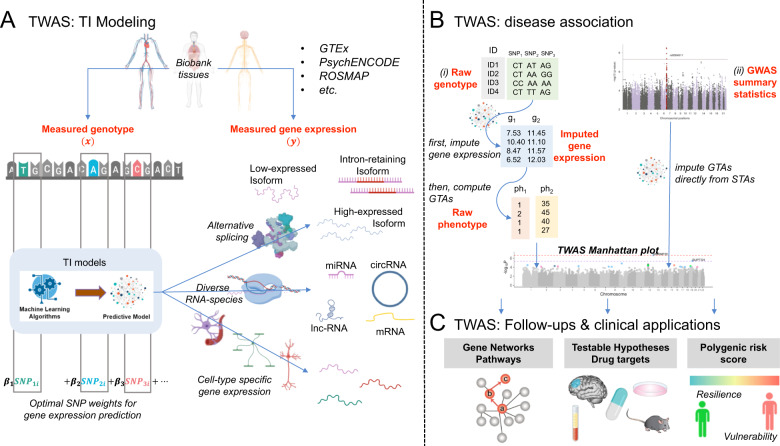
Genome-wide association studies (GWAS) associate genetic variants with traits. Neuropsychiatric traits have complex etiology, and GWAS have started to reveal their polygenic architecture, including multiple single nucleotide polymorphisms (SNPs) associated with each trait (SNP-trait association—STAs). Yet, GWAS-hits have generally small effect sizes and their biological interpretation has been challenging, since they are often noncoding, pleiotropic, and/or non-causative. The latter effect is due to nonrandom co-inheritance, linkage disequilibrium (LD), with nearby causative loci. These reasons necessitate methods for SNP-prioritization through fine mapping, and causal inference [1].
To this end, transcriptome-wide association studies (TWAS) leverage genetically regulated expression through genotype-based transcriptomic imputation (TI) to identify gene-trait associations (GTAs). TI predicts the expression of a variety of gene transcripts, including gene isoforms, based on typically cis-regulatory variants (Fig. 1a). Disease-association testing is accomplished by using: (i) raw genotype to impute first the transcriptome, and subsequently compute the GTAs; or (ii) GWAS summary statistics to directly impute GTAs from STAs (Fig. 1b).
Fig. 1. Anatomy of a transcriptome-wide association study (TWAS) of neuropsychiatric disease.
a Transcriptomic Imputation (TI) modeling: uses large reference panels (e.g., Genotype-Tissue Expression (GTEx) project, PsychENCODE Consortium, Religious Orders Study, and Memory and Aging Project (ROSMAP) Study) with measured genotype—“x” (i.e., single nucleotide polymorphisms—SNPs) and gene expression—“y” to train machine learning based TI predictive models of tissue- and/or cell-type-specific gene expression of diverse RNA-species (mRNA messenger RNA, lncRNA long noncoding RNA, miRNA micro-RNA, circRNA circular RNA, etc.) and their splice variants (e.g., low-expressed, intron-retaining and high-expressed isoforms). b Disease association: involves the use of SNP weights (β1, β2,…, βn) from the TI-models (i) to first impute the transcriptome (gene g1, g2,…, gn) from genotype, and then compute gene-trait associations (GTAs), which is the association of genes to phenotypes (e.g., categorical phenotype—ph1, continuous phenotype—ph2) or (ii) to directly impute GTAs from Genome-Wide Association Study (GWAS) summary statistics (containing SNP-trait associations—STAs). Both approaches result in tissue- and/or cell-type-specific GTAs represented in a TWAS Manhattan plot. Note that when TWAS summary statistics calculated on raw genotype/phenotype and GWAS summary statistics are combined, sample size based meta-analysis should be used. c. Follow-ups and clinical applications: TWAS findings can be understood in the context of dysregulated gene networks and pathways, and can lead to testable hypotheses that can be validated with experimental models or followed-up in human studies, and eventually lead to novel pathophysiological and neuropharmacological understanding of neuropsychiatric disease. Finally, TWAS-based polygenic risk scoring could be a novel avenue to map individuals’ vulnerability and resilience to psychiatric disease with clinical utility.
Compared to GWAS, TWAS benefit from less multiple testing burden, confer higher statistical power and deliver biological understanding of neuropsychiatric risk, given their results involve tissue/cell-type and directional specificity. Since neuropsychiatric TWAS relies on the GWAS sample size, it circumvents the need of disease-specific brain samples required to conduct well-powered differential gene expression analyses of disease. Moreover, since TI-models are trained on psychiatric controls, the resulting GTAs probe neuropsychiatric risk without confounding by reverse causality, as it is often true for brain transcriptomic studies [2].
Neuropsychiatric GTAs are often far from known GWAS-hits, and have implicated novel brain-based transcriptional mechanisms (e.g., alternative splicing), which have been validated in human studies and experimental models [3, 4]. For instance, our post-traumatic stress disorder TWAS, predicted SNRNP35 downregulation in prefrontal cortex (PFC), a region implicated in stress regulation. The function of SNRNP35 as a stress/glucocorticoid responsive U12-splicing regulator was confirmed in cell culture, mouse PFC and blood of war-exposed marines [4].
Experimental validation steps are essential for TWAS, since associations may be confounded [2]. Firstly, TI-modeling accuracy varies, hampered by the specific characteristics of the training dataset (e.g., sample size, demographic, and clinical information) and by high LD between eQTL-regions. Secondly, accuracy of GTAs and their biological specificity can be confounded by LD-based correlated predicted expression, gene co-regulation, and shared eQTLs across tissues/cell-types. Thirdly, ancestry mismatch between the TI-training and the GWAS can lead to false-positive/negative GTAs.
To mitigate these limitations, more accurate TI is necessary, by modeling nonadditive relationships between cis- and trans-regulation of gene expression, by distinguishing common- from rare-variant effects, by making TI-models focused on ancestry, sex, and life-stages, and by predicting RNA-species in brain cell types (e.g., [5]). GWAS-SNP imputation with appropriate reference panels would improve LD estimation and, consequently, GTAs. The improved GTAs in conjunction with their tissue-/cell-type-/directional- specificity can be understood in the context of affected pathways [6], and lead to testable mechanistic hypotheses, drug-target identification, and drug repurposing (Fig. 1c), which are lagging behind in neuropsychiatry. Finally, developing fine mapping, together with polygenic risk scoring for TWAS will advance personalized medicine efforts for these disorders.
Funding and disclosure
This study was supported by the 2019 Seed Grant from Silvio O. Conte Center for Stress Peptide Advanced Research, Education, & Dissemination (NIMH P50MH115874) to C.C., by a 2015 and a 2018 NARSAD Young Investigator grants from BBRF to N.P.D., a Jonathan Edward Brooking mental health research fellowship from McLean Hospital to N.P.D., an appointed KL2 award from Harvard Catalyst | The Harvard Clinical and Translational Science Center (National Center for Advancing Translational Sciences KL2TR002542, UL1TR002541) to N.P.D., and NIMH R21MH121909 to N.P.D. Over the past 3 years, N.P.D. has held a part-time paid position at Cohen Veteran Biosciences, has been a consultant for Sunovion Pharmaceuticals and is on the scientific advisory board for Sentio Solutions, Inc. for unrelated work. The remaining authors have nothing to disclose.
Author contributions
All authors wrote the original draft of the manuscript, prepared the figure and reviewed and edited all the manuscript versions.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Chris Chatzinakos, Foivos Georgiadis
References
- 1.Schaid DJ, Chen W, Larson NB. From genome-wide associations to candidate causal variants by statistical fine-mapping. Nat Rev Genet. 2018;19:491–504. doi: 10.1038/s41576-018-0016-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wainberg M, Sinnott-Armstrong N, Mancuso N, Barbeira AN, Knowles DA, Golan D, et al. Opportunities and challenges for transcriptome-wide association studies. Nat Genet. 2019;51:592–99. doi: 10.1038/s41588-019-0385-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gandal MJ, Zhang P, Hadjimichael E, Walker RL, Chen C, Liu S, et al. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science. 2018;362:eaat8127. https://science.sciencemag.org/content/362/6420/eaat8127/ [DOI] [PMC free article] [PubMed]
- 4.Huckins LM, Chatzinakos C, Breen MS, Hartmann J, Klengel T, da Silva Almeida AC, et al. Analysis of genetically regulated gene expression identifies a prefrontal PTSD gene, SNRNP35, specific to military cohorts. Cell Rep. 2020;31:107716. doi: 10.1016/j.celrep.2020.107716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li M, Santpere G, Imamura Kawasawa Y, Evgrafov OV, Gulden FO, Pochareddy S, et al. Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science. 2018;362:eaat7615. https://science.sciencemag.org/content/362/6420/eaat7615 [DOI] [PMC free article] [PubMed]
- 6.Chatzinakos C, Georgiadis F, Lee D, Na C, Vladimirov VI, Docherty A, et al. TWAS pathway method greatly enhances the number of leads for uncovering the molecular underpinnings of psychiatric disorders. Am J Med Genet B Neuropsychiatr Genet. In Press. [DOI] [PMC free article] [PubMed]