Abstract
Osteocytes, the bone cells embedded in the mineralized matrix, control bone modeling and remodeling through direct contact with adjacent cells and via paracrine and endocrine factors that affect cells in the bone marrow microenvironment or distant organs. Osteocytes express numerous G protein-coupled receptors (GPCRs) and mice lacking the stimulatory subunit of G-protein (Gsα) in osteocytes (Dmp1-GsαKO mice) have abnormal myelopoiesis, osteopenia and reduced adipose tissue. We previously reported that the severe osteopenia and the changes in adipose tissue present in these mice were mediated by increased sclerostin, which suppress osteoblast functions and promote browning of white adipocytes. Inversely, the myeloproliferation was driven by granulocyte colony-stimulating factor (G-CSF) and administration of neutralizing antibodies against G-CSF only partially restored the myeloproliferation, suggesting that additional osteocyte-derived factors might be involved. We hypothesized that osteocytes secrete Gsα-dependent factor(s) which regulate myeloid cells proliferation. To identify osteocyte-secreted proteins, we used the osteocytic cell line Ocy454 expressing or lacking Gsα expression (Ocy454-Gsαcont and Ocy454-GsαKO) to delineate the osteocyte “secretome” and its regulation by Gsα. Here we reported that factors secreted by osteocytes increased the number of myeloid colonies and promoted macrophage proliferation. The proliferation of myeloid cells was further promoted by osteocytes lacking Gsα expression. Myeloid cells can differentiate into bone-resorbing osteoclasts therefore we hypothesized that osteocyte-secreted factors might also regulate osteoclastogenesis in a Gsα-dependent manner. Conditioned medium (CM) from Ocy454 (both Gsαcont and GsαKO) significanlty increased the proliferation of bone marrow mononuclear cells (BMNC) and, at the same time, inhibited their differentiation into mature osteoclasts via a Gsα-dependent mechanism. Proteomics analysis of CM from Ocy454 Gsαcont and GsαKO cells identified neuropilin-1 (Nrp-1) and granulin (Grn) as osteocytic-secreted proteins upregulated in Ocy454-GsαKO cells compared to Ocy454-Gsαcont, whereas semaphorin3A was significantly suppressed. Treatment of Ocy454-Gsαcont cells with recombinant proteins or knockdown of Nrp-1 and Grn in Ocy454-GsαKO cells partially rescued the inhibition of osteoclasts, demonstrating that osteocytes control osteoclasts differentiation through Nrp-1 and Grn which are regulated by Gsα signalling.
Keywords: Osteocytes, Gsα, Osteoclast, Neuropilin-1, Macrophages, Myeloid cells
Introduction
Osteocytes are the most abundant cells in the adult skeleton, accounting for 90–95% of bone cells (1). They extend their dendritic processes within the canaliculi that connect the lacunae and form a complex lacuno-canalicular network. These dendritic processes allow osteocytes to contact and communicate with adjacent cells, with bone lining cells and osteoblasts on the endosteal and periosteal surfaces and with endothelial cells. Some of these processes extend into the bone marrow space allowing osteocytes to direclty communicate with cells in the bone marrow microenvironment (2–4). Osteocytes control bone homeostasis, muscle functions and adipogenesis through paracrine and endocrine factors, including sclerostin, Rank ligand (Rankl), prostaglandins and semaphorin 3A (Sema3A), as recently reported (5, 6).
Osteocytes express several G-protein coupled receptors, including the Parathyroid hormone (PTH) receptor type 1 (PTH1R), the prostaglandin receptors E2 and E4, and the β-adrenergic receptors (7–9). Heptahelical receptors, or GPCRs, signal via different heterotrimeric G proteins, each of which comprises an α-subunit, which binds a guanine nucleotide, and a tightly associated dimer comprised of Gβ and Gγ. Gα subunits are divided into four groups according to effector coupling and sequence similarity. The gene encoding the stimulatory subunit Gsα, Gnas, is a complex imprinted gene that generates multiple products through the use of alternative promoters and different first exons that splice onto a common second exon (exon 2). The major product encoded by Gnas is Gsα, the ubiquitously expressed Gα protien that is required for receptor-stimulated cAMP accumulation (for a review see (10)).
We have previously shown that mice lacking Gsα in osteocytes (Dmp1-Cre+;Gsαfl/fl, or Dmp1-GsαKO) have severe osteopenia characterized by a significant decrease in both trabecular and cortical bone, and an hematopoietic phenotype characterized by neutrophilia, thrombocytosis, and leukocytosis in bone marrow, spleen, and peripheral blood(11). The severe osteopenia present in the Dmp1-GsαKO mice is due to dramatic suppression of bone formation due to loss of endosteal and trabecular osteoblasts. Histomorphometric analysis of these animals also revealed a significant reduction in the number of osteoclasts, demonstrating an osteoclast defect as well (12). We identified granulocyte-colony stimulating factor (G-CSF) and sclerostin as two Gsα-dependent factors promoting the neutrophilia and the osteopenia, respectively, present in the Dmp1-GsαKO mice. Treatment of Dmp1-GsαKO mice with neutralizing antibodies against sclerostin and G-CSF only partially restored the skeletal and hematological phenotype, suggesting that additional osteocyte-derived factors might be involved. The main aim of this study was to identify osteocyte-derived secreted proteins (novel or known) regulated by Gsα and capable of controlling hematopoiesis and skeletal homeostasis that can provide new directions (and therapies) for the treatment and management of hematologic diseases and bone disorders. To gain insight into the cellular and molecular mechanisms by which osteocytes control myelopoiesis and skeletal homeostasis, we used the osteocytic cell line, Ocy454, to delineate the osteocyte secretome and the role played by Gsα signaling. Ocy454 cells express high Sost/sclerostin levels and respond to both PTH and mechanical forces. Importantly, as recently reported (13, 14), when Gsα was knocked out in these cells using the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and the CRISPR Associated (Cas) system (CRISPR/Cas9), there was a significant increase in Sost/sclerostin expression, phenocopying the in vivo findings (13). Interestingly, G-CSF expression was unchanged in Ocy454-GsαKO cells compared to Ocy454-Gsαcont suggesting that additional factors might be involved. By using a combination of in vitro and ex vivo models, we determined and reported here that osteocytes express and secrete several Gsα-dependent and -independent factors capable of supporting myelopoiesis, promoting macrophage proliferation and inhibiting osteoclast formation. We identified osteocyte-derived Neuropilin-1 (Nrp-1) and Granulin (Grn) as two Gsα-dependent factors regulating osteoclastogenesis. Also, we demonstrated that M-CSF secreted by osteocytes partially drives the increase in monocyte/macrophage present in the Dmp1-GsαKO mice.
Material and Methods
Mice
All animal experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of Boston University. Wild type mice C57BL/6N Cr were purchased from Charles River Laboratories whereas the Dmp1-cre:Gsαfl/fl and Gsαfl/fl were generated by mating Dmp1-Cre mice (Jackson #023047) with mice in which exon 1 of Gsα was flanked by LoxP sites (Gsαfl/fl kindly provided by Dr Weistein, NIH), as previously described (11, 15). Animals were housed at 4–5 animals per cage, subjected to 12 hr light/dark and fed ad libitum standard chow.
Cell cultures
The Ocy454 cell line was generated as previously reported (16). Cells were routinely maintained in non-collagen-coated flasks (Corning) in α-MEM containing 10% heat-inactivated Fetal Bovine Serum (FBS) (Gibco) and 1% antibiotic-antimycotic (AA) (Gibco)(complete medium); medium was changed twice a week. For all experiments, cells were plated at 1 × 105 cells/ml and allowed to proliferate at 33 ºC for two to three days (permissive temperature) and differentiate at 37 ºC (non-permissive temperature) for the subsequent time specified in each experiment. For PTH experiments, Ocy454-GsαKO and Ocy454-Gsαcont cells were plated at 1 × 105 cells/ml and allowed to reach confluence at 33 °C for three days. Cells were then cultured at 37 °C for 7 and 14 days. Cells were treated with 10 nM human parathyroid hormone (hPTH) (1–34) or 10 μM Forskolin (F6886, Sigma) for four hours prior to RNA isolation (RNeasy Plus Mini Kit, Qiagen).
Preparation of Ocy454-Gsα Control and KO conditioned media
Ocy454-GsαKO and Ocy454- Gsαcont clones were plated at 1 × 105 cells/ml in 6-well plates and were cultured in complete medium (alpha-MEM, 10% FBS, and 1% AA). Cells were allowed to proliferate at 33 ºC for three days prior to medium change and were then transferred to 37 ºC for six additional days to allow differentiation, with a media change on day four. On day six, medium was changed to low (αMEM, 0.1% FBS) or regular (αMEM, 10% FBS) serum-containing medium. Conditioned medium (CM) was collected 24 hours later, briefly centrifuged to eliminate cell debris, aliquoted, and stored at −20 ºC until use. Initial experiments demonstrated that control medium kept in the incubators (i.e 37 ºC and 5% CO2 for 24 hrs) was indistinguishable from fresh medium when used in both the colony formation assays and in the osteoclastogenesis therefore, for the majority of the experiments we used fresh complete medium.
Colony formation in Methylcellulose
Bone marrow (BM) cells were obtained from the long bone of 6–8 weeks-old wild type C57BL/6N mice. BM was flushed using phosphate-buffered solution (PBS)-2% FBS, followed by centrifugation at 1,200 rpm for 10 minutes at 4 ºC. Cells were then resuspended in 5 ml of αMEM, 10% FBS, and 1% antibiotic-antimycotic. Cells (2 × 104 cells) were cultured in MethoCult (STEMCELL Technologies) containing CM from Ocy454-GsαKO and Ocy454- Gsαcont cultures or complete medium alone (no treatment control). Two different methylcellulose media were used for the growth of myeloid cells: MethoCult M3534 and M3231. MethoCult M3534 contains growth factors and cytokines (FBS, BSA, insulin, transferrin, 2-mercaptoethanol, IL-3, and IL-6) that support the growth of myeloid cells whereas MethoCult M3231 lacks growth factors and cytokines. CM of Ocy454-GsαKO and Ocy454-Gsαcont CM or αMEM with 0.1% FBS (control medium) (600 μl) were mixed with either M3534 or M3231 (3 ml). These mixtures were placed in 35-mm culture dishes with a 2-mm grid (174926, Nalge Nunc), followed by incubation at 37 ºC. After seven days in culture, colonies were counted using a microscope (EVOS Cell Imaging System, Thermo Fisher). Colonies comprised of more than 25 cells were enumerated. Each experiment was done in duplicate or triplicate and repeated at least three times.
Bone marrow mononuclear cell isolation and differentiation
BM cells were flushed from the long bones (tibias and femurs) of 6–8 weeks-old C57BL/6N mice and cultured for 24–48 hours. Non-adherent cells were collected and separated by Ficoll Paque PLUS (General Electric Healthcare) to isolate the bone marrow mononuclear cell (BMNC) population. BMNC (2 × 104 cells/well) were plated in a 96-well plate, followed by treatment with 50 ng/ml of M-CSF (Shenandoah, Cat# 200–08) in complete medium (10% FBS) alone as control or in CM from Ocy454-GsαKO and Ocy454-Gsαcont (50% v/v). After three days in culture, 50 ng/ml of RANKL (Shenandoah, Cat# 200–04) was added, followed by incubation for additional days, as described in the results. TRAP staining and TRAP activity were determined at the end of the differentiation period as described below. Cells with three nuclei or more were scored as one osteoclast. In addition to M-CSF and RANKL treatments, BMNC were treated with recombinant mouse granulin (1000 ng/ml) (R&D, Cat# 2557-PG-050) or recombinant mouse neuropilin-1 (1000 ng/ml) (R&D, Cat# 5994-N1–05) in combination with in Ocy454-Gsαcont CM.
Resorption pit assay
BMNC (2 × 104 cells per well) were plated in a 96-well plate (Osteo Assay Surface, Corning) and treated with 50 ng/ml M-CSF in complete medium (10% FBS, control medium) or with CM from Ocy454-GsαKO or Ocy454-Gsαcont (50% v/v). After three days in culture, the BMNC were treated with 50 ng/ml RANKL, in addition to 50 ng/ml M-CSF. Treatment was repeated every three days. Culture medium was removed after 10 days and cells were removed with 100 μl of 10% bleach (10 minutes at room temperature). The plate was washed twice with 150 μl per well of distilled water and air-dried for one hour at room temperature. Pits were visualized using a microscope (EVOS Cell Imaging System, Thermo Fisher) and analyzed using Image J software.
F-actin ring staining
BMNC (2 × 104 cells per well) were plated in a 96-well plate and cultured as described above (Bone marrow mononuclear cell isolation and differentiation). After five days the medium was aspirated, and the plates were washed three times with PBS. Cells were fixed with 10% phosphate-buffered formalin (Fisher), washed twice with PBS, permeabilized with 0.5% Triton X-100 (Sigma-Aldrich) (diluted in PBS) for five minutes at room temperature, and then incubated with 0.5 μg/ml rhodamine phalloidin (Invitrogen) for 30 minutes in dark. DAPI (Thermo Fisher Scientific) was used to stain nuclei. Cells were washed with PBS and examined under fluorescence microscopy (BZ-X700, Keyence).
Apoptosis assay
Following five days of M-CSF and RANKL treatment, 100 μl of Caspase Glo® 3/7 reagent (Apo-Tox Glo™, Promega) was added to each well. The plate was rocked on a shaker at 500 rpm for 30 seconds then incubated at room temperature for 30 minutes. A microplate reader (Berthold, TriStar LB941) was used to measure the luminescence generated by the caspase activity.
Osteocyte-enriched bone explants (OEBE) preparation
OEBE were generated from femurs and tibia of 6-weeks-old Dmp1-Cre;Gsαfl/fl mice or control Gsαfl/fl mice, as described previously (11). Briefly, after removing the epiphyses and flushing out the BM, the explants were digested three times with collagenase type I and II (1:3 w/w) (Worthington) and once with 5 mM EDTA (Sigma) (20 min each) to completely remove endosteal and periosteal osteoblasts and BM cells. To maintain a constant volume/weight ratio among the wells, twenty μl of complete medium was added for every 10 mg of OEBE. OEBEs were plated in 12-well plates and cultured at 37 ºC in 5% CO2 for 24 hours prior to changing the medium, followed by five days of additional culture. CM was then collected and stored at −20 ºC. CM from OEBE was used to treat BMNC with M-CSF and RANKL, as described above in the Bone marrow mononuclear cell isolation and differentiation section. RNA was isolated from OEBE with TRIzol (Invitrogen), using a tissue homogenizer (TissueLyser II, Qiagen) and the Purelink RNA mini kit (Invitrogen, Thermo Fisher) according to the manufacturers’ recommendations. In a separate set of experiments, OEBEs were autoclaved (121 ºC for 30 min) to eliminate cells but still maintain the mineral and matrix component (non-viable OEBEs).
Mass spectrometric analysis
Phenol-free medium (Gibco) without FBS was used to prepare CM from both Ocy454-GsαKO and Ocy454-Gsαcont cells. Amicon columns (30K; Millipore) were washed with 0.1 N NaOH and then with Milli-Q water. Amicon columns were used to separate proteins in CM into > 30K and < 30K fractions. The < 30K fraction was placed in a C18 cartridge and sequentially eluted with 30–60% acetonitrile and 1% formic acid. Flow samples through the C18 cartridge were collected in 1.5-ml microcentrifuge tubes and placed in a speed vacuum for drying; the dried samples were then suspended in 30 μl of 0.1% RapiGest in 100 mM triethyl ammonium bicarbonate (TEAB). A solution of ethanol, acetone, and 0.1% acetic acid was chilled at −20 °C for one hour, then 1 ml of this solution was added to the > 30K fractions and these were kept at −20 °C overnight. The next day, the > 30K fractions were centrifuged (20,000 × g at 4 °C for 10 minutes). The tubes were inverted to decant the solution into new tubes without disturbing the pellets. The pellets were allowed to dry for three minutes and were then suspended in 30 μl of 0.1% RapiGest in 100 mM triethyl ammonium bicarbonate (TEAB) and mixed by vortexing. Both fractions were reduced with dithiothreitol (DTT) and then were alkylated with iodoacetamide and digested overnight with trypsin at 37 °C, then dried by speed vacuum and subjected to analysis by nano-HPLC MS/MS for protein identification and relative quantification of protein expression change. Label-free differential relative quantification was performed as described previously and details are provided in the supplemental material (17–19). Select results are shown in Table 1.
Table 1.
Secreted proteins identified as significantly changed in expression levels in Ocy454 GsαKO CM and Ocy454 GsαCont CM, as determined by mass spectrometry
| GSA-DEPENDENT SECRETED FACTORS | ||
|---|---|---|
| Proteins | P-values | Fold changes |
| CP | 0.017 | 13.6 |
| NRP-1 | 0.028 | 9.3 |
| β2M | 0.01 | 2.28 |
| GRN | 0.04 | 1.5 |
| Col3α1 | 0.047 | −17.4 |
| SEMA3D | 0.045 | −8.7 |
| SEMA3C | 0.005 | −5.2 |
| Col1α1 | 0.03 | −5.1 |
| Col1α2 | 0.02 | −4.8 |
| SEMA3A | 0.01 | −2.9 |
Label-Free LC-MS/MS
LC-MS/MS was performed using a nanoAcquity UPLC (Waters Corp.) coupled with a Q Exactive Orbitrap mass spectrometer (Thermo Fisher Scientific) equipped with a TriVersa NanoMate ion source (Advion). Separation was with 150 μm × 100 mm 1.7-μm BEH C18 material (Waters). A linear gradient of A:B solvents (1.5% ACN/0.1% FA: 98.5% ACN/0.1%FA) was used (3 to 55% B over 120 min) at a flow rate of 0.5 μl/min. Mass spectra were acquired in the positive-ion mode over m/z 400–2000 at 30,000 resolution (fwhm at ) m/z 400). MS/MS spectra were acquired on the top-10 precursors per duty cycle with signal intensities > 1 × e4, using HCD at 35 V. MS and MS/MS data analysis was carried out using Progenesis LCMS 4.1 (Nonlinear Dynamics/Waters) for label-free differential quantification, and Mascot 2.3.01 (Matrix Science) for protein identification. MS/MS data were searched against a UniProt Mouse proteome database (0589–10090) 08–2016 with 79,954 entries. Search parameters were as follows: tryptic cleavage with 4 missed sites, carbamidomethyl-cysteine as a fixed modification. Precursor mass error was set to 10 ppm and MS2 mass error was 0.05 Da. A protein false discovery rate of <5% was used. Label-free analysis was performed on all MS1 signals from the 2 groups of samples. ANOVA, PCA and hierarchical clustering were performed yielding 2 distinct groups of features which indicated an increase or decrease in protein expression related to the KO model. Final protein fold changes were restricted to those with > 1.5 change with p-values <0.05.
CRISPR/Cas 9 gene editing in Ocy454 cells
Two guided (g)RNAs were generated to target exon 1 and 2 of the Gsα gene and. gRNA (forward sequence: 5’-CCT CGG CAA CAG TAA GAC C-3’; reverse sequence: 5’-GAC CGA GGA CCA GCG CAA CG-3’) were subcloned into pSpCas9(BB)-2A-GFP (PX458; a gift from Feng Zhang) (Addgene plasmid # 48138) (15), which co-express Cas9, and eGFP and the construct was used to target Gsα in Ocy454 (16). A non-targeting gRNA was used to generate control cells. Ocy454 cells were plated at 1 × 105 cells/ml in 6-well plates and after 48 hours in culture, cells were transfected using FuGENE® (Promega). Transfection solution was prepared by mixing 3 μl of FuGENE®, 1 μg of plasmid, and 100 μl of serum-free medium for each well of a 6-well plate. After 48 hours high GFP (GFPhi) cells were sorted by FACS and single cells were plated in 96-well plates. Medium was changed twice per week during cell expansion. Cells were routinely maintained at 33ºC in non-collagen coated T-75 flasks. To account for clonal variability, three different knockout subclones (Ocy454-Gsα KO-1, KO-2 and KO-3) and three different control subclones (Ocy454-Gsα Control-1, Control-2 and Control-3) were used for the studies described here (17).
Quantitative real time PCR
Ocy454-GsαKO and Ocy454-GsαCon cells were plated at 1 × 105 cells per ml in 6-well plates and cultured in complete medium for 3 days at 33 ºC and for 7–14 days at 37 ºC. RNA was isolated at day 7 and 14 days. For BMNC, cells were plated at 1 × 105 in 12-well plates and treated with M-CSF (50 ng/ml) and Ocy454-GsαKO and Ocy454-GsαCon CM (50% v/v). RNA was extracted after 6 days in culture. For osteoclast differentiation experiments, BMNCs were plated at 1 × 105 in 12-well plates and treated with M-CSF and Ocy454-GsαKO and Ocy454- GsαCont CM for three days and with 50 ng/ml RANKL for an additional three and five days. RNA was extracted after three and five days of RANKL treatment. For all experiments, RNA was isolated using the RNAeasy kit according to the manufacturer’s recommendations (RNAeasy, Qiagen). RNA was quantified by UV absorbance (NanoDrop, Thermo Fisher). cDNA was synthesized from 0.5–1 μg of total RNA using qScripttm cDNA Super Mix (Quanta, Biosciences), followed by SYBR Green (Power SYBR, Thermo Fisher Scientific) quantitative real-time PCR (StepOnePlus, Life Technologies). Gene expression differences between samples were calculated using the comparative cycle threshold (CT) method 2 (−ΔΔCT). β-actin was used as reference. Experiments were run in triplicate unless otherwise indicated.
Cell proliferation assays
BMNs were plated in 96-well plates at 2 × 104 cells/well and treated either with CM from Ocy454-GsαKO and Ocy454-GsαCont or complete medium (as an experimental control), with or without M-CSF (50 ng/ml). CM and complete medium were changed every three days. After 6, 24, 48, 72, 96, and 168 hours (7 days) in culture, cells were frozen at −80 ºC and then assayed for cell proliferation. Reagents were prepared according to the CYQUANT Cell Proliferation Assay protocol (Invitrogen). Plates were thawed at room temperature and 200 μL of fluorescent reagent was added to each well, followed by incubation for 15 minutes at room temperature with light protection. From each well, 150 μL were transferred to a black 96-well plate (Corning) and fluorescence was measured using a microplate reader at 490 nm excitation and 533 nm emission. Bacterial DNA was used to generate a standard curve to determine the DNA concentration (ng/ml) in each sample.
Flow cytometry
BMNC (1 × 105 per well) were cultured in 12-well plates and treated with CM from Ocy454-GsαKO, Ocy454- GsαCont or complete media (as an experimental control). After four days in culture, BMNC were stained for 20 minutes with fluorescence-conjugated antibodies against CD11b, F4/80, and Gr1 (1:200 dilution for all) (eBioscience). After staining, cells were washed with 2% FBS-PBS and analyzed by flow cytometry using LSRII (BD eBioscience).
Lentivirus production
ShRNAs were purchased from Sigma-Aldrich. HEK293T cells were plated at 2.2 × 105 cells/ml in 6-well plates in high-glucose DMEM (Gibco) and 10% FBS without antibiotics. Cells were transfected the next day when approximately 80% confluent. In each well, 100 μl of serum-free DMEM, 500 ng of pLKO1 puro plasmid containing the gene of interest, 500 ng of psPAX2 (a gift from Dr. Trono, Addgene #12260) and 50 ng of pMD2.G (a gift from Dr. Trono, Addgene #12259) were transfected using 3 μl of FuGENE® HD transfection reagent (Promega). Twenty-four hours after the transfection, media were replaced with DMEM + 10% FBS with antibiotic-antimycotic. After 48 hours, medium was collected from each well, spun down at 1,000 rpm, 4 °C for 10 minutes, and virus supernatant was collected, aliquoted, and stored at −80 °C.
Lentivirus infection
Ocy454-GsαKO and Ocy454- GsαCont cells were plated at 1 × 105 cells/mL in 12-well plates 24 hours before infection. Cells were infected at 80% confluency with 100 μl of lentivirus in the presence of 10μg/ml polybrene. Culture media were changed 24 hours after the infection and 5μl/ml of puromycin was added to the medium. Medium (containing puromycin) were replaced every three days thereafter. After day 10 in culture, the cells were trypsinized and transferred to T-25 flask. CM were collected as described above.
TRAP staining
For TRAP staining, culture media were aspirated from the 96-well plates and cells were washed three times with PBS. Cells were fixed in 10% formalin (10–15 minutes, at room temperature), fixed in acetone and methanol (50:50) and then incubated with TRAP staining solution (0.1 M sodium acetate, 0.1 M of sodium tartrate, 0.1 mg/ml naphthol AS-MX phosphate, 0.6 mg/ml red-violet LB salt in PBS) at 37 ºC for 20–30 minutes. Plates were washed with water and allowed to dry. Osteoclasts were counted under the microscope (EVOS Cell Imaging System, Thermo Fisher). A cell with three or more nuclei was scored as a positive osteoclast. Occupancy of surface area by osteoclasts was calculated using Image J software after manually tracing each osteoclast in each well.
TRAP activity
Cell culture supernatant (30 μl) was transferred into a 96-well plate and TRAP staining solution was added to each well (170 μl/well). The plate was incubated at 37ºC for up to two hours in the dark and the OD values were read at 540 nm. TRAP solution alone was used as a reference and subtracted from each sample reading.
Protein extraction and immunoblotting
Whole cell lysates from cell culture were prepared using lysis buffer containing 50 mM Tris base, 1 mM EDTA, 1.5 mM MgCl2, 150 mM NaCl, 1% Triton X-100, and 10% glycerol supplemented with protease and phosphatase inhibitors (Sigma, cat# P5726 and cat# P0044, respectively). Protein concentration was determined using the Bradford protein assay (Protein Assay Dye Reagent, Bio-Rad). Bovine serum albumin (BSA) (New England Biolabs) was used to generate a standard curve. Approximately 15–20 μg of protein were separated on a 10% polyacrylamide gel (Fisher Scientific) and transferred to a PVDF membrane (Bio-Rad). The membranes were blocked with blocking buffer (TBST) supplemented with 5% BSA (Sigma) for one hour and incubated overnight at 4 °C with anti-Gsα antibody (diluted 1:2000) (Millipore, cat# 06–237). Following overnight incubation, membranes were washed three times with TBST, and hybridized with HRP-conjugated secondary antibody (anti-rabbit IgG, diluted 1:2000, Cell Signaling, #7074) in blocking buffer for one hour at room temperature. The membranes were developed using enhanced chemiluminescence or SuperSignal™ West Femto (Thermo Fisher Scientific) and the signals were detected using a charge-coupled device (CCD) camera on G: Box (Syngene).
Statistical analysis
Data are presented as mean ± standard error of the mean (SEM). Statistically significant differences between groups and treatments were determined by the student t-test or ANOVA using Graph Pad Prism software. P value and fold change are reported for each experiment. Each experiment was conducted in triplicate and performed at least three times, unless otherwise specified in the figure legends.
Results
Characterization of Ocy45- GsαKO cells
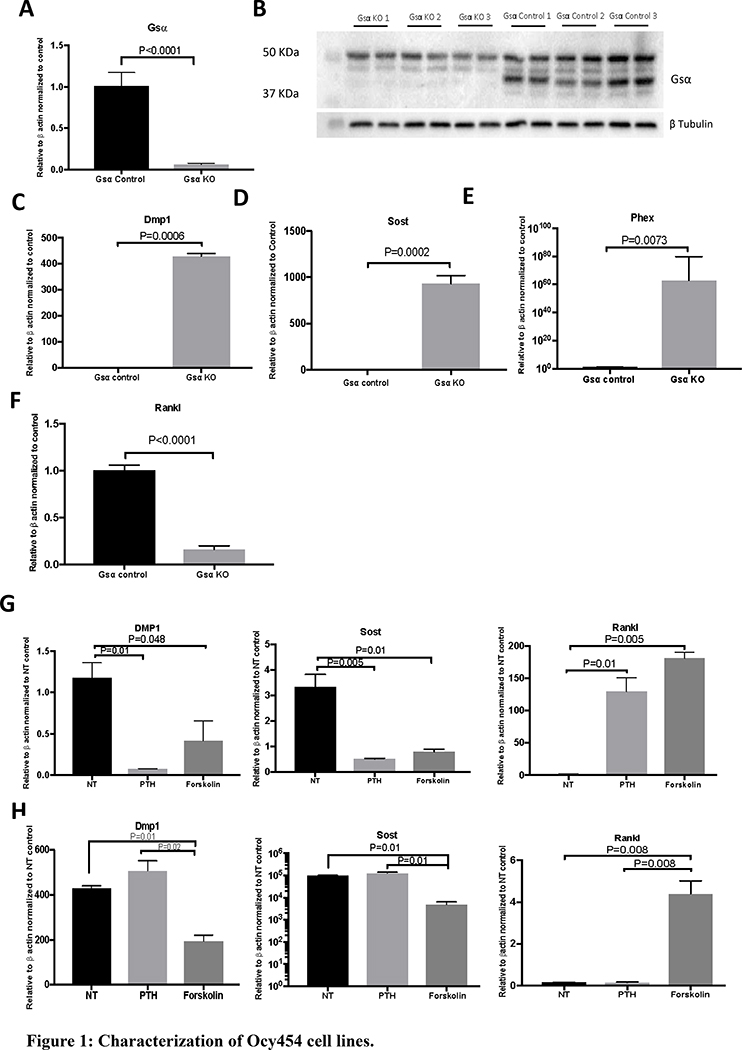
The Ocy454 cell line was derived as previously reported (13). Gnas was knocked out using the CRISPR/Cas9 technique and single cell colonies were isolated. To account for clonal variability, three different knockout subclones (Ocy454-Gsα KO-1, KO-2, and KO-3) and three control subclones (Ocy454-Gsα Control-1, Control-2, and Control-3) were studied. Gsα deletion in Ocy454-GsαKO subclones was confirmed at both protein and transcript level (Fig. 1A–1B). Deletion of Gsα in the Ocy454 cell line resulted in an increase of Dmp1 (428 fold), Phex (62 fold), Sost (600 fold) expression and 84% decrease in expression of Rankl (Fig. 1C–1F). Functional inactivation of Gsα was further analyzed upon treatment with PTH and forskolin. As expected, Ocy454-Gsαcont clones responded to PTH with a significant suppression of Dmp1 and Sost and increased Rankl expression (Fig 1G). In the absence of Gsα expression, there was no response to PTH treatment; this result confirmed the successful deletion of Gsα (Fig. 1H). The response to forskolin (positive cotrol) was similar in the two cell types, Ocy454-GsαKO and Ocy454-Gsαcont cells (Fig. 1G–1H). Deletion of Gsα did not affect cell proliferation: there was no difference in DNA concentration between Ocy454-Gsαcont and Ocy454-GsαKO cells (Fig. S1A).
Figure 1: Characterization of Ocy454 cell lines.
CRISPR/Cas9 was used to delete Gsα expression in Ocy454 cells. A) Realtime qPCR for Gsα expression in Ocy454-Gsα control and KO cells. B) Western Blot analysis of Ocy454 Gsαcont (Control 1, 2 and 3) and Ocy454 GsαKO (KO 1, 2 and 3) clones. The 37-KDa band corresponding to Gsα was not present in KO clones. Loading was normalized with tubulin. Realtime qPCR showed a significant increase in Dmp1 (C), Sost (D) and Phex (E) expression in KO cells compared to control, whereas Rankl (F) was significantly suppressed. Gene regulation in response to treatment with PTH (100 nM) and Forskolin (10 μM) in Ocy454 Gsα control (G) and KO (H) cells. Realtime data are expressed as relative to beta actin and normalized to control. Data are expressed as mean ± SEM. Each experiment was run in duplicate or triplicate samples and repeated at least three times. (One-way ANOVA)
Osteocytes secrete factors supporting myeloid cell proliferation in vitro
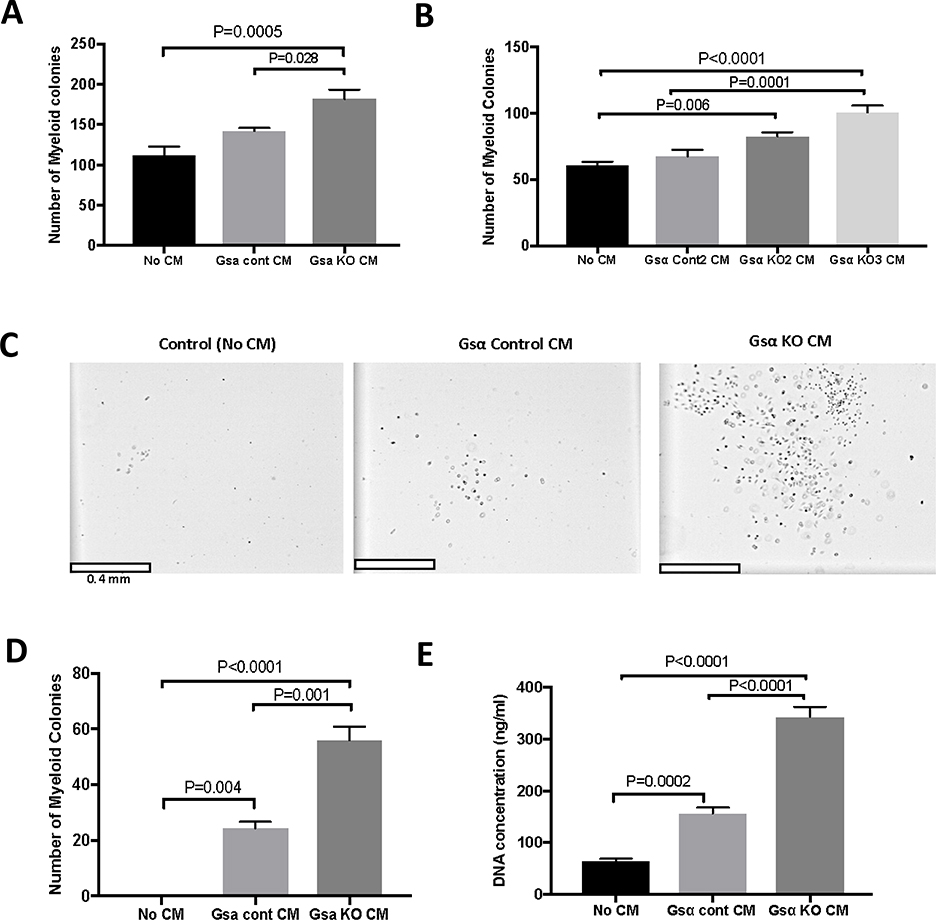
Mice lacking expression of Gsα in osteocytes have increased numbers of myeloid cells in bone marrow, spleen, and peripheral blood. Ex vivo studies using conditioned medium (CM) from osteocyte-enriched bone explants (OEBE) from Dmp1-GsαWT and Dmp1-GsαKO animals demonstrated that osteocytes secrete factor(s) that support myeloid proliferation (11). To study the molecular mechanisms of osteocyte-mediated myeloid cell proliferation, we used an in vitro model in which media from Ocy454-Gsαcont and Ocy454-GsαKO cells were used to induce myeloid cell proliferation. When supportive methylcellulose medium (M3534, containing insulin and citokines) was used, there was a 1.6 and 1.3 fold increase in the number of myeloid colonies after treatment with Ocy454-GsαKO CM as compared to medium alone or CM from Ocy454-Gsαcont respectively (Fig. 2A). We then tested if similar effects were obtained when using additional Ocy454-GsαKO clones. As shown in Fig. 2B, Ocy454-GsαKO2 and Ocy454-GsαKO3 significanlty increased the number of myeloid colonies by 1.4 and 1.7 fold respectively, as compared to medium alone. Furrhermore, Ocy454-GsαKO3 also significanlty increase by 1.5 fold the number of colonies as compared to Ocy454-GsαCont2, whereas Ocy454-GsαKO2 did not, suggesting some variability among clones. Next, we examined whether osteocyte-secreted factors present in the CM could promote myeloid cell proliferation in non-supportive conditions (i.e, in the absence of added insulin, stem cell factor, interleukin 3 and interleukin 6). Consistent with previous results, there was a significant 23 fold increase in the number of myeloid cells when CM from Ocy454-Gsαcont was added to non-supportive cultures (M3231) and the number of colonies was further increased 2.3 fold upon treatment with CM from Ocy454-GsαKO cells (Fig. 2C and 2D). CM from Ocy454-GsαKO cells significantly increased myeloid cell DNA synthesis compared to Ocy454-Gsαcont or medium control (Fig. 2E) by 2.2 and 5.3 fold respectively, demonstrating that osteocytes control myeloid cell proliferation in a Gsα-dependent manner.
Figure 2: Osteocytes support myeloid cell proliferation through Gsα-dependent secreted factor(s).
A) Quantification of myeloid colonies grown in myeloid-supporting medium and treated with conditioned medium (CM) from Ocy454-Gsα control (Gsα cont CM) and Ocy-454GsαKO (GsαKO CM) cells. B) Treatment with CM from one additional control and two additional Ocy-454GsαKO clones using supporting myeloid medium. C) Microscopic images of myeloid cells after CM treatment in non-supporting medium. D) Quantification of the number of myeloid colonies after treatment with CM from Ocy454-Gsα control and Ocy-454GsαKO cells in non-supporting medium. E) Proliferation of myeloid cells after CM treatment compared to non-treatment control. Each experiment was run in duplicate or triplicate samples and repeated at least three times. (One-way ANOVA)
Osteocytes promote proliferation of bone marrow mononuclear cells (BMNC) and macrophages
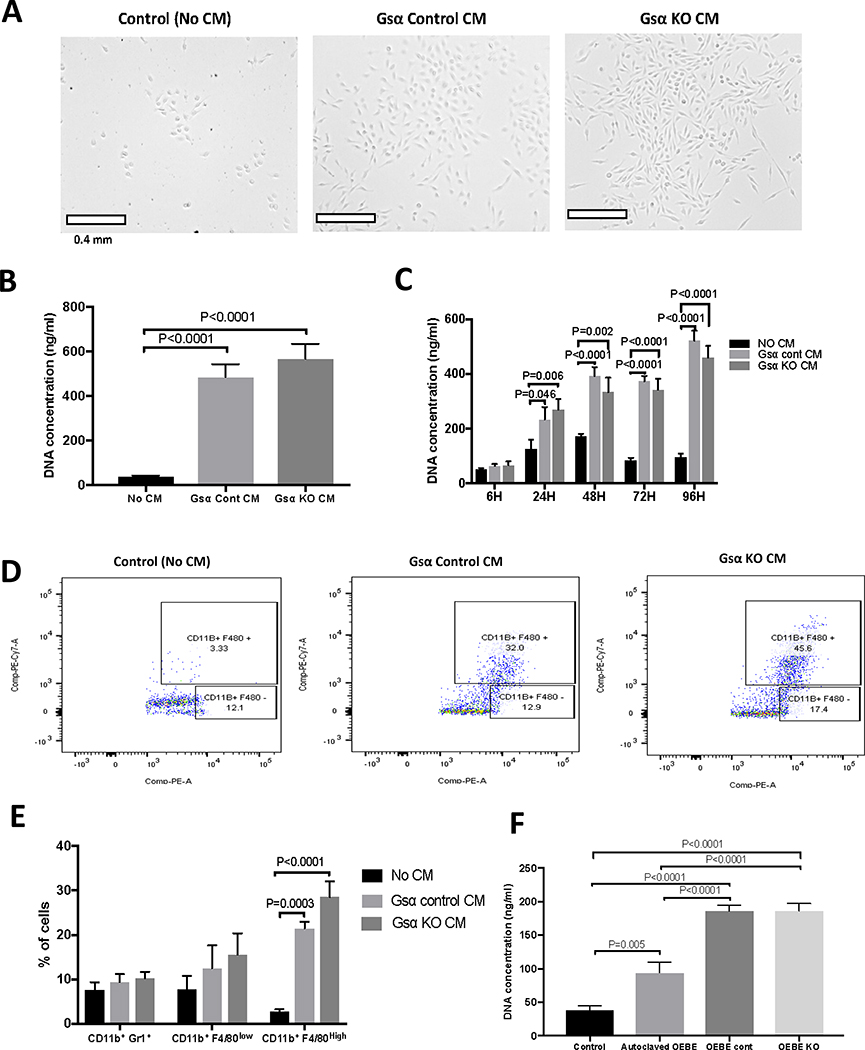
As reported previously, mice lacking Gsα in osteocytes have increased numbers of myeloid cells and marked osteopenia (11). Progenitor cells in the myeloid lineage comprise macrophages which differentiate into osteoclasts upon stimulation with M-CSF and RANKL. We hypothesized that osteocytes secrete factors that regulate BMNC proliferation and that these factors are dependent on Gsα signalling. To test this hypothesis we treated BMNCs with CM from Ocy454-Gsαcont and Ocy454-GsαKO or with medium alone. As shown in Fig. 3, CM from both the investigated cell lines, Ocy454-Gsαcont and Ocy454-GsαKO, significantly induced cell proliferation, as demonstrated by a 12–14 fold increase in DNA synthesis (Fig. 3A–B) in BMNC, as compared to the medium alone. Time-course analysis showed that BMNC proliferation was already increased by 2 fold after 24 hours of treatment with CM, and increased even further (5 fold) over time (Fig. 3C). Flow cytometric analysis demonstrated a 8–10 fold increase in the number of CD11b+F4/80high cells (macrophages) after treatment with CM from Ocy454-Gsαcont and Ocy454-GsαKO, as compared to the no-treatment control. CD11b+/F4/80Low cells (monocytes) and CD11b+/Gr1+ cells (granulocytes) were not affected by treatment with CM (Fig. 3D). Taken together these results demonstrated that Ocy454 cells secrete factors which promote macrophage proliferation and that these factors are not regulated by Gsα signaling (Gsα-independent). Next, we investigate if the same effect, i.e macrophage proliferation, could be induced by CM from bone explanst (i.e OEBEs) from mice lacking Gsα in osteocytes (DMP1-Cre; Gsαfl/fl or Dmp1GsαKO) or littermate controls (Gsαfl/fl or Dmp1GsαWT) (11). CM from OEBE-Dmp1GsαWT and OEBE- Dmp1GsαKO were used to treat BMNC isolated from bone marrow of wild-type mice. CM from non-viable OEBE (OEBEs exposed to 121 C for 30 min to eliminate the cellular component) was also used as a control to assess the effects of factors released from the bone matrix independent of cellular activities. BMNC proliferation was increased by 4–5 fold after treatment with CM from OEBE-Dmp1GsαWT and OEBE-Dmp1GsαKO. There was no difference in BMNC or macrophage proliferation upon treatment with CM from Dmp1GsαWT and Dmp1GsαKO OEBEs, demonstrating an osteocyte-mediated, Gsα-independent effect. Interestingly, CM from non-viable OEBE was also able to support BMNC proliferation, as compared to no treatment control (Fig. 3E), suggesting that the bone matrix can release factors or denatured proteins, independently of cellular activities. These results demonstrate that osteocytes, and possibly some osteoblasts or other cells still present in the OEBEs, secrete factors that support proliferation of macrophages and these factors are not dependent on Gsα signaling.
Figure 3: Osteocytes promote BMNC proliferation via secreted factor(s) independent of Gsα signaling.
A) Microscopic images of BMNC after treatment with regular medium (Control, No CM) or with CM from Ocy454-Gsα control (Gsα control CM) and Ocy-454 GsαKO (Gsα KO CM) cells. B) CM from control and KO cells promotes cell proliferation, as shown by a 12–14 fold increase in DNA concentration . C) Time course of BMNC cells proliferation upon treatment with regular medium (black bars, no CM) or with CM from Ocy454-Gsα control (light grey bars) and Ocy454-GsαKO (dark grey bars) cells. D-E) Flow cytometric analysis of BMNC treated with regular medium (Control, No CM) or CM from Ocy454-Gsα control and Ocy454-GsαKO cells. BMNC were stained with anti-CD11b and F4/80 fluorescently conjugated antibodies prior to flow cytometry. F) Proliferation of BMNC after treatment with CM from OEBEs (ex vivo bone explants) from Dmp1-GsαWT (OEBE cont) or Dmp1-GsαKO (OEBE KO) animals. To test non-cell effects, bones were autoclaved to eliminate the cellular components prior to collection of CM. Error bars represent mean ± SEM. Each experiment was run in duplicate or triplicate samples and repeated at least three times. (One-way ANOVA).
Osteocytes suppress osteoclast differentiation via secreted factor(s)
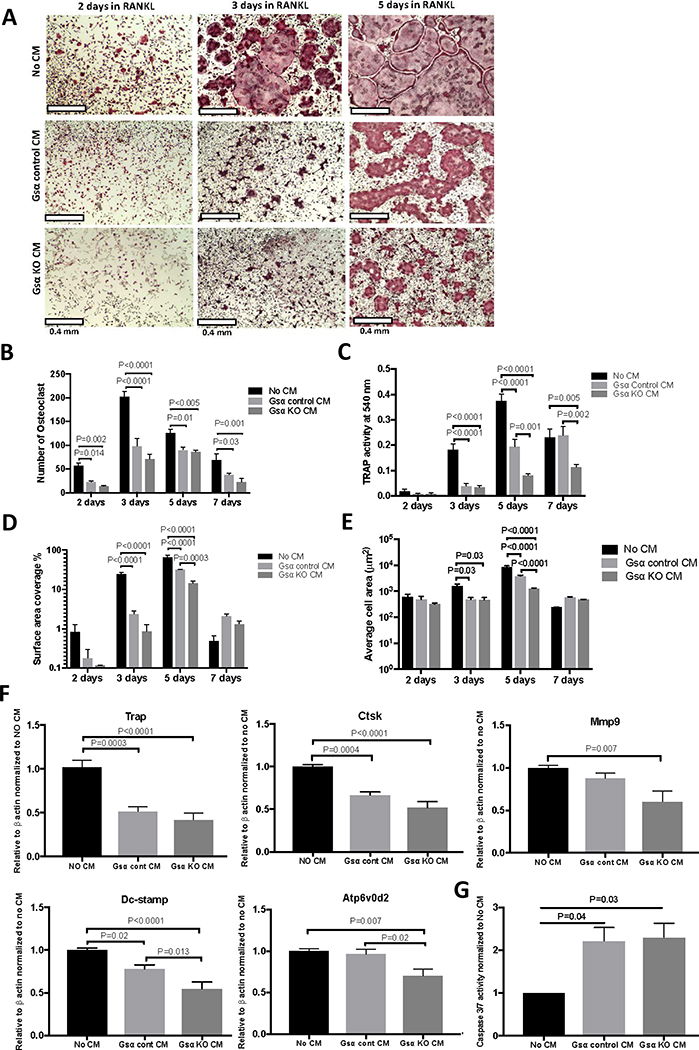
Osteoclasts are terminally differentiated macrophages derived from the myeloid lineage. We hypothesize that osteocyte-secreted factors also affect osteoclast differentiation. BMNC were treated with CM from Ocy454-Gsαcont or Ocy454-GsαKO cells in the presence of M-CSF (50 ng/ml) and RANKL (50 ng/ml). Time-course experiments demonstrated that, after two days of treatment with M-CSF and RANKL (M+R), there was a significant 60–80% decrease in the osteoclast number upon treatment with CM from Ocy454-Gsαcont and Ocy454-GsαKO cells respectively (Fig. 4A and B), whereas TRAP activity, percentage of surface area covered by osteoclasts, and average cell area were unaffected (Fig. 4C–F). After three days of M+R treatment, CM from both Ocy454-Gsαcont and Ocy454-GsαKO cells significantly suppressed osteoclast number, TRAP activity, the surface area covered by osteoclasts, and the average cell area, when compared to no CM treatment (Fig. 4A–E). After five days of M+R treatment, CM from Ocy454-Gsαcont and Ocy454-GsαKO significanlty reduced the number of osteoclasts (Fig. 4B), TRAP activity (Fig. 4C), the surface area covered by osteoclasts (Fig. 4D), and the average cell area (Fig. 4E), when compared to medium alone (no CM). In addition, CM from Ocy454-GsαKO further reduced TRAP activity (Fig. 4C), the surface area covered by osteoclasts (Fig. 4D), and the average cell area (Fig. 4E), when compared to Ocy454-Gsαcont CM, indicating that these effects are, in part, regulated by Gsα signaling. After seven days of M+R treatment, despite the presence of osteoclasts apoptosis, as demonstrated by a decrease in the number of osteoclasts (Fig. 4B) and the surface area (Fig. 4D), there was still a decrease in both the number of osteoclasts (in both cont and KO) and in TRAP activity (in KO cells)(Fig. 4C) as compared to controls. Next we evaluate gene expression in osteoclasts, upon treatment with M+R in the presence or absence of CM. Expression of Trap, Ctsk, and Dc-stamp were significantly decreased upon treatment with CM from Ocy454-Gsαcont and Ocy454-GsαKO, as compared to medium alone after three days of M+R treatment (Fig. S1 B). In addition, Mmp9, Dc-stamp, and Atp6v0d2 expression was further reduced upon treatment with Ocy454-GsαKO CM, as compared to CM from Ocy454-Gsαcont or medium alone after five days of treatment with M+R (Fig. 4F), demonstrating Gsα dependent mechanism, similarly to what observed in Fig. 4C–E. Osteoclast apoptosis was also significantly increased in cells treated with CM from Ocy454-Gsαcont and Ocy454-GsαKO cells, as compared to control (Fig. 4G). Taken together, these results demonstrated that osteocytes secrete factors which inhibit osteoclast differentiation (even in the presence of M-CSF and RANKL) and promote their apoptosis and that Gsα signaling further controls the expression Dc-stamp, and Atp6v0d2.
Figure 4: Osteocytes suppress osteoclast differentiation and promote apoptosis via Gsα-dependent secreted factor(s).
BMNCs were treated with M-CSF (50 ng/ml) and RANKL (50 ng/ml) (M+R) for various time (2,3,5 and 7 days), in the presence of regular medium (No CM, black bars) or CM from Ocy454-Gsαcont (light grey bars) or Ocy454-GsαKO (dark grey bars) cells. A) TRAP staining, B) Number of multinucleated TRAP+ osteoclasts, C) TRAP activity, D) Percentages of surface area covered by osteoclasts and E) Average cells area after 2, 3, 5 and 7 days of treatment with M+R. F) Realtime qPCR for osteoclast-specific genes Trap, Ctsk, Mmp9, Dc-stamp and Atp6v0d2 after 5 days of treatment with M+R. G) Quantification of apoptotic cells measured by Caspase3/7 activity in BMNC treated with regular medium (No CM) or CM from control and KO cells. Error bars represent mean ± SEM. Each experiment was run triplicate samples and repeated at least three times. (One-way ANOVA).
Osteocytes inhibit osteoclast function
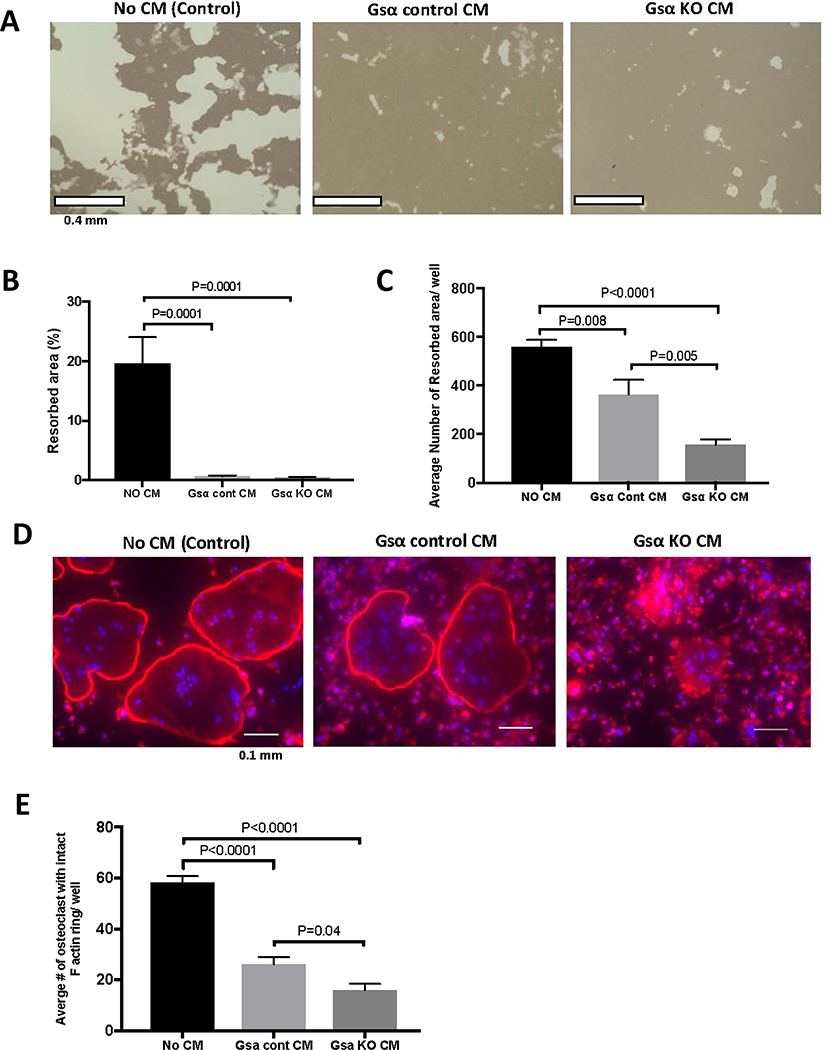
Next, we examined several functional parameters of osteoclasts upon treatment with Ocy454-Gsαcont or Ocy454-GsαKO CM. As shown in Fig. 5, there was a significant decrease in pit formation in cells treated with CM from Ocy454-Gsαcont and Ocy454-GsαKO, as compared to the medium control (Fig. 5A–C). The percentage of the resorbed area was not significantly different between Ocy454-Gsαcont or Ocy454-GsαKO CM treatment (Fig. 5B), however, the number of the resorbed areas was significantly higher in cells treated with Ocy454-Gsαcont CM, as compared to Ocy454-GsαKO CM (Fig. 5C). Formation of F-actin rings in osteoclasts is essential for proper function. There was a significant decrease in the number of osteoclasts with an intact F-actin ring when cells were treated with CM from Ocy454-Gsαcont and the number was further reduced by treatment with CM from Ocy454-GsαKO cells (Fig. 5D and 5E), demonstrating that factors released from osteocytes affect osteoclast differentiation and functions.
Figure 5: Osteocytes inhibit osteoclast functions via Gsα-dependent secreted factors.
CM from Ocy454-Gsα control and GsαKO cells suppresses osteoclast function, as shown by a reduction in the resorption area. BMNCs were seeded into Osteo-assay plates and resorption pits were analyzed after 10 days of treatment with CM and M-CSF and RANKL. A) Microscopic images of resorption area induced by osteoclasts. Quantification of B) the percentages of resorbed area and C) the average number of resorbed area in each well. Regular medium (No CM, black bars), CM from Ocy454-Gsαcont (light grey bars) or Ocy454-GsαKO (dark grey bars) cells. D) Rhodamine-phalloidin staining of F-actin rings in osteoclasts treated with regular medium (No CM), CM from Ocy454-Gsα control or CM from Ocy454-GsαKO cells. E) Quantification of the number of osteoclasts with an intact F-actin ring. (One-way ANOVA). Error bars represent mean ± SEM. Each experiment was repeated at least 3 times.
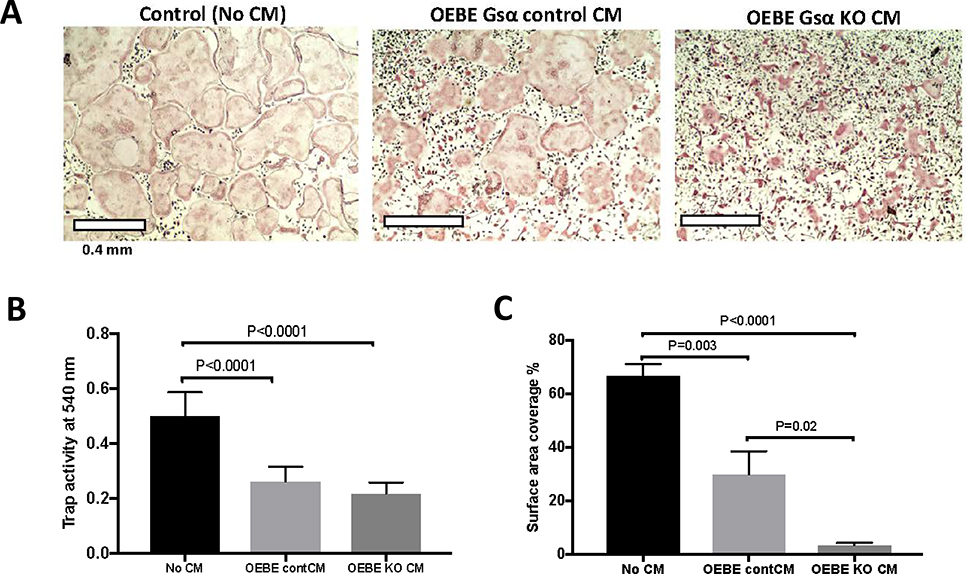
To further validate our in vitro findings in an ex vivo model, we isolated OEBE from Dmp1-GsαKO and littermate controls (Dmp1-GsαWT) (11). There was a significant decrease in osteoclast differentiation, as indicated by a significant decrease in osteoclast number, TRAP activity, and surface area covered by osteoclasts when BMNC were treated with CM from OEBE-Dmp1GsαWT and OEBE-Dmp1GsαKO, in the presence of M+R, as compared to medium alone (Fig. S2A–C). Furthermore, after five days of treatment with M+R, there was a significant decrease in the percentage of surface area covered by osteoclasts between cells treated with CM from Dmp1GsαKO, when compared to Dmp1GsαWT OEBE (Fig. 6C). Taken together these results demosntrate that osteocytes within OEBEs secrete factor(s) that suppress osteoclast differentiation and this effect is partially Gsα-dependent. (Fig. 6C).
Figure 6 : Osteocytes-enriched bone explants suppress osteoclast differentiation via Gsα-dependent secreted factors.
A) TRAP staining of osteoclasts after treatment with medium alone (No CM) or CM from Dmp1GsαWT and Dmp1GsαKO OEBEs. B) TRAP activity and C) percentages of surface area covered by osteoclasts. Regular medium (No CM, black bars), CM from Dmp1-GsαWT (light grey bars) or Dmp1-GsαKO (dark grey bars) OEBEs (One-way ANOVA). Error bars represent mean ± SEM. Each experiment was repeated at least 3 times.
Identification of osteocyte-secreted factors
To identify factors secreted by osteocytes, we performed comparative proteomic analysis of cell lysates and CM from Ocy454-GsαKO and Ocy454-Gsαcont cells. Numerous proteins (3,748 proteins in total) were detected in the CM of these cells. We focused our initial screening on secreted proteins that were significantly different between Ocy454-GsαKO and Ocy454-Gsαcont cells. As a result, several secreted proteins known to control myeloid proliferation or differentiation were identified, as shown in Table 1. Among the top upregulated secreted proteins we identified Ceruloplasmin (CP) (13.6-fold), neuropilin-1 (NRP-1) (9.3-fold), β2-microglobulin (β2M) (2.28-fold), and granulin (GRN) (1.5-fold). In contrast, Collagen 3α1 (COL3α1) and several semaphorins (SEMA 3A, C and D) were significantly decreased in CM from Ocy454-GsαKO, when compared to Ocy454-Gsαcont CM (Table 1). Additional proteins known to have a role in myeloid proliferation or osteoclast differentiation, such as M-CSF, osteopontin (OPN), macrophage migration inhibitory factor (MIF), OPG, and TGF-β were identified in the CM of these cells but the levels were not significantly different between Ocy454-GsαKO and Ocy454-Gsαcont.
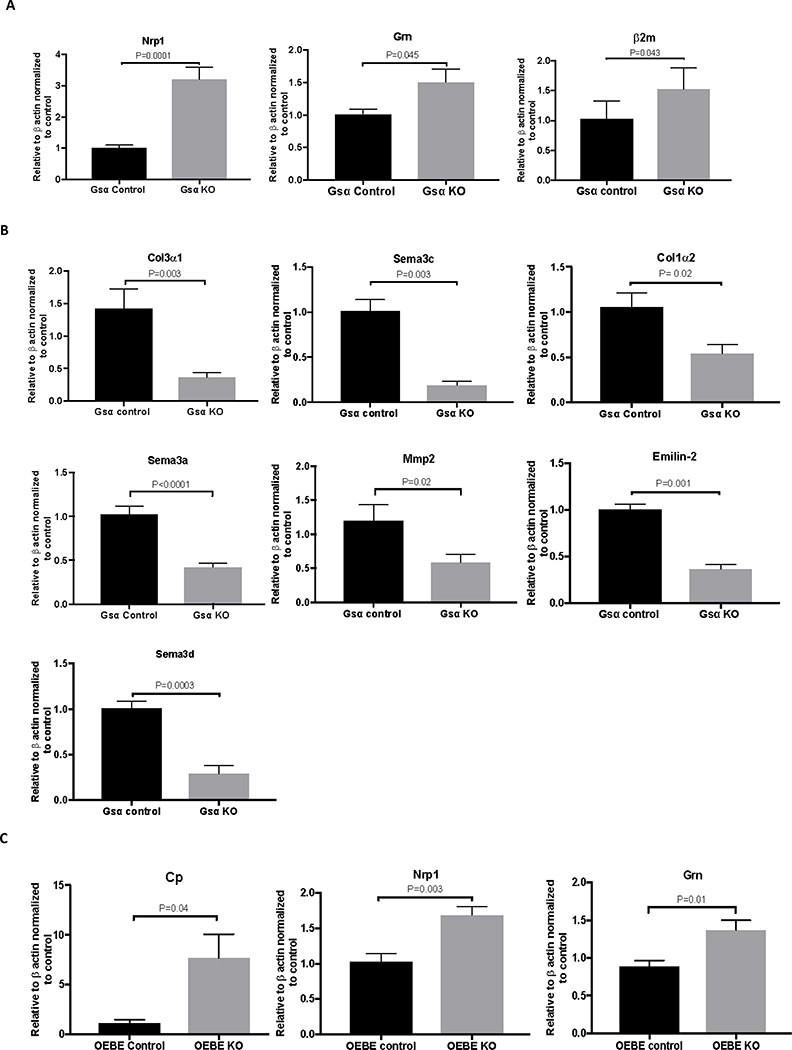
Consistent with proteomic analysis, mRNA expression of Nrp1, Grn, and β2m was also significantly increased in Ocy454-GsαKO cells (Fig. 7A), whereas expression levels of Col3α1, Sema3c, Col1α2, Sema3a, Mmp2, Emilin2, and Sema3d were significantly decreased in Ocy454-GsαKO cells, as compared to Ocy454-Gsαcont cells (Fig. 7B). Expression levels of Timp2, Mif, Opn, Igsf10, Col1α1, Mcsf, and Tgf-β were not significantly different between the two cell types (Fig. S3A). Expression levels of Cp, Nrp1, Grn, Opg, and Sema3c were similarly regulated in OEBE-Dmp1GsαKO and OEBE- Dmp1GsαWT (Fig. 7C), whereas Mcsf, Mif, Mmp2, Igsf10, Opn, Sema3a, Col1α1, Col1α2, Col3α1, Timp2, Emilin2, and Tgf-β were not significantly different between the two groups (Fig. S3B). Taken together these results identified several transcripts and proteins which are differentially regulated by Gsα-signaling in osteocytes.
Figure 7: Gene expression of identified secreted factors in Ocy454-Gsαcont and Ocy454-GsαKO cells and Dmp1-GsαWT or Dmp1-GsαKO OEBEs.
A) mRNA expression of Nrp-1, Grn and β2m was significantly increased in Ocy454 GsαKO cells (grey bars) compared to Ocy454-Gsα Control cells (balck bars). B) mRNA expression of Col3α1, Sema3c, Col1α2, Sema3a, Mmp2, Emillin2, and Sema3d was significantly reduced in Ocy454 GsαKO cells (grey bars) compared to Ocy454-Gsα Control cells (black bars). C) mRNA expression of Nrp-1, Grn and CP was significantly increased in OEBEs from Dmp1-GsαKO animals (grey bars) compared to Dmp1-GsαControl (black bars) (unpaired t-test). Error bars represent mean ± SEM. Each experiment was repeated at least 3 times.
Bioinformatics analysis of Ocy454-Gsαcont and Ocy454-GsαKO cell lysates
Proteomic analysis of Ocy454-Gsαcont and Ocy454-GsαKO cell lysates was then performed to identify proteins differentially regulated in these cells. Ingenuity Pathway Analysis (IPA, Qiagen) identified proteins in Ocy454-GsαKO cells that were significantly upregulated when compared to Ocy454-Gsαcont cells, including β2M and GRN, among others (Suppl Table 1).
The effect of osteocyte-secreted factors on BMNCs and osteoclasts
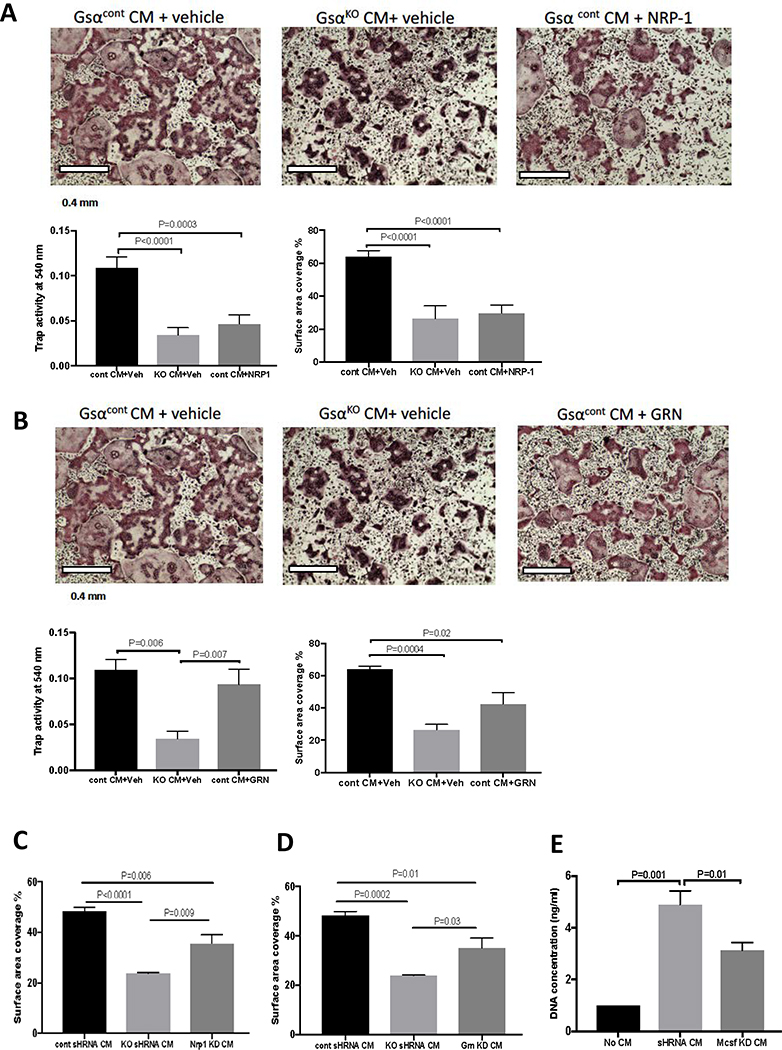
Nrp, Grn and β2M expressions were upregulated in CM from Ocy454-GsαKO cells compared to that from Ocy454-Gsαcont therefore we investigated the effects of these factors on BMNC and osteoclast proliferation and differentiation. We used two complementary approaches; we supplemented Ocy454-Gsαcont CM with each of these proteins, to recapitulate the GsαKO phenotyp, and we generated Ocy454-GsαKO cells in which the corresponding gene of interest was silenced by shRNA. When recombinant mouse Nrp-1 (1 μg/ml) was added to osteoclast cultures in the presence of Ocy454-Gsαcont CM, there was a significant decrease in the TRAP activity (Fig. 8A) and osteoclast areas (Fig. 8A), demonstrating that NRP-1 partially inhibits osteoclastogenesis. Similarly, treatment of BMNC with recombinant GRN (1 μg/ml) and Ocy454-Gsαcont CM induced a significant decrease in osteoclast area, when compared to Ocy454-Gsαcont CM treatment alone (Fig. 8B), whereas TRAP activity was not significantly changed. These results suggest that GRN only partially suppresses osteoclast activity and other proteins might also be involved.
Figure 8: Effects of osteocyte-derived secreted NRP-1 and GRN on osteoclasts differentiation.
A) BMNCs were treated with CM from Ocy454-Gsαcont (black bars), Ocy454-GsαKO (light grey bars) or Ocy454-Gsαcont in the presence of recombinant mouse neuropilin1 (NRP-1) (1000 ng/ml) (dark grey bars) or vehicle (black and grey bars). NRP-1 inhibited osteoclasts formation in BMNC treated with Ocy454-Gsαcont CM (and vehicle) as shown by a significant decrease in TRAP-positive cells, TRAP activity and the percentage of surface area covered by osteoclasts as compared to Ocy454-Gsαcont CM + vehicle treated cells (black bars compared to dark grey bars). B) Treatment with recombinant mouse GRN (1 μg/ml) partially reduced the number of osteoclasts, as shown by decreased percentage of surface area covered by osteoclasts upon treatment with GRN, whereas it had no effect on TRAP activity . D) Knock-down (KD) of Grn in Ocy454-GsαKO cells (Grn KD, dark grey bar) partially restored osteoclasts number, as shown by increased surface area. C) Knock-down (KD) of NRP1 in Ocy454-GsαKO cells (Nrp1 KD, dark grey bar) partially restored osteoclast numbers, as shown by increased surface area compared to cells treated with CM from Ocy454-GsαKO. E) Treatment of BMNC with CM from Ocy454-Mcsf-KD cells showed a significant decrease in proliferation, when compared with CM from Ocy454shRNA control (light grey bar). (One-way ANOVA). Error bars represent mean ± SEM. Each experiment was repeated at least 3 times.
Next, shRNAs targeting Grn, Nrp-1, β2m and Mcsf were used to knock down each of these genes in Ocy454-GsαKO cells. Scrambled shRNA was used as a control. Real-time PCR demonstrated successful suppression of these genes, as demonstrated by a significant decrease in Grn (80%), β2m (90%), and Nrp-1 (50%), Mcsf (70%) expression, as compared to control cells (Fig. S4). Osteoclasts treated with CM from Nrp-1 knockdown (KD) or Grn-KD Ocy454-GsαKO cells showed a significant increase in the surface area covered by osteoclasts (Fig. 8C and D), but no changes in the TRAP activity, as compared to the cells treated with CM from Ocy454-GsαKO cell (Fig. S5A,B). CM from β2m-KD cells showed no changes in osteoclasts TRAP activity, when compared to Ocy454-GsαKO CM treatment (Fig. S5C), suggesting that this factor does not control osteoclast differentiation or proliferation.
Lastly, we investigated the role of these proteins in BMNCs proliferation. There were no significant changes in BMNC proliferation when cells were treated with CM from Grn, Nrp-1 and β2m KD cells, as compared to CM from shRNA controls. In contrast, treatment of BMNC with CM from Mcsf-KD cells showed a significant decrease in proliferation, when compared to the shRNA control, demonstrating that osteocytes contribute to BMNC proliferation, at least in part, through M-CSF secretion (Fig. 8D).
Discussion
Mutations in the GNAS locus, which contains the gene encoding the Gsα subunit, are associated with several human diseases, including Albright hereditary osteodystrophy (AHO), fibrous dysplasia, McCune-Albright syndrome (MAS), progressive osseous heteroplasia (POH), pseudohypoparathyroidism, and thyroid and pituitary gland tumors (20). Osteocytes express several GPCRs, including the PTH1R, the prostaglandin receptors E2-E4, the β2 adrenergic receptors and possibly others. In mice, deletion of Gsα in osteocytes (Dmp1-GsαKO mice) induces severe osteopenia associated with increased myeloid cells in the bone marrow, spleen, and peripheral blood. The osteopenia is associated with increased Sost/sclerostin expression; treatment with anti-sclerostin antibodies partially rescued the skeletal phenotype, but not the myelopoiesis, thereby suggesting that additional mechanisms control the proliferation of myeloid cells (11, 12).
Due to the complexity of the physiological settings and the difficulty of studying osteocytes in vivo, we used an in vitro (Ocy454 osteocytic cells) and ex vivo (OEBEs) models (13) to investigate, at the molecular level, the mechanisms by which osteocytes control myelopoiesis. Here we report that osteocytes in vitro and ex vivo secrete Gsα-dependent and independent factors which support myeloid cell proliferation and osteoclast differentiation and function. This finding is consistent with previous in vivo and ex vivo results (21) and supports the hypothesis that osteocytes directly regulate myeloid cells by providing soluble paracrine factors that are regulated by Gsα signaling. Moreover, osteocytes secrete factor(s) which promote macrophage proliferation and inhibit osteoclast formation through Gsα-dependent and independent mechanisms. Interstingly, whereas osteocyte-secreted factor(s) promote macrophage proliferation and inhibit osteoclast differentiation quite rapidly (between 24 hr and 2 days, see Fig. 3C and Fig. 4A–D) and independently of Gsα-signaling, the effects mediated by Gsα require at least 5 days of treatment with RANKL and M-CSF to become evident. One possible explanation is that Gsα-dependent factors control later stages of osteoclastogenesis, like cell fusion and adicification of the resorption pit, as suggested by the marked distruption of the actin ring (Fig. 5D) and by the decrease in expression of Dc-Stamp and Atp6v0d2. Further studies will be needed to further elucidate these effects.
Many cytokines and hormones have been reported to increase proliferation of myeloid cells, including G-CSF, M-CSF, GM-CSF, IL-1, IL-3, IL-6, erythropoietin, thrombopoietin, vascular endothelial growth factor (VEGF), and others (22–27). Serum G-CSF was elevated in Dmp1-GsαKO mice and in CM of their OEBE, suggesting that G-CSF secreted by osteocytes was promoting, in part, the myelopoiesis (21). However, the origin of G-CSF remained unclear. To investigate whether osteocytes were the source of G-CSF, we sought to measure its level in CM of Ocy45 using both proteomic and ELISA methodologies, but we could not detect any, suggesting that the G-CSF involved in myelopoiesis derives from other cells that are regulated, directly or indirectly, by osteocytes. Osteoblasts, monocytes, fibroblasts and endothelial cells have all been found to secrete G-CSF (28–32). We hypothesized that osteocytes might secrete factors other than G-CSF that control myelopoisies. To test this hypothesis we used CM from Ocy454-Gsαcont and Ocy454-GsαKO cells to treat myeloid progenitors. We reported here, for the first time, that osteocytes in vitro, support proliferation of myeloid cells through a Gsα-dependent mechanism. We can speculate that signaling via Gsα either inhibit a pro-myelogenic factor or induce an anti-myelogenic one. Indeed, among the proteins identified in our proteomic analysis, few have been associated with myeloid cells, such as CP and Nrp1 and further studies will be needed to elucidate their role.
Recent studies provided strong evidence that osteocytes serve as a master regulator of bone hemostasis through their secreted factors sclerostin and RANKL (3, 33, 34). Studies in mice in which osteocytes were eliminated using the diphtheria receptor suggested that dying osteocytes release osteoclastogenic cues. Here we demonstrated, using both in vitro and ex vivo models, that osteocytes secrete factors which promote monocyte/macrophage proliferation and, at the same time, restrain osteoclast differentiation and functions. Mice lacking Gsα in osteocytes (Dmp1-GsαKO) have a significant decrease in both trabecular and cortical bone and histomorphometric analysis showed that the osteopenia was mostly driven by more than 90% decrease in osteoblast number and activity. Interstingly, osteoclast numbers and activity was also reduced in these mice, suggesting that osteocytes control bith osteoblasts and osteoclasts through Gsα-dependent mechanisms. Here, we identified neuropilin-1 and granulin as two important Gsα-dependent osteocyte-derived factors capable of controlling osteoclasts. We also demonstrated that BMNC and monocyte/macrophage proliferation is regulated by factors secreted by osteocytes, including M-CSF.
Interestingly, CM from cell-deprived OEBE was still able to significantly increase BMNC proliferation, suggesting that proteins secreted by osteoblasts or present in the bone matrix, such as TGF-β, BMPs, OPN, IGFs, VEGF, PDGF, Collagen I, and BSP (35–37), could be released in the culture and support cell proliferation. In addition, potential release of mineral ions from the bones, including calcium and phosphate, might also contribute to cell growth and proliferation. It has been shown (and previously reported) that collagen undergoes denaturation and degradation when heated above 60°C. In our experiment, bone were autoclaved for 30 min at 121°C, a condition that has been shown to denature collagen into a gelatin solution (38). We can speculate that when OEBEs are autoclaved, the cellular component is inactivated, the proteins are denatured and collagen is partially transformed into gelatin. OEBEs can then release gelatin in the CM and gelatin has been reported to promote not only cell adhesion, but also cell proliferation (39). Future studies will be needed to elucidate this effect.
Our results are in agreement with a previous study on isolated rat calvaria organ cultures that suggested that osteocytes were constitutively negative regulators of osteoclastic activity and that the inhibitory activity was lost upon osteocyte apoptosis (40). Here we showed that, when BMNC were treated with conditioned medium from Ocy454 cells or OEBEs, there was a significant suppression of osteoclast differentiation and maturation. Functional assays indicated a significant and dramatic inhibition of bone resorption upon treatment with CM from osteocytes. No difference in the size of the osteoclast resorption area between Ocy454-Gsαcont CM and Ocy454-GsαKO CM treatments was detected, suggesting a Gsα independent mechanism; however, a significant decrease in the number of resorption areas was observed with Ocy454-GsαKO CM treatment, demonstrating that there was a decrease in the number of active osteoclasts and/or a defect in osteoclast activity. The F-actin ring is required for proper osteoclast function and osteocyte-secreted factors alter the osteoclast cytoskeleton and inhibit F-actin ring formation, leading to the defect in podosomes and in osteoclast activity. These findings indicated that osteocytes and associated Gsα signaling play a role in controlling osteoclastogenesis (40–42). For example, a previous study reported that viable osteocytes secrete factors that suppress osteoclast activity, whereas apoptotic osteocytes activate osteoclasts and trigger bone resorption (40). Similarly, in vivo depletion of osteocytes, using the diphtheria toxin receptor, increased the number of osteoclasts (43). Our data are in agreement with such previous studies that established the concept that osteocytes control osteoclastogenesis. Consistent with these results, we found that osteocytes secrete factor(s) that promote(s) osteoclast apoptosis. In addition, mechanically stimulated osteocytes secrete MEPE which upregulate OPG and decrease the RANKL/OPG ratio, leading to osteoclast inhibition (41).
It has been reported that osteocytes are the primary source of RANKL in adult mice and that genetic deletion of RANKL in osteocytes results in osteopetrosis (34). However, our proteomic analysis did not identify RANKL as a secreted factor. One possible explanation for this discrepancy is that the amount of RANKL secreted by Ocy454 cells may be below the detection limit of the mass spectrometry method used in this investigation. Additional studies are needed to elucidate the role of osteocyte’s RANKL on osteoclastogenesis. When Gsα was ablated from Ocy454 cells, there was a significant increase in OPG expression, whereas RANKL was downregulated, significantly reducing the RANKL/OPG ratio. Interestingly, although RANKL was added to the osteoclast cultures, the decrease in osteoclast formation and function remained, suggesting that osteoclast inhibition is mediated by other, or additional factors.
To delineate the osteocyte secretome and to identify Gsα-dependent factors capable of controlling myelopoiesis and osteoclastogenesis, we performed proteomic analysis of CM and cell lysates from both Ocy454-Gsαcont cells and Ocy454-GsαKO cells. Several Gsα-dependent and -independent factors were identified. Among proteins we identified, NRP-1, GRN, β2M, and CP were significantly increased in Ocy454-GsαKO CM, when compared to their levels in Ocy454-Gsαcont CM.
NRP-1 is a transmembrane glycoprotein that plays a role in the development of the nervous system and angiogenesis. NRP-1 acts as a receptor for SEMA3A protein and some isoforms of VEGF (44). Increased levels of NRP-1 have been found to be associated with human tumors, including brain, prostate, lung, colon, and breast cancers, as well as with tumor metastasis (45, 46). The semaphorin family of proteins, which was initially identified in neuronal development and guidance, was recently shown to be associated with bone hemostasis. Semaphorin 3 is the secreted form of the semaphorin family proteins, which are further divided into several subclasses, including SEMA3A, SEMA3B, SEMA3C, SEMA3D, SEMA3E, SEMA3F, and SEMA3G. Plexins (PLXN A, B, C, D) and neuropilins (NRP-1 and NRP-2) are the receptors for semaphorins (47, 48). A previous study showed that SEMA3A binds to NRP-1 and blocks RANKL-induced osteoclast differentiation by inhibiting Rho and immunoreceptor tyrosine-based activation motif (ITAM) signaling pathways. The same study showed that SEMA3A also supports osteoblast differentiation by stimulating the Wnt signaling pathway (5). Furthermore, SEMA3A-KO and NRP-1 knock-in mice presented with severe osteopenia both in cortical and trabecular bone, associated with an increase in osteoclastogenesis (5). In our study, we identified NRP-1 as an osteocyte Gsα-dependent secreted protein capable of inhibiting osteoclastogenesis when added to Ocy454-Gsαcont CM. GRN was also identified as a secreted factor which belongs to a group of glycoproteins with 7.5 repeats of a highly conserved 12-cysteine granulin/epithelin motif. GRN is expressed in a variety of tissues, including hematopoietic cells, macrophages, skeletal muscles, neurons, adipocytes, and chondrocytes (49–52). It has been shown that GRN is involved in inflammation, tumorigenesis, epithelial proliferation, wound healing, and insulin resistance (49, 53–55).
We also identified β2M as a Gsα-dependent secreted factor. β2M is a part of type I and type II major histocompatibility complexes and CD1 complexes that are expressed by most cell types and play roles in immunity. It was previously reported that increased CP serum levels were associated with acute and chronic inflammation and with rheumatoid arthritis (56, 57). Interestingly, Dmp1-GsαKO mice have increased numbers of neutrophils, suggesting the occurrence of inflammation in these mice.
Osteocyte-secreted factors significantly decreased in Ocy454-GsαKO CM, as compared to Ocy454-Gsαcont CM, include COL3α1, SEMA3D, SEMA3C, COL1α1, COL1α2, SEMA3A, IGSF-10, MMP2, EMILIN2, and TIMP-2. The decrease in secreted collagens (COL1α1, COL1α2, and COL3α1) in Ocy454-GsαKO CM is consistent with our previous observations that Dmp1-GsαKO mice exhibit an osteopenic phenotype. SEMA3A is expressed by osteoblasts and osteocytes and plays a role in bone remodeling by enhancing osteoblast differentiation as well as inhibiting osteoclast formation. It has been reported that SEMA3A-deficient mice are osteopenic due to suppressed osteoblast-mediated bone formation and increased osteoclast-mediated bone resorption. Furthermore, treatment with CM from OPG knockout osteoblasts inhibited osteoclast formation, suggesting that osteoblasts secrete SEMA3A and other factors that inhibit osteoclast formation independently of OPG (5). In our study, we found SEMA3A, SEMA3D, and SEMA3C to be osteocyte-secreted proteins. Future studies are needed to define the role of semaphorins in osteocytes.
Osteocytes also produce other secreted proteins, including M-CSF, OPN, MIF, TGF-β, and OPG. M-CSF is important for the proliferation and survival of osteoclast precursors and mice lacking M-CSF display an osteopetrotic phenotype and hematopoietic abnormalities (58, 59).
One of the limitations in this work was that deletion of a G protein that transduces the signal through multiple receptors may yield changes in multiple signaling pathways. Future studies will be needed to identify the different signaling pathways and receptors associated with them.
In summary, we demonstrated that osteocytes secrete factors controlling myeloid cell proliferation in a Gsα-dependent manner. We showed that osteocytes secrete factors that promote BMNC and macrophage proliferation in a Gsα-independent manner and we demonstrated that osteoclast differentiation and function were suppressed by Gsα-dependent osteocyte-mediated secreted factor(s). We identified NRP-1 as a novel osteocyte-secreted factor capable of inhibiting osteoclastogenesis. In addition, we found that M-CSF secreted by osteocytes is responsible in part for BMNC proliferation. Future studies are needed to determine the role of osteocyte-mediated NRP-1 and the other identified secreted factors in myelopoiesis and osteoclastogenesis.
Supplementary Material
Acknowledgments
The authors wish to thanks Dr. Marc Wein (Mass General Hospital, Boston MA) for providing gRNAs for CRISPR/Cas9. This work was partially supported by NIH grant R01 AR060221 to PDP, by a pilot and Feasibility Grant from CTSI, Boston University (NIH grant UL1 TR001430), and NIH grant P41 GM104603 to CEC. This work was supported by the Boston University Flow Cytometry Core Facility.
Grant Supporters: This work was partially supported by NIH grant R01 AR060221 to PDP, by a pilot and Feasibility Grant from CTSI, Boston University (NIH UL1 TR001430), and NIH grant P41 GM104603 to CEC. This work was supported by the Boston University Flow Cytometry Core Facility. The authors have no competing financial conflict-of-interest.
Nonstandard Abbrevations
- BM
Bone Marrow
- BMNC
Bone Marrow Mononuclear Cells
- BSA
Bovine Serum Albumin
- cAMP
cyclic adenosine mono-phosphate
- Cas9
CRISPR Associated (Cas) system
- CM
Conditioned medium
- CRISPR
Clustered Regularly Interspaced Short Palindromic Repeats
- DAPI
4′,6-diamidino-2-phenylindole
- Dmp1
Dentin Matrix Protein 1
- EDTA
ethylenediaminetetraacetic acid disodium salt dihydrate
- FBS
Fetal Bovine Serum
- G-CSF
Granulocyte Colony-stimulating Factor
- Grn
Granulin
- GPCR
G protein-coupled receptors
- IL-3
Interleukin 3
- IL-6
Interleukin 6
- M-CSF
Macrophage Colony-stimulating Factor
- Nrp-1
neuropilin-1
- OEBE
Osteocyte Enriched Bone Explant
- OPG
Osteoprotegerin
- OPN
Osteopontin
- PBS
Phosphate Buffered Solution
- PTH
Parathyroid Hormone
- PTH1R
Parathyroid Hormone 1 receptor
- RANKL
receptor-activator of NF-kB ligand
- Sema3A
Semaphorin 3A
- TRAP
Tartrate Resistant Acid Phospahatse
Footnotes
Supplemental Data: Supplementary Methods, Supplementary Figures 1–8.
References
- 1.Marotti G (1996) The structure of bone tissues and the cellular control of their deposition. Ital J Anat Embryol 101, 25–79 [PubMed] [Google Scholar]
- 2.Aarden EM, Burger EH, and Nijweide PJ (1994) Function of osteocytes in bone. J Cell Biochem 55, 287–299 [DOI] [PubMed] [Google Scholar]
- 3.Bonewald LF (2011) The amazing osteocyte. J Bone Miner Res 26, 229–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dallas SL, Prideaux M, and Bonewald LF (2013) The osteocyte: an endocrine cell … and more. Endocr Rev 34, 658–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayashi M, Nakashima T, Taniguchi M, Kodama T, Kumanogoh A, and Takayanagi H (2012) Osteoprotection by semaphorin 3A. Nature 485, 69–74 [DOI] [PubMed] [Google Scholar]
- 6.Hayashi M, Nakashima T, Yoshimura N, Okamoto K, Tanaka S, and Takayanagi H (2019) Autoregulation of Osteocyte Sema3A Orchestrates Estrogen Action and Counteracts Bone Aging. Cell Metab 29, 627–637.e625 [DOI] [PubMed] [Google Scholar]
- 7.Powell WF Jr., Barry KJ, Tulum I, Kobayashi T, Harris SE, Bringhurst FR, and Pajevic PD (2011) Targeted ablation of the PTH/PTHrP receptor in osteocytes impairs bone structure and homeostatic calcemic responses. J Endocrinol 209, 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cherian PP, Cheng B, Gu S, Sprague E, Bonewald LF, and Jiang JX (2003) Effects of mechanical strain on the function of Gap junctions in osteocytes are mediated through the prostaglandin EP2 receptor. J Biol Chem 278, 43146–43156 [DOI] [PubMed] [Google Scholar]
- 9.Yao Q, Liang H, Huang B, Xiang L, Wang T, Xiong Y, Yang B, Guo Y, and Gong P (2017) Beta-adrenergic signaling affect osteoclastogenesis via osteocytic MLO-Y4 cells’ RANKL production. Biochem Biophys Res Commun 488, 634–640 [DOI] [PubMed] [Google Scholar]
- 10.Jüppner H, and Bastepe M (2006) Different mutations within or upstream of the GNAS locus cause distinct forms of pseudohypoparathyroidism. J Pediatr Endocrinol Metab 19 Suppl 2, 641–646 [DOI] [PubMed] [Google Scholar]
- 11.Fulzele K, Krause DS, Panaroni C, Saini V, Barry KJ, Liu X, Lotinun S, Baron R, Bonewald L, and Feng JQ (2012) Myelopoiesis is regulated by osteocytes through Gsα-dependent signaling. Blood, blood-2012–2006-437160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fulzele K, Dedic C, Lai F, Bouxsein M, Lotinun S, Baron R, and Divieti Pajevic P (2018) Loss of Gsα in osteocytes leads to osteopenia due to sclerostin induced suppression of osteoblast activity. Bone 117, 138–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spatz JM, Wein MN, Gooi JH, Qu Y, Garr JL, Liu S, Barry KJ, Uda Y, Lai F, Dedic C, Balcells-Camps M, Kronenberg HM, Babij P, and Pajevic PD (2015) The Wnt Inhibitor Sclerostin Is Up-regulated by Mechanical Unloading in Osteocytes in Vitro. J Biol Chem 290, 16744–16758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wein MN, Spatz J, Nishimori S, Doench J, Root D, Babij P, Nagano K, Baron R, Brooks D, Bouxsein M, Pajevic PD, and Kronenberg HM (2015) HDAC5 controls MEF2C-driven sclerostin expression in osteocytes. J Bone Miner Res 30, 400–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fulzele K, Lai F, Dedic C, Saini V, Uda Y, Shi C, Tuck P, Aronson JL, Liu X, Spatz JM, Wein MN, and Divieti Pajevic P (2017) Osteocyte-Secreted Wnt Signaling Inhibitor Sclerostin Contributes to Beige Adipogenesis in Peripheral Fat Depots. J Bone Miner Res 32, 373–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalajzic I, Staal A, Yang WP, Wu Y, Johnson SE, Feyen JH, Krueger W, Maye P, Yu F, Zhao Y, Kuo L, Gupta RR, Achenie LE, Wang HW, Shin DG, and Rowe DW (2005) Expression profile of osteoblast lineage at defined stages of differentiation. J Biol Chem 280, 24618–24626 [DOI] [PubMed] [Google Scholar]
- 17.Yao C, Behring JB, Shao D, Sverdlov AL, Whelan SA, Elezaby A, Yin X, Siwik DA, Seta F, Costello CE, Cohen RA, Matsui R, Colucci WS, McComb ME, and Bachschmid MM (2015) Overexpression of Catalase Diminishes Oxidative Cysteine Modifications of Cardiac Proteins. PLoS One 10, e0144025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao C, Yu J, Taylor L, Polgar P, McComb ME, and Costello CE (2015) Protein Expression by Human Pulmonary Artery Smooth Muscle Cells Containing a. Int J Mass Spectrom 378, 347–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Behring JB, Kumar V, Whelan SA, Chauhan P, Siwik DA, Costello CE, Colucci WS, Cohen RA, McComb ME, and Bachschmid MM (2014) Does reversible cysteine oxidation link the Western diet to cardiac dysfunction? FASEB J 28, 1975–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weinstein LS, Chen M, and Liu J (2002) Gs(alpha) mutations and imprinting defects in human disease. Ann N Y Acad Sci 968, 173–197 [DOI] [PubMed] [Google Scholar]
- 21.Fulzele K, Krause DS, Panaroni C, Saini V, Barry KJ, Liu X, Lotinun S, Baron R, Bonewald L, Feng JQ, Chen M, Weinstein LS, Wu JY, Kronenberg HM, Scadden DT, and Divieti Pajevic P (2013) Myelopoiesis is regulated by osteocytes through Gsalpha-dependent signaling. Blood 121, 930–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heyworth CM, Vallance SJ, Whetton AD, and Dexter TM (1990) The biochemistry and biology of the myeloid haemopoietic cell growth factors. J Cell Sci Suppl 13, 57–74 [DOI] [PubMed] [Google Scholar]
- 23.Kampen KR, Ter Elst A, and de Bont ES (2013) Vascular endothelial growth factor signaling in acute myeloid leukemia. Cell Mol Life Sci 70, 1307–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Souza LM, Boone TC, Gabrilove J, Lai PH, Zsebo KM, Murdock DC, Chazin VR, Bruszewski J, Lu H, Chen KK, and et al. (1986) Recombinant human granulocyte colony-stimulating factor: effects on normal and leukemic myeloid cells. Science 232, 61–65 [DOI] [PubMed] [Google Scholar]
- 25.Yang YC, Ciarletta AB, Temple PA, Chung MP, Kovacic S, Witek-Giannotti JS, Leary AC, Kriz R, Donahue RE, Wong GG, and et al. (1986) Human IL-3 (multi-CSF): identification by expression cloning of a novel hematopoietic growth factor related to murine IL-3. Cell 47, 3–10 [DOI] [PubMed] [Google Scholar]
- 26.Cozzolino F, Rubartelli A, Aldinucci D, Sitia R, Torcia M, Shaw A, and Di Guglielmo R (1989) Interleukin 1 as an autocrine growth factor for acute myeloid leukemia cells. Proc Natl Acad Sci U S A 86, 2369–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piacibello W, Sanavio F, Brizzi MF, Garetto L, Severino A, Aronica MG, Dragonetti G, Aglietta M, and Pegoraro L (1997) Megakaryocyte growth and development factor (MGDF)-induced acute leukemia cell proliferation and clonal growth is associated with functional c-mpl. Leukemia 11, 531–540 [DOI] [PubMed] [Google Scholar]
- 28.Taichman RS, and Emerson SG (1994) Human osteoblasts support hematopoiesis through the production of granulocyte colony-stimulating factor. J Exp Med 179, 1677–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaushansky K, Lin N, and Adamson JW (1988) Interleukin 1 stimulates fibroblasts to synthesize granulocyte-macrophage and granulocyte colony-stimulating factors. Mechanism for the hematopoietic response to inflammation. J Clin Invest 81, 92–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zsebo KM, Yuschenkoff VN, Schiffer S, Chang D, McCall E, Dinarello CA, Brown MA, Altrock B, and Bagby GC Jr. (1988) Vascular endothelial cells and granulopoiesis: interleukin-1 stimulates release of G-CSF and GM-CSF. Blood 71, 99–103 [PubMed] [Google Scholar]
- 31.Koeffler HP, Gasson J, Ranyard J, Souza L, Shepard M, and Munker R (1987) Recombinant human TNF alpha stimulates production of granulocyte colony-stimulating factor. Blood 70, 55–59 [PubMed] [Google Scholar]
- 32.Cannistra SA, Vellenga E, Groshek P, Rambaldi A, and Griffin JD (1988) Human granulocyte-monocyte colony-stimulating factor and interleukin 3 stimulate monocyte cytotoxicity through a tumor necrosis factor-dependent mechanism. Blood 71, 672–676 [PubMed] [Google Scholar]
- 33.Heino TJ, Kurata K, Higaki H, and Vaananen HK (2009) Evidence for the role of osteocytes in the initiation of targeted remodeling. Technol Health Care 17, 49–56 [DOI] [PubMed] [Google Scholar]
- 34.Nakashima T, Hayashi M, Fukunaga T, Kurata K, Oh-Hora M, Feng JQ, Bonewald LF, Kodama T, Wutz A, Wagner EF, Penninger JM, and Takayanagi H (2011) Evidence for osteocyte regulation of bone homeostasis through RANKL expression. Nature Medicine 17, 1231–1234 [DOI] [PubMed] [Google Scholar]
- 35.Holt DJ, and Grainger DW (2012) Demineralized bone matrix as a vehicle for delivering endogenous and exogenous therapeutics in bone repair. Adv Drug Deliv Rev 64, 1123–1128 [DOI] [PubMed] [Google Scholar]
- 36.Wildemann B, Kadow-Romacker A, Haas NP, and Schmidmaier G (2007) Quantification of various growth factors in different demineralized bone matrix preparations. J Biomed Mater Res A 81, 437–442 [DOI] [PubMed] [Google Scholar]
- 37.Wildemann B, Kadow-Romacker A, Pruss A, Haas NP, and Schmidmaier G (2007) Quantification of growth factors in allogenic bone grafts extracted with three different methods. Cell Tissue Bank 8, 107–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Köhler P, Kreicbergs A, and Strömberg L (1986) Physical properties of autoclaved bone. Torsion test of rabbit diaphyseal bone. Acta Orthop Scand 57, 141–145 [DOI] [PubMed] [Google Scholar]
- 39.Kim AY, Kim Y, Lee SH, Yoon Y, Kim WH, and Kweon OK (2017) Effect of Gelatin on Osteogenic Cell Sheet Formation Using Canine Adipose-Derived Mesenchymal Stem Cells. Cell Transplant 26, 115–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gu G, Mulari M, Peng Z, Hentunen TA, and Vaananen HK (2005) Death of osteocytes turns off the inhibition of osteoclasts and triggers local bone resorption. Biochem Biophys Res Commun 335, 1095–1101 [DOI] [PubMed] [Google Scholar]
- 41.Kulkarni RN, Bakker AD, Everts V, and Klein-Nulend J (2010) Inhibition of osteoclastogenesis by mechanically loaded osteocytes: involvement of MEPE. Calcif Tissue Int 87, 461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heino TJ, Hentunen TA, and Vaananen HK (2002) Osteocytes inhibit osteoclastic bone resorption through transforming growth factor-beta: enhancement by estrogen. J Cell Biochem 85, 185–197 [DOI] [PubMed] [Google Scholar]
- 43.Tatsumi S, Ishii K, Amizuka N, Li M, Kobayashi T, Kohno K, Ito M, Takeshita S, and Ikeda K (2007) Targeted ablation of osteocytes induces osteoporosis with defective mechanotransduction. Cell Metab 5, 464–475 [DOI] [PubMed] [Google Scholar]
- 44.Gagnon ML, Bielenberg DR, Gechtman Z, Miao HQ, Takashima S, Soker S, and Klagsbrun M (2000) Identification of a natural soluble neuropilin-1 that binds vascular endothelial growth factor: In vivo expression and antitumor activity. Proc Natl Acad Sci U S A 97, 2573–2578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parikh AA, Fan F, Liu WB, Ahmad SA, Stoeltzing O, Reinmuth N, Bielenberg D, Bucana CD, Klagsbrun M, and Ellis LM (2004) Neuropilin-1 in human colon cancer: expression, regulation, and role in induction of angiogenesis. Am J Pathol 164, 2139–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bachelder RE, Crago A, Chung J, Wendt MA, Shaw LM, Robinson G, and Mercurio AM (2001) Vascular endothelial growth factor is an autocrine survival factor for neuropilin-expressing breast carcinoma cells. Cancer Res 61, 5736–5740 [PubMed] [Google Scholar]
- 47.Verlinden L, Vanderschueren D, and Verstuyf A (2016) Semaphorin signaling in bone. Mol Cell Endocrinol 432, 66–74 [DOI] [PubMed] [Google Scholar]
- 48.Kang S, and Kumanogoh A (2013) Semaphorins in bone development, homeostasis, and disease. Semin Cell Dev Biol 24, 163–171 [DOI] [PubMed] [Google Scholar]
- 49.Matsubara T, Mita A, Minami K, Hosooka T, Kitazawa S, Takahashi K, Tamori Y, Yokoi N, Watanabe M, Matsuo E, Nishimura O, and Seino S (2012) PGRN is a key adipokine mediating high fat diet-induced insulin resistance and obesity through IL-6 in adipose tissue. Cell Metab 15, 38–50 [DOI] [PubMed] [Google Scholar]
- 50.Feng JQ, Guo FJ, Jiang BC, Zhang Y, Frenkel S, Wang DW, Tang W, Xie Y, and Liu CJ (2010) Granulin epithelin precursor: a bone morphogenic protein 2-inducible growth factor that activates Erk1/2 signaling and JunB transcription factor in chondrogenesis. FASEB J 24, 1879–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Daniel R, He Z, Carmichael KP, Halper J, and Bateman A (2000) Cellular localization of gene expression for progranulin. J Histochem Cytochem 48, 999–1009 [DOI] [PubMed] [Google Scholar]
- 52.Okura H, Yamashita S, Ohama T, Saga A, Yamamoto-Kakuta A, Hamada Y, Sougawa N, Ohyama R, Sawa Y, and Matsuyama A (2010) HDL/apolipoprotein A-I binds to macrophage-derived progranulin and suppresses its conversion into proinflammatory granulins. J Atheroscler Thromb 17, 568–577 [DOI] [PubMed] [Google Scholar]
- 53.He Z, and Bateman A (1999) Progranulin gene expression regulates epithelial cell growth and promotes tumor growth in vivo. Cancer Res 59, 3222–3229 [PubMed] [Google Scholar]
- 54.He Z, and Bateman A (2003) Progranulin (granulin-epithelin precursor, PC-cell-derived growth factor, acrogranin) mediates tissue repair and tumorigenesis. J Mol Med (Berl) 81, 600–612 [DOI] [PubMed] [Google Scholar]
- 55.Guerra RR, Kriazhev L, Hernandez-Blazquez FJ, and Bateman A (2007) Progranulin is a stress-response factor in fibroblasts subjected to hypoxia and acidosis. Growth Factors 25, 280–285 [DOI] [PubMed] [Google Scholar]
- 56.Deshmukh VK, Raman PH, Dhuley JN, and Naik SR (1985) Role of ceruloplasmin in inflammation: increased serum ceruloplasmin levels during inflammatory conditions and its possible relationship with anti-inflammatory agents. Pharmacol Res Commun 17, 633–642 [DOI] [PubMed] [Google Scholar]
- 57.Conforti A, Franco L, Menegale G, Milanino R, Piemonte G, and Velo GP (1983) Serum copper and ceruloplasmin levels in rheumatoid arthritis and degenerative joint disease and their pharmacological implications. Pharmacol Res Commun 15, 859–867 [DOI] [PubMed] [Google Scholar]
- 58.Wiktor-Jedrzejczak W, Bartocci A, Ferrante AW Jr., Ahmed-Ansari A, Sell KW, Pollard JW, and Stanley ER (1990) Total absence of colony-stimulating factor 1 in the macrophage-deficient osteopetrotic (op/op) mouse. Proc Natl Acad Sci U S A 87, 4828–4832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harris SE, MacDougall M, Horn D, Woodruff K, Zimmer SN, Rebel VI, Fajardo R, Feng JQ, Gluhak-Heinrich J, Harris MA, and Abboud Werner S (2012) Meox2Cre-mediated disruption of CSF-1 leads to osteopetrosis and osteocyte defects. Bone 50, 42–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.