Abstract
Fungal growth and development depend on adaptation to the particular pH of their environment. Ambient pH sensing implies the activation of the pacC signaling pathway, which then acts as a critical regulator for different physiological conditions. The PacC transcription factor may also be associated with the control of salt stress tolerance. In a pH-dependent manner, salinity stress is surpassed by changes in gene expression and coordinated activation of other signaling pathways, thus permitting survival in the challenging environment. In this study, we assessed the regulatory role of Trichophyton interdigitale PacC in response to pH variation and salinity stress. By employing gene expression analysis, we evaluated the influence of PacC in the modulation of salt stress–related genes, including the transcription factors crz1, egr2, and the MAP kinase hog1 in the dermatophyte T. interdigitale. In our analysis, we also included the evaluation of a potassium/sodium efflux P-type ATPase aiming to identify the role of PacC on its ion pumping activity. Here we demonstrated that salinity stress and buffered pH conditions might affect the pacC gene modulation in the dermatophyte T. interdigitale.
Keywords: Dermatophyte, Salt stress, P-type ATPases, Gene expression regulation
Introduction
Transcription factors (TFs) comprise one of the largest functional gene groups in most genomes. They can regulate a target gene with or without TF–DNA interaction independently or by integrating cooperative networks, through the combined actions of several other transcription factors in co-regulatory mechanisms [1, 2].
In fungi, gene expression regulation by pH is exerted by the C2H2 zinc finger transcription factor Rim101/PacC, which acts as a positive or negative regulator of alkaline or acid-expressed genes [3]. PacC is itself pH regulated and is induced in alkaline environmental conditions [4, 5]. PacC regulates genes by directly binding to a specific DNA sequence (consensus 5′-GCCARG-3′) as an effector or by collaborating with other genes in transcriptional networks [3, 4, 6, 7]. The occurrence of cross-talk between PacC and different signaling pathways may occur, among other purposes, to activate compensatory functions for the proper cellular responses to environmental changes.
The pacC gene is required for the import and export of essential substrates, to control ion homeostasis [8], and to participate in the direct regulation of salt tolerance by controlling efflux systems. It also interferes with the modulation of efflux-associated genes in a time-dependent manner [9–11]. PacC mutants present with impaired growth under salt stress, with severe growth impairment under alkaline pH conditions [7, 12, 13]. Thus, evidence of the existence of pacC-controlled salt efflux systems, the consequent developmental defects observed after pacC deletion, and the associated networks under PacC regulation indicate a noteworthy role of PacC in salt tolerance.
In this study, we evaluated the regulatory effect of pacC on the transcriptional profiles of genes required during salinity stress, namely, hog1 mitogen-activated protein (MAP) kinase, the transcription factors crz1 and egr2, and a potassium/sodium efflux ATPase, to assess the regulatory role of Trichophyton interdigitale PacC in salt stress associated with pH modulation. In our results, PacC exerted a pH-dependent regulatory effect in the transcription factor egr2, acting as a co-regulator. The transcription of the other evaluated genes occurred independently of PacC in most of the cases. Finally, we highlighted the occurrence of salt-conditioned transcription of pacC in buffered conditions, thus expanding the understanding of PacC-dependent regulation under salt stress.
Material and methods
Strains and culture conditions
T. interdigitale strain H6 (ATCC MYA 3108) and its mutant ∆pacC [14] were used in this study. The wild-type strain was grown at 28 °C on malt extract agar (MEA; 2% glucose, 2% malt extract, 0.1% peptone, 2% agar, pH 5.7), while the ∆pacC strain was grown on MEA supplemented with 400 μg/mL hygromycin B. Growth in the presence of NaCl was evaluated in agar plates containing this salt in concentrations ranging from 0.5 to 0.8 M in Sabouraud dextrose broth (SDB; 2% (w/v) glucose, 1% (w/v) peptone). Radial growth of the two strains was evaluated in buffered and non-buffered salt stress medium at pH 5.0, 5.7, 7.0, or 8.0. The media was buffered with 50 mM sodium citrate (pH 5.0), 50 mM MES (pH 5.7 and 7.0), or 50 mM Tris–HCl (pH 8.0). For growth in liquid medium, 106 conidia were germinated into 100 mL of SDB media and incubated at 28 °C for 96 h under agitation. The mycelia of individual flasks were aseptically filtered and transferred into new flasks according to the following conditions. In summary, they are 50 mL of SDB and 0.5 M NaCl, pH adjusted to 5.7 or 8.0, and incubated at 28 °C under agitation in short exposure times for 30 min, 3 h, or 6 h, or mycelia were transferred into 50 mL of SDB buffered to pH 4.0 or pH 8.0 with sodium citrate or Tris–HCl, respectively, with and without 0.5 M NaCl, and incubated at 28 °C under agitation for 3 h. After incubation, the mycelia from each experiment were frozen in liquid nitrogen and stored at − 80 °C until RNA extraction.
RNA isolation and cDNA synthesis
Total RNA was isolated using the Illustra RNAspin Mini RNA Isolation Kit (GE Healthcare, Chicago, IL, USA). First-strand cDNA synthesis was performed using a high-capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA, USA). RNA extraction and cDNA synthesis were performed according to the manufacturer’s recommendations on three replicates from all experimental conditions.
Primer design and qRT-PCR
Specific primer pairs for each gene were designed using the software Primer3 (the designed oligonucleotide primer pairs had an approximately 50% GC content that yielded an expected amplicon size of around 150 bp with a melt temperature of 60 °C). Real-time PCR reactions were performed using Power SYBR Green PCR Master Mix in a StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The thermal conditions for qRT-PCR were 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min. DNA-dependent RNA polymerase II (rpb2) was used as an internal control [15]. All reactions were performed in triplicate. Table 1 provides a list of the primer sequences and PCR efficiency. The 2−ΔΔct relative expression quantification method was used to calculate relative expression levels [16]. One-way ANOVA followed by Tukey’s ad hoc test was carried out for statistical analysis using the GraphPad Prism v. 5.1 (GraphPad Software, La Jolla, CA, USA).
Table 1.
List of primers used in qRT-PCR analysis
| ID | Gene product name | Primers (5′-3′) | Efficiency (%) | Concentration (nM) |
|---|---|---|---|---|
| H101_05158 | C2H2 transcription factor Crz1, putative (T. benhamiae) |
FW: GCACAGTATCCTGCACTACA REV: CTCTTCATCCCCAAATCCAA |
98.31 | 300 |
| H101_06519 | potassium/sodium efflux P-type ATPase, fungal-type |
FW: GCAGCTGGATTTGTGACTTC REV: GCTACAATGCCCCATTCCCA |
94.76 | 400 |
| H101_01964 | C2H2 transcription factor Egr2, putative (T. verrucosum) |
FW: CCAATCTCGCCGACACTCAA REV: GCCGTCTTTCATCCTCTGCA |
100.06 | 400 |
| H101_07360 | pH-response transcription factor pacC/RIM101 |
FW: CCCCCATGGGCAATCTG REV: TGCTCCAGGAATTGGTCAATA |
98.98 | 400 |
| H101_07478 | CMGC/MAPK/P38 protein kinase Hog1 |
FW: AACCAAGCAACATCCTCATC REV: TTGCCATGTCAGCATGATCT |
102.53 | 500 |
| H101_04190 | DNA-dependent RNA polymerase II subunit (T. mentagrophytes) |
FW: TGCAGGAGCTGGTGGAAGA REV: GCTGGGAGGTACTGTTTGATCGA |
94.99 | 300 |
Results
Exposure to sodium chloride
Sensitivity of T. interdigitale H6 and the ∆pacC strains to NaCl stress was evaluated in SDB agar plates. In Fig. 1, the radial growth highlighted the increased sensitivity of the mutant to the salt, mainly in the highest NaCl concentration. The plates of the propagated mycelia were photographed after 6 days.
Fig. 1.
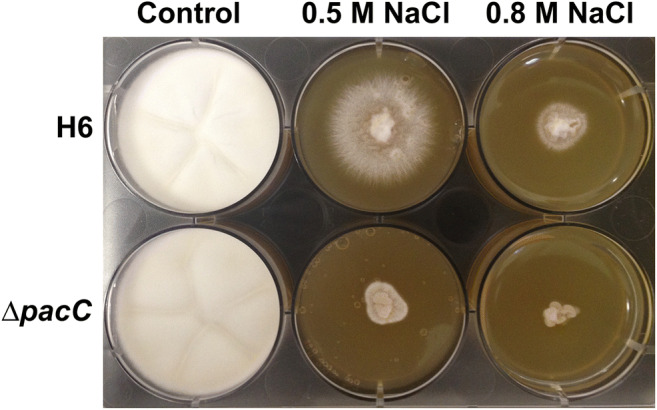
Stress induced by NaCl. Trichophyton interdigitale H6 and ΔpacC knockout strains grown for 6 days in SDB media, in the presence or absence of NaCl
To better observe the effect of buffering and pH variation in the wild-type and the mutant pacC strains, both the strains were cultivated in SDB agar plates in acidic (pH 5.0), neutral (pH 7.0), and alkaline pH (pH 8.0) and in the natural pH of the culture media (pH 5.7) (Fig. 2). Growth conditions considerably restricted T. interdigitale development, especially that of the ΔpacC mutant strain. The most adaptable condition was the natural pH of the SDB media, with a particular positive effect on growth at the buffered pH of 5.7.
Fig. 2.
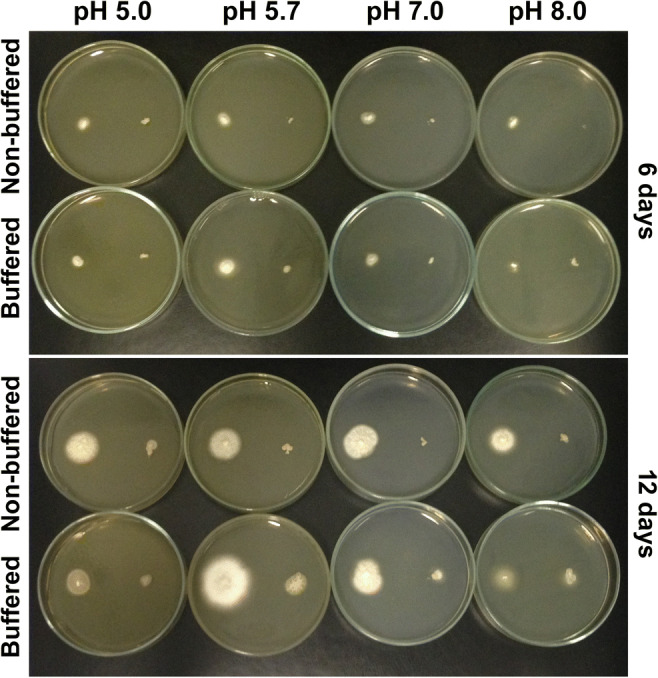
Radial growth in a salt stress medium at different pH levels. In each plate, T. interdigitale (wild type) is grown on the left and its ΔpacC mutant is grown on the right. Strains were grown in SDB medium supplemented with 0.5 M NaCl in buffered and non-buffered medium at pH 5.0, 5.7, 7.0, or 8.0. The plates were incubated at 28 °C for 6 or 12 days
pacC expression profiling under stress
The transcription level of the pacC gene was evaluated in each of the tested stress conditions using 96-h grown SDB mycelia of T. interdigitale H6 as the control. The expression analysis aimed to correlate between the observed growth effects and gene modulation. The overall profile showed no significant expression in response to the NaCl or pH variation challenges, except for the buffered conditions supplemented with NaCl (Fig. 3). Under acidic pH conditions, buffering and salinity stress together caused severe inhibition of pacC expression. On the other hand, buffering the growth media to achieve an alkaline pH combined with saline stress resulted in induction of pacC.
Fig. 3.
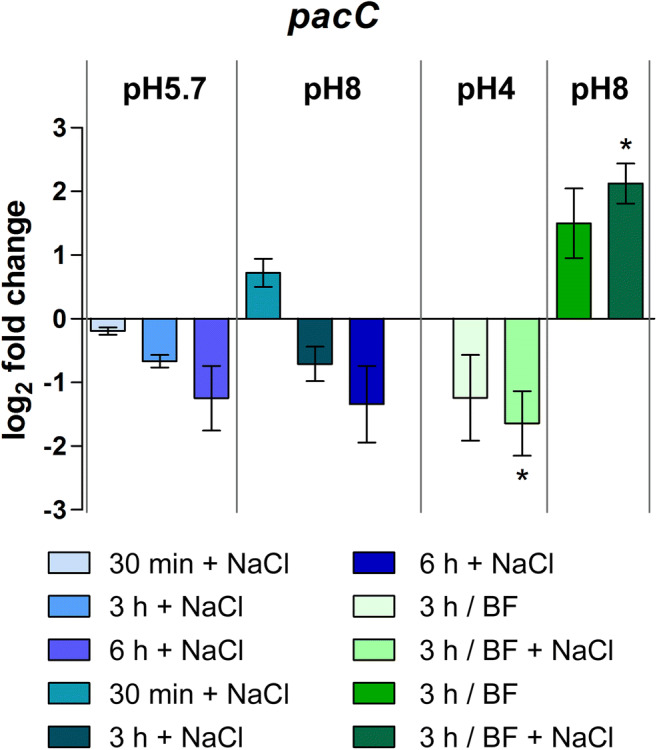
pacC gene expression profiling. Relative expression of the pacC gene represented as log2-fold change evaluated using Trichophyton interdigitale H6 (wild type) 96 h-grown SDB mycelia as the reference sample after normalization with the rpb2 endogenous gene. Values are the average and standard deviation of three independent experiments. Asterisks indicate statistical significance determined by ANOVA followed by Tukey’s ad hoc test (∗P < 0.05). BF buffered
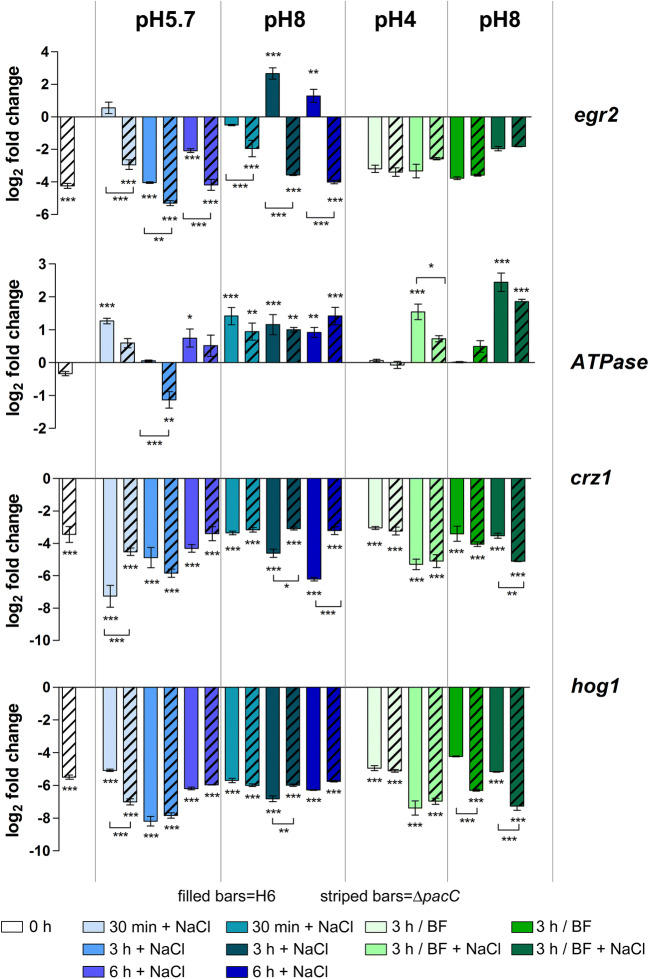
The relative expression of genes required during salinity stress was then evaluated by attempting to establish regulatory relationships between their expression and the salt stress effect in T. interdigitale. A potassium/sodium efflux P-type ATPase, the MAP kinase hog1, and two transcription factors, crz1 and egr2, expressed in the ΔpacC mutant strain were compared to the control condition (wild-type T. interdigitale H6 mycelia, 96–h grown in SDB). The ATPase gene was preferentially induced, whereas egr2, crz1, and hog1 were mainly repressed. Furthermore, the transcription of the ATPase, crz1, and hog1 occurred mostly independent of pacC (Fig. 4).
Fig. 4.
Gene expression profiling. Relative expression of genes represented as log2-fold change evaluated using Trichophyton interdigitale H6 (wild type) 96-h grown SDB mycelia as the reference sample after normalization with the rpb2 endogenous gene. Values are the average and standard deviation of three independent experiments. Asterisks indicate statistical significance determined by ANOVA followed by Tukey’s ad hoc test (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001). Statistical significance was either evaluated by comparing the wild-type and mutant strains by condition (paired comparison) using the wild type as the reference (asterisks indicated below the lines). BF buffered
Egr2 was not responsive to buffered conditions, whereas buffering only affected the ATPase in the presence of NaCl. Under buffered conditions, crz1 expression was only responsive to PacC absence at pH 8.0 under salinity stress; at the same pH, hog1 was sensitive to buffering in a non-NaCl-dependent manner.
Discussion
Stress-associated responses of pacC
Radial colony growth is profoundly affected by ∆pacC mutation in different NaCl concentrations as compared to the control strain. The inhibitory growth effect was accentuated as the concentration of NaCl raised. This growth pattern indicated a direct relationship between PacC and T. interdigitale phenotype. Our results demonstrated that, in some way, the tolerance to high salt concentrations is dependent on pacC. However, we observed that the transcriptional profile of pacC was differentially modulated during salinity stress solely in the buffered conditions. A repressive effect was observed in the buffered acidic pH, and the gene was induced at the buffered alkaline pH (Fig. 3). These results suggested that the effort to develop in buffered conditions required the transcriptional activation of pacC.
We further conducted radial growth tests in SDB agar plates using buffered and non-buffered salt stress medium at different pH levels (acidic, neutral, and alkaline). We also conducted these tests at pH 5.7, the recommended pH of the culture media. Variation hampered ∆pacC radial growth by inhibiting mycelial growth. With the exception of pH 5.7, buffering either restricted the development of the wild strain, with visible phenotypical growth defects. Growth conditions at pH 8.0 were the most challenging among the evaluated ones and buffering considerably affected development. These observations were similar to the ones obtained previously when both the strains were cultivated in minimal medium [17].
The functional activity of PacC has been supposed to occur at both acidic and alkaline ambient pH, with the highest transcriptional levels of pacC generally achieved under alkaline conditions. Its transcription levels are lowered under acid growth conditions [4, 5]. In this work, pacC transcriptional accumulation was not observed during NaCl stress independently of the pH, nor in the absence of salt at buffered conditions. This result shows that although pacC is not differentially expressed, it alters the behavior of downstream regulatory targets depending on the cultivation stress imposed.
A non-significant transcriptional accumulation of pacC was observed for the fungus Beauveria bassiana at the earlier stages of cultivation under saline conditions; however, pacC gradually accumulated over time following the alkalization of the culture medium [18]. In this study, we evaluated the role of pacC in the initial stages of T. interdigitale development in salt stress conditions. We suppose that in later time points, the modulatory activation of pacC may increase, following the pattern observed in the literature.
PacC can act both as an activator and as a repressor of transcription
The environmental stress conditions resulted in the overall repression of egr2 (Fig. 4). An inducible effect was only identified under alkaline conditions in the wild-type strain after 3 and 6 h of salt exposure. This induction decreased over time, which suggested an adaptive response to the alkaline culture media. In Saccharomyces cerevisiae, Nrg1/Egr2 is involved in pH-responsive gene regulation and ion tolerance [19]. Here, the significant induction of Egr2 implies an inducible PacC activity as a response to culture pH. The absence of pacC makes egr2 behave similar to that under acidic pH condition.
The Egr2-repressive profile observed in acidic pH was accentuated in the mutant strain compared with that in the wild-type strain. In neutral–alkaline environments, PacC/Rim101 is a negative regulator of nrg1/egr2 expression [6]. Here, we confirm that PacC cooperates with Egr2, as repressors, in response to acidic pH, suggesting that they act as transcriptional co-repressors in response to salt. Since egr2 is no longer differentially expressed in buffered media, we believe that the modulation pattern is also ambient pH dependent.
The transcriptional response of a P-type ATPase, putatively active in the extrusion of Na+ and K+, was evaluated. The pumping activity of ATPases is crucial for growth in unsuitable conditions such as at high external pH values or under toxic concentrations of cations, as it protects cells from extreme environmental stress [20, 21]. In our study, pumping activity was not activated in the ∆pacC strain in the 96-h grown SDB condition. The activation is dependent on NaCl supporting the ATPase activity. Under acidic conditions, T. interdigitale extrusion activity was activated in the initial period, and the pumping activity weakened after 3 and 6 h of salinity stress. Buffering induces ATPase expression within 3 h, the deletion of pacC compromises cation transport, and alkaline conditions require ATPase activation independently of pacC.
A high number of cations ATPases were identified in dermatophyte genomes [22]. Antifungals upregulated some sodium ATPase genes in Trichophyton rubrum, which suggest their participation in the resistance to these drugs [23–25]. In T. interdigitale H6, sodium ATPases, including the ATPase analyzed in the present study, were overexpressed after growth in a keratinocyte serum-free medium or at alkaline pH values [23, 24]. These findings revealed that dermatophytes activate ATPase genes in response to non-specific stress conditions [25], and they act as both drug and cation pumps in challenging conditions.
Transcription of a few stress-responsive genes is not under pacC control
In our study, we observed a repression pattern of the transcription factor crz1, and in most cases, it was independent of PacC. In Candida albicans, a complex mechanism is activated in response to the environmental pH, integrating Rim101 and calcineurin/Crz1 signaling pathways, which results in the adaptation to alkaline pH [6, 26]. Here, crz1 expression was not regularly regulated by pacC, suggesting the independence between both pathways. After growing the strains for 3 h under alkaline conditions, the deletion of pacC resulted in an opposite transcriptional pattern when comparing the buffered and non-buffered conditions. In the non-buffered condition, crz1 was more deeply repressed in the wild-type strain when compared with the mutant strain. Thus, the lack of pacC attenuates the repressive effect of crz1. This repressive effect persisted, with a more significant difference between the wild-type and mutant strains at 6 h of salinity stress. When buffered, the alkaline environment more profoundly suppressed crz1 expression in the mutant strain. This enhanced repression of crz1 in ∆pacC in the buffered condition suggests that PacC attenuates crz1 repression as a response to the culture buffering.
The transcription of hog1 was repressed independent of the experimental conditions and the presence of PacC (Fig. 4). The repressive profile was likewise observed in the ∆pacC 96 h-grown SDB condition. Hog1 is a mitogen-activated protein kinase activated by high osmolarity to phosphorylate different transcriptional regulators, which influences gene expression intimately in response to osmotic stress. A direct relationship between NaCl sensitivity and repression of hog1 was established in S. cerevisiae; however, the particular role of this regulatory event was not clarified [27]. Here, we demonstrated that in buffered alkaline conditions, the repression of hog1 was accentuated in response to pacC deletion independent of NaCl presence, suggesting a PacC-dependent regulatory activity. An inverse pattern was observed in the non-buffered alkaline condition, at the same exposure time (3 h), suggesting that, as for crz1, PacC accentuates hog1 repression as an effect of pH fluctuation.
The complemented strain carrying the deleted gene would provide further evidence about the essential role of PacC. However, the difficulty of genetic manipulation in dermatophytes is caused by the unusually low transformation frequencies that diminish the chances to succeed in gene deletion by restricting the number of null mutants and the construction of complement strains as well [28]. These challenging factors have so far hindered the genetic analysis of dermatophyte fungi.
In summary, our study demonstrated that salinity stress and buffered pH conditions influence pacC modulation. The phenotypic effect was not associated with pacC transcriptional modulation; however, its influence may be related to the regulation exercised over other transcriptional regulators, especially under alkaline pH, and thus, over transcriptional networks.
Acknowledgments
We thank V. M. de Oliveira, M. Mazucato, and M. D. Martins for technical assistance.
Funding information
This study was funded by the Research Support Foundation of São Paulo State (FAPESP; Grant No. 14/03847-7, and Fellowship No. 10/15017-8 to LGS, 18/11319-1 to MPM, and 09/08411-4 to NTAP), National Council for Scientific and Technological Development (CNPq, Grant Nos. 305797/2017-4 and 304989/2017-7), Coordination for the Improvement of Higher Education Personnel (CAPES, Finance Code 001), and Foundation for Support to Teaching, Research, and Assistance (FAEPA/HCFMRP-USP).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Larissa Gomes da Silva and Maíra Pompeu Martins contributed equally to this work.
References
- 1.Kim J, Choi M, Kim JR, Jin H, Kim VN, Cho KH. The co-regulation mechanism of transcription factors in the human gene regulatory network. Nucleic Acids Res. 2012;40(18):8849–8861. doi: 10.1093/nar/gks664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dai Z (2019) Transcription factors indirectly regulate genes through nuclear colocalization. Cells 8(7). 10.3390/cells8070754 [DOI] [PMC free article] [PubMed]
- 3.Ment D, Alkan N, Luria N, Bi FC, Reuveni E, Fluhr R, Prusky D. A role of AREB in the regulation of PACC-dependent acid-expressed-genes and pathogenicity of Colletotrichum gloeosporioides. Mol Plant-Microbe Interact. 2015;28(2):154–166. doi: 10.1094/MPMI-09-14-0252-R. [DOI] [PubMed] [Google Scholar]
- 4.Tilburn J, Sarkar S, Widdick DA, Espeso EA, Orejas M, Mungroo J, Penalva MA, Arst HN., Jr The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid- and alkaline-expressed genes by ambient pH. EMBO J. 1995;14(4):779–790. doi: 10.1002/j.1460-2075.1995.tb07056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rossi A, Cruz AH, Santos RS, Silva PM, Silva EM, Mendes NS, Martinez-Rossi NM. Ambient pH sensing in filamentous fungi: pitfalls in elucidating regulatory hierarchical signaling networks. IUBMB Life. 2013;65(11):930–935. doi: 10.1002/iub.1217. [DOI] [PubMed] [Google Scholar]
- 6.Kullas AL, Martin SJ, Davis D. Adaptation to environmental pH: integrating the Rim101 and calcineurin signal transduction pathways. Mol Microbiol. 2007;66(4):858–871. doi: 10.1111/j.1365-2958.2007.05929.x. [DOI] [PubMed] [Google Scholar]
- 7.O’Meara TR, Xu W, Selvig KM, O’Meara MJ, Mitchell AP, Alspaugh JA. The Cryptococcus neoformans Rim101 transcription factor directly regulates genes required for adaptation to the host. Mol Cell Biol. 2014;34(4):673–684. doi: 10.1128/MCB.01359-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martins MP, Martinez-Rossi NM, Sanches PR, Gomes EV, Bertolini MC, Pedersoli WR, Silva RN, Rossi A. The pH signaling transcription factor PAC-3 regulates metabolic and developmental processes in pathogenic fungi. Front Microbiol. 2019;10:2076. doi: 10.3389/fmicb.2019.02076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruiz A, Arino J. Function and regulation of the Saccharomyces cerevisiae ENA sodium ATPase system. Eukaryot Cell. 2007;6(12):2175–2183. doi: 10.1128/EC.00337-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caracuel Z, Casanova C, Roncero MI, Di Pietro A, Ramos J. pH response transcription factor PacC controls salt stress tolerance and expression of the P-type Na+-ATPase Ena1 in Fusarium oxysporum. Eukaryot Cell. 2003;2(6):1246–1252. doi: 10.1128/ec.2.6.1246-1252.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lanza M, Haro R, Conchillo LB, Benito B. The endophyte Serendipita indica reduces the sodium content of Arabidopsis plants exposed to salt stress: fungal ENA ATPases are expressed and regulated at high pH and during plant co-cultivation in salinity. Environ Microbiol. 2019;21:3364–3378. doi: 10.1111/1462-2920.14619. [DOI] [PubMed] [Google Scholar]
- 12.Wu Y, Yin Z, Xu L, Feng H, Huang L. VmPacC is required for acidification and virulence in Valsa mali. Front Microbiol. 2018;9:1981. doi: 10.3389/fmicb.2018.01981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arechiga-Carvajal ET, Ruiz-Herrera J (2005) The RIM101/pacC homologue from the basidiomycete Ustilago maydis is functional in multiple pH-sensitive phenomena, Eukaryot Cell. 4(6):999–1008. 10.1128/EC.4.6.999-1008.2005 [DOI] [PMC free article] [PubMed]
- 14.Ferreira-Nozawa MS, Silveira HC, Ono CJ, Fachin AL, Rossi A, Martinez-Rossi NM. The pH signaling transcription factor PacC mediates the growth of Trichophyton rubrum on human nail in vitro. Med Mycol. 2006;44(7):641–645. doi: 10.1080/13693780600876553. [DOI] [PubMed] [Google Scholar]
- 15.Jacob TR, Peres NT, Persinoti GF, Silva LG, Mazucato M, Rossi A, Martinez-Rossi NM. rpb2 is a reliable reference gene for quantitative gene expression analysis in the dermatophyte Trichophyton rubrum. Med Mycol. 2012;50(4):368–377. doi: 10.3109/13693786.2011.616230. [DOI] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Martinez-Rossi NM, Persinoti GF, Peres NT, Rossi A. Role of pH in the pathogenesis of dermatophytoses. Mycoses. 2011;55(5):381–387. doi: 10.1111/j.1439-0507.2011.02162.x. [DOI] [PubMed] [Google Scholar]
- 18.Zhou YH, Hou L, Zhang YJ, Fan YH, Luo ZB, Jin D, Zhou QS, Li YJ, Wang Y, Pei Y. Expression and promoter characterization of BbPacC, a pH response transcription factor gene of the entomopathogenic fungus Beauveria bassiana. Microbiology. 2014;160(Pt 2):353–361. doi: 10.1099/mic.0.071159-0. [DOI] [PubMed] [Google Scholar]
- 19.Lamb TM, Mitchell AP. The transcription factor Rim101p governs ion tolerance and cell differentiation by direct repression of the regulatory genes NRG1 and SMP1 in Saccharomyces cerevisiae. Mol Cell Biol. 2003;23(2):677–686. doi: 10.1128/MCB.23.2.677-686.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Serrano R, Ruiz A, Bernal D, Chambers JR, Arino J. The transcriptional response to alkaline pH in Saccharomyces cerevisiae: evidence for calcium-mediated signalling. Mol Microbiol. 2002;46(5):1319–1333. doi: 10.1046/j.1365-2958.2002.03246.x. [DOI] [PubMed] [Google Scholar]
- 21.Benito B, Garciadeblas B, Rodriguez-Navarro A. Potassium- or sodium-efflux ATPase, a key enzyme in the evolution of fungi. Microbiology. 2002;148(Pt 4):933–941. doi: 10.1099/00221287-148-4-933. [DOI] [PubMed] [Google Scholar]
- 22.Martinez DA, Oliver BG, Graser Y, Goldberg JM, Li W, Martinez-Rossi NM, Monod M, Shelest E, Barton RC, Birch E, Brakhage AA, Chen Z, Gurr SJ, Heiman D, Heitman J, Kosti I, Rossi A, Saif S, Samalova M, Saunders CW, Shea T, Summerbell RC, Xu J, Young S, Zeng Q, Birren BW, Cuomo CA, White TC. Comparative genome analysis of Trichophyton rubrum and related dermatophytes reveals candidate genes involved in infection. MBio. 2012;3(5):e00259–e00212. doi: 10.1128/mBio.00259-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peres NT, Sanches PR, Falcao JP, Silveira HC, Paiao FG, Maranhao FC, Gras DE, Segato F, Cazzaniga RA, Mazucato M, Cursino-Santos JR, Aquino-Ferreira R, Rossi A, Martinez-Rossi NM. Transcriptional profiling reveals the expression of novel genes in response to various stimuli in the human dermatophyte Trichophyton rubrum. BMC Microbiol. 2010;10:39. doi: 10.1186/1471-2180-10-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silveira HC, Gras DE, Cazzaniga RA, Sanches PR, Rossi A, Martinez-Rossi NM. Transcriptional profiling reveals genes in the human pathogen Trichophyton rubrum that are expressed in response to pH signaling. Microb Pathog. 2010;48(2):91–96. doi: 10.1016/j.micpath.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 25.Martins MP, Franceschini AC, Jacob TR, Rossi A, Martinez-Rossi NM. Compensatory expression of multidrug-resistance genes encoding ABC transporters in dermatophytes. J Med Microbiol. 2016;65(7):605–610. doi: 10.1099/jmm.0.000268. [DOI] [PubMed] [Google Scholar]
- 26.Wang H, Liang Y, Zhang B, Zheng W, Xing L, Li M. Alkaline stress triggers an immediate calcium fluctuation in Candida albicans mediated by Rim101p and Crz1p transcription factors. FEMS Yeast Res. 2011;11(5):430–439. doi: 10.1111/j.1567-1364.2011.00730.x. [DOI] [PubMed] [Google Scholar]
- 27.Galgoczy DJ, Cassidy-Stone A, Llinas M, O’Rourke SM, Herskowitz I, DeRisi JL, Johnson AD. Genomic dissection of the cell-type-specification circuit in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2004;101(52):18069–18074. doi: 10.1073/pnas.0407611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamada T, Makimura K, Hisajima T, Ishihara Y, Umeda Y, Abe S. Enhanced gene replacements in Ku80 disruption mutants of the dermatophyte, Trichophyton mentagrophytes. FEMS Microbiol Lett. 2009;298(2):208–217. doi: 10.1111/j.1574-6968.2009.01714.x. [DOI] [PubMed] [Google Scholar]