effect-oriented” and “dose-oriented” mapping of the data demonstrates synergism between the two small molecule inhibitors. Similarly, the Bliss independence model showed a synergistic effect greater than 20%, in a dosage range of 1–2 μM for CBP30, and 1–10 μM for CARM1iTP, respectively (Figure 5D). Thus, the combined loss of CARM1 and CBP/p300 activity significantly impairs DLBCL growth.
Figure 5. CARM1 and CBP/p300 small molecule inhibition display synergistic effects in human DLBCL cells.
A) Cellular viability of U2932 line treated with the indicated concentrations of CBP30 and CARM1iTP. All combinations of 0–20uM drug were tested, and measurements were taken at day 6 of treatment. n=3. B and C) Combination Index analysis of the effect-oriented (B), and dose-oriented (C) maps to evaluate synergism between CARM1iTP and CBP30i, as described previously (65). B) The combination index (CI) values were calculated using CompuSyn software (ComboSyn, Inc., Paramus, NJ, USA) as a function of the fraction of cells affected (Chou-Talalay plot). Values below 1 indicate synergistic interactions, while values above 1 indicate antagonistic interactions. (C) Normalized isobolograms (Chou-Chou plots) show the synergism for the indicated drug concentrations, the diagonal line corresponding to an additive effect. Data points falling on the lower left of this line indicate synergism, while those falling on the upper right indicate antagonism. The Key at the bottom represents the combination treatments used in μMs, with ‘CA’ representing CARM1iTP and ‘CB’ representing CBP30. Each symbol represents the mean of three biological replicates tested for each combination of CBP30 and CARM1iTP indicated. D) Bliss independence model detailing percent inhibition (left) and Bliss score (right) for each combination of CBP30 and CARM1iTP tested. Each value represents the mean of three biological replicates.
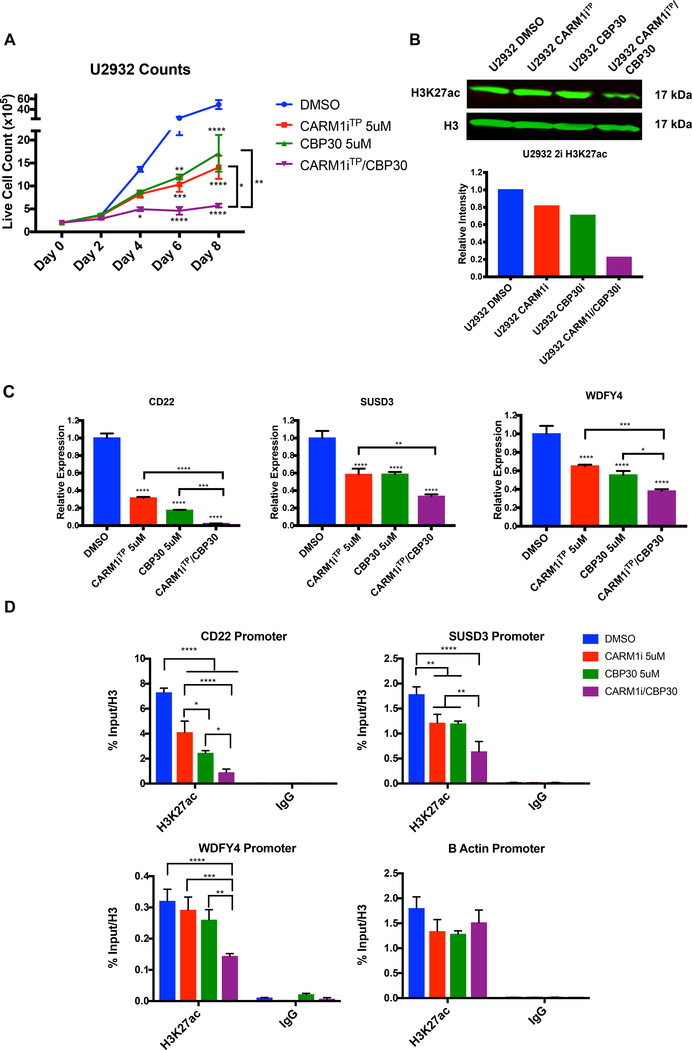
CARM1 inhibition further reduces CBP-dependent histone acetylation in CBP30-treated DLBCL cells
Recent studies in both murine and human BCLs revealed that CBP loss of function preferentially affects H3K27ac, leading to aberrant transcriptional silencing (41, 48). Since CARM1 is well known to methylate the CBP/p300 complex and positively modulate its HAT activity (12–15), we reasoned that the synergy between CARM1iTP and CBP30 would translate in further reduction of CBP/p300- dependent H3K27ac. To test this, we treated U2932 cells with 5uM of CARM1iTP and CBP30, either alone or in combination. As predicted, we observed a synergistic decrease in cell growth (Figure 6A). This was accompanied by a synergistic reduction in H3K27ac levels detected by western blot (Figure 6B). We next chose the three candidate genes from the “CBP core target” gene list that were downregulated in our RNA-seq dataset upon CARM1iTP treatment alone (Figure 2D&E) to interrogate by RT-qPCR whether CARM1iTP and CBP30 have a synergistic effect in downregulating these genes. As shown in Figure 6C, we observed downregulation of these genes by single treatments with either CARM1iTP or CBP30; but when used in combination, the downregulation was significantly greater. Finally, we also observed that the combination treatment further decreased the levels of H3K27ac at promoters of these specific genes (Figure 6D, all primers used in RNA and ChIP studies are listed in Supplementary Table 4). Taken together, our data show that inhibition of CARM1 activity synergizes with inhibition of CBP/p300 HAT function in DLBCL cells.
Figure 6. CARM1 and CBP/p300 inhibition synergistically reduces CBP-dependent histone acetylation and CBP target gene expression in human DLBCL cells.
A) Graph representing reduction of U2932 cell growth in DMSO, CARM1iTP, CBP30, and two-inhibitor treated cells. Error bars represent SEM, n=3. Statistical significance was calculated using a two-way ANOVA. B) Western blot analysis and quantification of H3K27ac levels in DMSO, CARM1iTP, CBP30, or two-inhibitor treated cells. C) RT-qPCR analysis of transcriptional changes in three CBP target genes upon treatment with DMSO, CARM1iTP, CBP30, or both inhibitors. Statistical significance was calculated using a one-way ANOVA. Error bars represent SEM, n=3. D) ChIP-qPCR analysis of H3K27ac levels at the promoters of the three CBP target genes listed in C. B Actin promoter is shown as an endogenous control with no significant changes in H3K27ac. Error bars represent standard deviation, n=4 separate ChIP-qPCR experiments from 2 biological replicates. All experiments were performed on the U2932 cell line, treated for 6 days with DMSO, 5uM of CARM1iTP, 5uM of CBP30, or 5uM each of both inhibitors. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Therapeutic potential of CARM1 inhibition against CREBBP/EP300 mutated DLBCLs
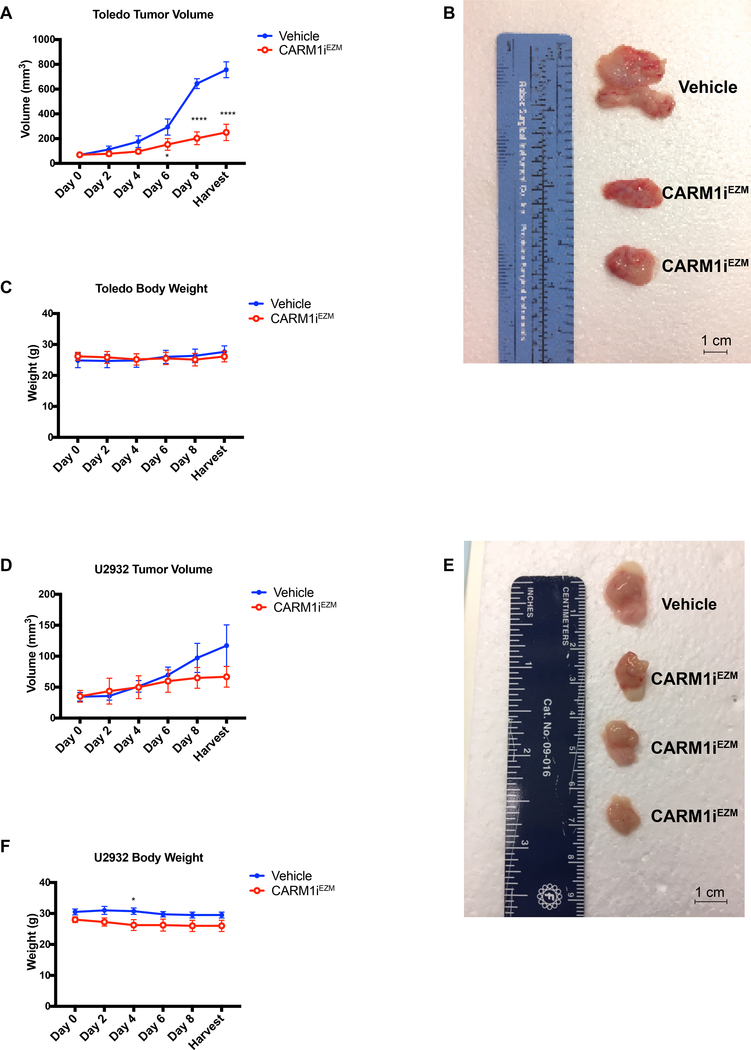
In order to determine the impact of CARM1 inhibition against DLBCLs harboring mutations in CREBBP/EP300 in a preclinical model, we tested the action of the bioavailable CARM1 inhibitor EZM2302 (CARM1EZM) in mice bearing human lymphoma xenografts. One DLBCL line carrying mutations in both CREBBP/EP300, Toledo; and one WT for these genes, U2932, were used to establish xenografts in NOD-SCID mice. For each cell line, control cohorts were treated with vehicle (0.5% methylcellulose) and experimental cohorts with 150mg/Kg CARM1EZM twice daily. Dosing of this compound was chosen based on previous reports of antitumoral effects with little or no toxicity (37, 38). Treatment was initiated when tumors reached approximately 100 mm3. As shown in Figure 7A&B, CARM1EZM treatment led to a significant reduction in the growth of Toledo xenograft tumors, without any significant loss in body weight (Figure 7C). The same treatment performed in U2932 injected animals led to a slight reduction in tumor growth (Figure 7D&E), although not enough to reach statistical significance. We confirmed the efficacy of CARM1EZM in inhibiting CARM1 activity in vivo by performing western blots for methylated PABP1 (Supplementary Figure 4). Thus, we conclude that the in vivo growth of CREBBP/EP300 mutated DLBCLs depends on CARM1 activity, further confirming the feasibility of using small molecule inhibitors of CARM1 as a method for treating BCLs with mutations in CREBBP/EP300.
Figure 7. In vivo therapeutic potential of CARM1 inhibition against human DLBCL with lesions in CREBBP/EP300.
A) Tumor volume of established Toledo Xenografts injected into the flanks of NOD-SCID mice, monitored over a 9-day treatment protocol with Vehicle (0.5% methylcellulose) or EZM2302 (CARM1iEZM) 150 mg/Kg twice a day. Error bars indicate SEM, n=16. B) Representative picture of Vehicle vs CARM1iEZM-treated tumor morphology from Toledo Xenografts. Bar in bottom right corner represents 1cm. C) Body weight of NOD-SCID Toledo recipients during 9-day treatment with Vehicle or CARM1iEZM. Error bars indicate SEM, n=16. D) Tumor volume of established U2932 Xenografts injected into the flanks of NOD-SCID mice, monitored over a 9-day treatment protocol with Vehicle (0.5% methylcellulose) or CARM1iEZM as above. Error bars indicate SEM, n=12. E) Representative picture of Vehicle vs CARM1iEZM-treated tumor morphology from U2932 Xenografts. Bar in bottom right corner represents 1cm. F) Body weight of NOD-SCID U2932 recipients during 9-day treatment with Vehicle or CARM1iEZM. Error bars indicate SEM, n=12.
Discussion
CREBBP and EP300 are among the most frequently mutated genes in human lymphomas and our studies demonstrate that inhibition of CARM1 activity causes synthetic lethality by further attenuating the remaining activity of the HAT complex towards histone acetylation and transcriptional activation. Thus, DLBCLs can survive the partial loss of CBP and/or p300, but they cannot survive the subsequent loss of CARM1.
CARM1 has been shown to methylate many different proteins, including histones, splicing factors and chromatin bound factors(19). Thus, enzymatic inhibition of this methyltransferase can result in pleiotropic effects. Consistent with this, inhibition of CARM1 activity affected many different genes and pathways involved in Cell Cycle and DNA Replication, Recombination and Repair (Figure 2). Although different CARM1-dependent mechanisms may have an effect on DLBCL growth, our study specifically identifies CARM1 as a positive regulator of CBP/p300 activity in DLBCL cells that is essential to maintain the remaining HAT activity of the complex in cells harboring CREBBP and/or EP300 genetic lesions. CARM1 inhibition affected H3K27ac genome wide, in both promoter and putative enhancer regions that have been shown to be bound by CBP and dependent on its HAT activity in human B cells. CARM1 is recruited to gene promoters (19) and its activity has also been mapped to enhancer regions (20, 39). Our studies suggest that CARM1 may be present at promoter and enhancer regions bound by CBP, although the lack of ChIP-seq validated CARM1 antibodies does not allow to directly map CARM1 binding sites genome wide.
Although we cannot enumerate the list of genes responsible for the observed synthetic lethality in our studies, our data suggest that these are genes that require a redundant activity of either CBP or p300 that is dependent on CARM1 methylation and essential for the survival of BCL cells. A recent study (49) revealed shared epigenetic programs of CBP and p300 in mouse GCB cells and indicated that CBP and p300 must have a common transcriptional program for which they can partially substitute for eachother, since deletion of both CBP and p300 was incompatible with GC formation in mice. Previous studies have already shown a strong negative selection against CBP/p300-negative cells (50, 51). It is likely that at least a subset of these essential genes in the GC that are dependent on CBP or p300 require CARM1 activity to be expressed.
Both CBP and p300 are well known substrates for CARM1 methylation (12–15). A comprehensive study on estrogen receptor α transcriptional regulation identified five arginine residues that are methylated in vivo by CARM1, resulting in in gene-selective association of distinct meCBP species (12). Further studies will be required to determine which CARM1-methylated CBP residue(s) is important for DLBCL growth and sensitivity to CARM1 inhibition.
Previous studies describe the development and testing of small molecule inhibitors targeting the enzymatic activity of PRMTs in hematological cancers, namely of PRMT5 (52–55). These drugs have been proven as potent therapeutic targets and the first in-human study for PRMT5 inhibition in advanced solid tumors and Non-Hodgkin lymphoma is now underway (NCT02783300). CARM1 inhibition with EZM2302 has been shown to have anti-cancer effects in multiple myeloma and acute myeloid leukemia (37, 38) without inducing toxic effects in mice. Our data now suggest that CARM1 inhibition is a potent strategy to target CREBBP/EP300 mutated BCLs.
In addition to GC-derived BCLs, approximately 10–15% of non-small cell lung cancers and small cell lung cancers harbor loss-of-function mutations in CREBBP (56, 57) and recent genome wide studies revealed that these aberrations are also present in other types of human cancer including leukemia (18%) and bladder cancer (15–27%) (8, 9, 58–61). Recurrent missense mutations in CREBBP tend to cluster around the HAT domain encoding region. Specifically, mutations affecting the amino-acid residues p.Gly1411, p.Trp1472 and p.His1487 are frequently observed and known to impede the HAT and/or transcriptional co-activator activity of the complex(9, 57). Moreover, protein-truncating mutations and deletions are also common (9, 57, 62, 63). Thus, therapeutic strategies that specifically kill CREBBP/EP300 deficient tumors hold potential for personalized medicine and studies are warranted to investigate the efficacy of CARM1 inhibition in slowing the growth of CREBBP/EP300 mutated cancers other than DLBCL. In addition, the potential of combination therapies using CBP/p300 inhibitors together with CARM1 inhibitors in cancers other than DLBCL also requires further investigation. Although very infrequently, inactivating mutations in both CREBBP and EP300 are seen in some instances (64), suggesting that alternative mechanisms may at times compensate for the lack of HAT activity and potentially play a role in resistance to long-term paralog treatments. Possible mechanisms of resistance to CARM1i treatments in CREBBP/EP300 mutated BCLs will be the focus of future studies.
In summary, we found that CREBBP/EP300 inactivating mutations render lymphoma cells vulnerable to inhibition of CARM1 resulting in a further attenuation of HAT activity, reduced expression of CBP/p300 target genes and synthetic lethality. Together with our preclinical xenograft models, these findings leverage the use of CARM1 inhibitors as potential targets in GC-BCLs and other cancers.
Supplementary Material
Acknowledgments
We thank Manu Sebastian Ph.D., Abhinav Jain Ph.D., Michelle Barton Ph.D., Carlos Perez, and members of the M.A.S. laboratory for technical support and helpful discussions. Core facilities were supported by the NIH Grant P30CA16672 and Cancer Prevention Research Institute of Texas (CPRIT) Grants RP120348, RP170002 and RP170628. This work was supported by CPRIT Recruitment of First-time Tenure-Track Faculty award RR160097 (to H.X.), NIH grant GM126421 (to M.T.B.) and aAndrew Sabin Family Fellow Award, American Society of Hematology Junior Faculty Scholar Award and CPRIT First-time Tenure-Track Faculty award RR150039 (to M.A.S). M.A.S. and H.X. are CPRIT Scholars in Cancer Research.
Footnotes
Competing Interests
The authors declare no conflict of interests.
References
- 1.Al-Tourah AJ, Gill KK, Chhanabhai M, Hoskins PJ, Klasa RJ, Savage KJ, et al. Population-based analysis of incidence and outcome of transformed non-Hodgkin’s lymphoma. J Clin Oncol. 2008;26(32):5165–9. [DOI] [PubMed] [Google Scholar]
- 2.Guo L, Lin P, Xiong H, Tu S, Chen G. Molecular heterogeneity in diffuse large B-cell lymphoma and its implications in clinical diagnosis and treatment. Biochim Biophys Acta. 2018;1869(2):85–96. [DOI] [PubMed] [Google Scholar]
- 3.Klein U, Dalla-Favera R. Germinal centres: role in B-cell physiology and malignancy. Nat Rev Immunol. 2008;8(1):22–33. [DOI] [PubMed] [Google Scholar]
- 4.MacLennan IC. Germinal centers. Annu Rev Immunol. 1994;12:117–39. [DOI] [PubMed] [Google Scholar]
- 5.Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol. 2012;30:429–57. [DOI] [PubMed] [Google Scholar]
- 6.Green MR, Kihira S, Liu CL, Nair RV, Salari R, Gentles AJ, et al. Mutations in early follicular lymphoma progenitors are associated with suppressed antigen presentation. Proc Natl Acad Sci U S A. 2015;112(10):E1116–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, Sougnez C, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci U S A. 2012;109(10):3879–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476(7360):298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pasqualucci L, Dominguez-Sola D, Chiarenza A, Fabbri G, Grunn A, Trifonov V, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471(7337):189–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pasqualucci L, Trifonov V, Fabbri G, Ma J, Rossi D, Chiarenza A, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43(9):830–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J, Grubor V, Love CL, Banerjee A, Richards KL, Mieczkowski PA, et al. Genetic heterogeneity of diffuse large B-cell lymphoma. Proc Natl Acad Sci U S A. 2013;110(4):1398–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ceschin DG, Walia M, Wenk SS, Duboe C, Gaudon C, Xiao Y, et al. Methylation specifies distinct estrogen-induced binding site repertoires of CBP to chromatin. Genes Dev. 2011;25(11):1132–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chevillard-Briet M, Trouche D, Vandel L. Control of CBP co-activating activity by arginine methylation. EMBO J. 2002;21(20):5457–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee YH, Coonrod SA, Kraus WL, Jelinek MA, Stallcup MR. Regulation of coactivator complex assembly and function by protein arginine methylation and demethylimination. Proc Natl Acad Sci U S A. 2005;102(10):3611–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu W, Chen H, Du K, Asahara H, Tini M, Emerson BM, et al. A transcriptional switch mediated by cofactor methylation. Science. 2001;294(5551):2507–11. [DOI] [PubMed] [Google Scholar]
- 16.Bedford MT, Frankel A, Yaffe MB, Clarke S, Leder P, Richard S. Arginine methylation inhibits the binding of proline-rich ligands to Src homology 3, but not WW, domains. J Biol Chem. 2000;275(21):16030–6. [DOI] [PubMed] [Google Scholar]
- 17.Yang Y, Lu Y, Espejo A, Wu J, Xu W, Liang S, et al. TDRD3 is an effector molecule for arginine-methylated histone marks. Mol Cell. 2010;40(6):1016–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, et al. Regulation of transcription by a protein methyltransferase. Science. 1999;284(5423):2174–7. [DOI] [PubMed] [Google Scholar]
- 19.Bedford MT, Richard S. Arginine methylation an emerging regulator of protein function. Mol Cell. 2005;18(3):263–72. [DOI] [PubMed] [Google Scholar]
- 20.Lupien M, Eeckhoute J, Meyer CA, Krum SA, Rhodes DR, Liu XS, et al. Coactivator function defines the active estrogen receptor alpha cistrome. Mol Cell Biol. 2009;29(12):3413–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yadav N, Lee J, Kim J, Shen J, Hu MC, Aldaz CM, et al. Specific protein methylation defects and gene expression perturbations in coactivator-associated arginine methyltransferase 1-deficient mice. Proc Natl Acad Sci U S A. 2003;100(11):6464–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim J, Lee J, Yadav N, Wu Q, Carter C, Richard S, et al. Loss of CARM1 results in hypomethylation of thymocyte cyclic AMP-regulated phosphoprotein and deregulated early T cell development. J Biol Chem. 2004;279(24):25339–44. [DOI] [PubMed] [Google Scholar]
- 23.Yadav N, Cheng D, Richard S, Morel M, Iyer VR, Aldaz CM, et al. CARM1 promotes adipocyte differentiation by coactivating PPARgamma. EMBO Rep. 2008;9(2):193–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito T, Yadav N, Lee J, Furumatsu T, Yamashita S, Yoshida K, et al. Arginine methyltransferase CARM1/PRMT4 regulates endochondral ossification. BMC Dev Biol. 2009;9:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Brien KB, Alberich-Jorda M, Yadav N, Kocher O, Diruscio A, Ebralidze A, et al. CARM1 is required for proper control of proliferation and differentiation of pulmonary epithelial cells. Development. 2010;137(13):2147–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim D, Lee J, Cheng D, Li J, Carter C, Richie E, et al. Enzymatic activity is required for the in vivo functions of CARM1. J Biol Chem. 2010;285(2):1147–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Y, Bedford MT. Protein arginine methyltransferases and cancer. Nat Rev Cancer. 2013;13(1):37–50. [DOI] [PubMed] [Google Scholar]
- 28.El Messaoudi S, Fabbrizio E, Rodriguez C, Chuchana P, Fauquier L, Cheng D, et al. Coactivator-associated arginine methyltransferase 1 (CARM1) is a positive regulator of the Cyclin E1 gene. Proc Natl Acad Sci U S A. 2006;103(36):13351–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hong H, Kao C, Jeng MH, Eble JN, Koch MO, Gardner TA, et al. Aberrant expression of CARM1, a transcriptional coactivator of androgen receptor, in the development of prostate carcinoma and androgen-independent status. Cancer. 2004;101(1):83–9. [DOI] [PubMed] [Google Scholar]
- 30.Kim YR, Lee BK, Park RY, Nguyen NT, Bae JA, Kwon DD, et al. Differential CARM1 expression in prostate and colorectal cancers. BMC Cancer. 2010;10:197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osada S, Suzuki S, Yoshimi C, Matsumoto M, Shirai T, Takahashi S, et al. Elevated expression of coactivator-associated arginine methyltransferase 1 is associated with early hepatocarcinogenesis. Oncol Rep. 2013;30(4):1669–74. [DOI] [PubMed] [Google Scholar]
- 32.Mann M, Cortez V, Vadlamudi R. PELP1 oncogenic functions involve CARM1 regulation. Carcinogenesis. 2013;34(7):1468–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang L, Zhao Z, Meyer MB, Saha S, Yu M, Guo A, et al. CARM1 methylates chromatin remodeling factor BAF155 to enhance tumor progression and metastasis. Cancer Cell. 2014;25(1):21–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karakashev S, Zhu H, Wu S, Yokoyama Y, Bitler BG, Park PH, et al. CARM1-expressing ovarian cancer depends on the histone methyltransferase EZH2 activity. Nat Commun. 2018;9(1):631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bao J, Di Lorenzo A, Lin K, Lu Y, Zhong Y, Sebastian MM, et al. Mouse Models of Overexpression Reveal Distinct Oncogenic Roles for Different Type I Protein Arginine Methyltransferases. Cancer Res. 2019;79(1):21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakayama K, Szewczyk MM, Dela Sena C, Wu H, Dong A, Zeng H, et al. TP-064, a potent and selective small molecule inhibitor of PRMT4 for multiple myeloma. Oncotarget. 2018;9(26):18480–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Drew AE, Moradei O, Jacques SL, Rioux N, Boriack-Sjodin AP, Allain C, et al. Identification of a CARM1 Inhibitor with Potent In Vitro and In Vivo Activity in Preclinical Models of Multiple Myeloma. Sci Rep. 2017;7(1):17993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greenblatt SM, Man N, Hamard PJ, Asai T, Karl D, Martinez C, et al. CARM1 Is Essential for Myeloid Leukemogenesis but Dispensable for Normal Hematopoiesis. Cancer Cell. 2018;33(6):1111–27 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng D, Vemulapalli V, Lu Y, Shen J, Aoyagi S, Fry CJ, et al. CARM1 methylates MED12 to regulate its RNA-binding ability. Life Sci Alliance. 2018;1(5):e201800117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee J, Bedford MT. PABP1 identified as an arginine methyltransferase substrate using high-density protein arrays. EMBO Rep. 2002;3(3):268–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J, Vlasevska S, Wells VA, Nataraj S, Holmes AB, Duval R, et al. The CREBBP Acetyltransferase Is a Haploinsufficient Tumor Suppressor in B-cell Lymphoma. Cancer Discov. 2017;7(3):322–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107(50):21931–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andersen CL, Asmar F, Klausen T, Hasselbalch H, Gronbaek K. Somatic mutations of the CREBBP and EP300 genes affect response to histone deacetylase inhibition in malignant DLBCL clones. Leuk Res Rep. 2012;2(1):1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hashwah H, Schmid CA, Kasser S, Bertram K, Stelling A, Manz MG, et al. Inactivation of CREBBP expands the germinal center B cell compartment, down-regulates MHCII expression and promotes DLBCL growth. Proc Natl Acad Sci U S A. 2017;114(36):9701–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haery L, Lugo-Pico JG, Henry RA, Andrews AJ, Gilmore TD. Histone acetyltransferase-deficient p300 mutants in diffuse large B cell lymphoma have altered transcriptional regulatory activities and are required for optimal cell growth. Mol Cancer. 2014;13:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hammitzsch A, Tallant C, Fedorov O, O’Mahony A, Brennan PE, Hay DA, et al. CBP30, a selective CBP/p300 bromodomain inhibitor, suppresses human Th17 responses. Proc Natl Acad Sci U S A. 2015;112(34):10768–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Han K, Jeng EE, Hess GT, Morgens DW, Li A, Bassik MC. Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions. Nat Biotechnol. 2017;35(5):463–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang Y, Ortega-Molina A, Geng H, Ying HY, Hatzi K, Parsa S, et al. CREBBP Inactivation Promotes the Development of HDAC3-Dependent Lymphomas. Cancer Discov. 2017;7(1):38–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meyer SN, Scuoppo C, Vlasevska S, Bal E, Holmes AB, Holloman M, et al. Unique and Shared Epigenetic Programs of the CREBBP and EP300 Acetyltransferases in Germinal Center B Cells Reveal Targetable Dependencies in Lymphoma. Immunity. 2019;51(3):535–47 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kasper LH, Fukuyama T, Biesen MA, Boussouar F, Tong C, de Pauw A, et al. Conditional knockout mice reveal distinct functions for the global transcriptional coactivators CBP and p300 in T-cell development. Mol Cell Biol. 2006;26(3):789–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu W, Fukuyama T, Ney PA, Wang D, Rehg J, Boyd K, et al. Global transcriptional coactivators CREB-binding protein and p300 are highly essential collectively but not individually in peripheral B cells. Blood. 2006;107(11):4407–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chan-Penebre E, Kuplast KG, Majer CR, Boriack-Sjodin PA, Wigle TJ, Johnston LD, et al. A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nat Chem Biol. 2015;11(6):432–7. [DOI] [PubMed] [Google Scholar]
- 53.Kaushik S, Liu F, Veazey KJ, Gao G, Das P, Neves LF, et al. Genetic deletion or small-molecule inhibition of the arginine methyltransferase PRMT5 exhibit anti-tumoral activity in mouse models of MLL-rearranged AML. Leukemia. 2018;32(2):499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tarighat SS, Santhanam R, Frankhouser D, Radomska HS, Lai H, Anghelina M, et al. The dual epigenetic role of PRMT5 in acute myeloid leukemia: gene activation and repression via histone arginine methylation. Leukemia. 2016;30(4):789–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu X, Fernando TM, Lossos C, Yusufova N, Liu F, Fontan L, et al. PRMT5 interacts with the BCL6 oncoprotein and is required for germinal center formation and lymphoma cell survival. Blood. 2018;132(19):2026–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.George J, Lim JS, Jang SJ, Cun Y, Ozretic L, Kong G, et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524(7563):47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kishimoto M, Kohno T, Okudela K, Otsuka A, Sasaki H, Tanabe C, et al. Mutations and deletions of the CBP gene in human lung cancer. Clin Cancer Res. 2005;11(2 Pt 1):512–9. [PubMed] [Google Scholar]
- 58.Gui Y, Guo G, Huang Y, Hu X, Tang A, Gao S, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet. 2011;43(9):875–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mullighan CG, Zhang J, Kasper LH, Lerach S, Payne-Turner D, Phillips LA, et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature. 2011;471(7337):235–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Okosun J, Bodor C, Wang J, Araf S, Yang CY, Pan C, et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat Genet. 2014;46(2):176–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilson BG, Helming KC, Wang X, Kim Y, Vazquez F, Jagani Z, et al. Residual complexes containing SMARCA2 (BRM) underlie the oncogenic drive of SMARCA4 (BRG1) mutation. Mol Cell Biol. 2014;34(6):1136–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peifer M, Fernandez-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet. 2012;44(10):1104–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.So CK, Nie Y, Song Y, Yang GY, Chen S, Wei C, et al. Loss of heterozygosity and internal tandem duplication mutations of the CBP gene are frequent events in human esophageal squamous cell carcinoma. Clin Cancer Res. 2004;10(1 Pt 1):19–27. [DOI] [PubMed] [Google Scholar]
- 64.Ogiwara H, Sasaki M, Mitachi T, Oike T, Higuchi S, Tominaga Y, et al. Targeting p300 Addiction in CBP-Deficient Cancers Causes Synthetic Lethality by Apoptotic Cell Death due to Abrogation of MYC Expression. Cancer Discov. 2016;6(4):430–45. [DOI] [PubMed] [Google Scholar]
- 65.Gao G, Zhang L, Villarreal OD, He W, Su D, Bedford E, et al. PRMT1 loss sensitizes cells to PRMT5 inhibition. Nucleic Acids Res. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.