Abstract
With the high-frequency use or abuse of antifungal drugs, the crisis of drug-resistant fungi continues to increase worldwide; in particular, the infection of drug-resistant Candida albicans brings the great challenge to the clinical treatment. Therefore, to decelerate the spread of this resistance, it is extremely urgent to facilitate the new antifungal targets with novel drugs. Phosphopantetheinyl transferases PPTases (Ppt2 in Candida albicans) had been identified in bacterium and fungi and mammals, effects as a vital enzyme in the metabolism of organisms in C. albicans. Ppt2 transfers the phosphopantetheinyl group of coenzyme A to the acyl carrier protein Acp1 in mitochondria for the synthesis of lipoic acid that is essential for fungal respiration, so making Ppt2 an ideal target for antifungal drugs. In this study, 110 FDA-approved drugs were utilized to investigate the Ppt2 inhibition against drug-resistant Candida albicans by the improved fluorescence polarization experiments, which have enough druggability and structural variety under the novel strategy of drug repurposing. Thereinto, eight agents revealed the favourable Ppt2 inhibitory activities. Further, broth microdilution assay of incubating C. albicans with these eight drugs showed that pterostilbene, procyanidine, dichlorophen and tea polyphenol had the superior MIC values. In summary, these findings provide more valuable insight into the treatment of drug-resistant C. albicans.
Electronic supplementary material
The online version of this article (10.1007/s42770-020-00318-w) contains supplementary material, which is available to authorized users.
Keywords: Phosphopantetheinyl transferases PPTases, Ppt2 inhibitor, Drug-resistant Candida albicans
Introduction
Invasive fungal infections (IFIs) and their serious drug resistance have become one of the most serious healthy issues for global public health in recent decades, especially in immunocompromised hosts including patients with transplant, AIDS, cancer or other immunomodulatory therapies [1]. More seriously, IFIs are regarded as human “hidden killers”due to the high incidence and mortality. Of particular concern is the high rate of mortality associated with IFIs, which often exceeds 50% despite the availability of several antifungal drugs [2]. Candida albicans, one of the most common species of fungi that is responsible for IFIs, accounts for 20–40% of the mortality in the nosocomial infections in total [3]. In China, C. albicans accounts for 46.20% and 57.80% of all Candida infections in large hospitals in Beijing and Shanghai [4, 5]. However, current available antifungal drugs, such as polyenes, azoles, allylamines, echinocandins and pyrimidines, elicit unwanted side effects such as nephrotoxicity induced by amphotericin B, visual impairment caused by voriconazole and congestive heart failure caused by itraconazole [6, 7]. In addition, the combination of rifampicin and voriconazole can reduce the concentration of voriconazole and increase the concentration of rifampicin to the level of toxicity. Furthermore, the widespread and irrational use of antifungal drugs has led to many changes in the composition and susceptibility of pathogenic fungi. Under the drug selection pressure, more and more susceptive strains are inhibited or killed, whereas drug-resistant strains are gradually favoured, thereby increasing the emergence of resistant fungi. There have been important changes in the epidemiology of invasive candidiasis, with an overall shift toward Candida spp. other than C. albicans, particularly Candida glabrata, with reduced susceptibility to the azole antifungals [8]. In the CHIF-NET study, 12.2% of the C. glabrata sensu stricto isolates were fluconazole resistant and 17.8% had non-wild-type susceptibility to voriconazole. Seven Candida tropicalis strains were cross-resistant to fluconazole and voriconazole. Resistance to fluconazole and voriconazole was seen in 31.9% of the uncommon Candida including Candida pelliculosa and Candida quercitrusa [9].
In this “post-antibiotic era”, the development of conventional antifungal drugs cannot keep up with the pace of resistant fungal mutations, and it is urgent to find out antifungal drugs with new treatment targets and strategies to avoid these known resistance mechanisms.
Phosphopantetheinyl transferases PPTases are fundamental in the metabolism of bacteria, fungi and mammals [10]. They transfer the phosphopantetheinyl moiety of CoA to an inactive carrier protein, further render it active. In fungi, phosphopantetheinyl transferases PPTases are divided into three types, including (1) Domain-type enzymes are involved in the biosynthesis of fatty acids in the cytoplasm; (2) Sfp-type enzymes participate in the biosynthesis of lysine and activate α-amino acetyl reductase to synthesize polyketides and non-ribosomal peptides [11]; (3) AcpS-type enzymes (Ppt2 in C. albicans) are involved in the biosynthesis of fatty acids in mitochondria. In addition, AcpS-type enzymes as a vital cofactor for the synthesis of lipoic acid can transfer the phosphopantetheinyl group of coenzyme A to the inactive acyl carrier protein Acp1 in mitochondria effectively, while lipoic acid represents enough importance as another coenzyme for mitochondrial respiration in fungi. Recent studies disclosed that C. albicans cells in which the PPT2 gene is blocked are unable to grow on synthetic-dropout (SD) medium. Importantly, in contrast with all Sfp-type PPTases in mammals, Ppt2 in C. albicans is AcpS-type which is absent from humans, making it the promising antifungal drug target [12].
Drug repurposing (also known as drug repositioning) as a practical drug discovery technology can accelerate the novel drug development process by aiming to apply old drugs (marketed drugs or FDA-approved drugs) to new diseases. In addition, the druggability and structural variety of old drugs, along with the well-established toxicology and pharmacology, fascinate the researchers to pay more attention to find new usage by screening the “old drug bank” [13]. Furthermore, it is reported that drug repurposing can save about 40% of research and development costs [14]. In this study, 110 FDA-approved drugs were selected for investigating the Ppt2 inhibitory activities.
In the present study, we successfully improved an assay suitable for high-throughput screening of potential Ppt2p inhibitors for use as antifungals based on Dobb’s documented methods [12]. The experiment principle of the fluorescence polarization (FP) drug screening is that the transfer of the 4’PPT subunit (labelled with fluorescent dye BODIPY-BTMR) from the small molecule CoA to the macromolecule Acp1 leads to an increase in the FP value. Then, we screened 110 FDA-approved drugs, eight compounds in them were found to inhibit C. albicans Ppt2 by more than 50%. We measured IC50 values and MIC values in vitro for these eight compounds, and pterostilbene, procyanidine, dichlorophen and tea polyphenol were found to be the more potential Ppt2 inhibitors.
Results
Fluorescence polarization assay
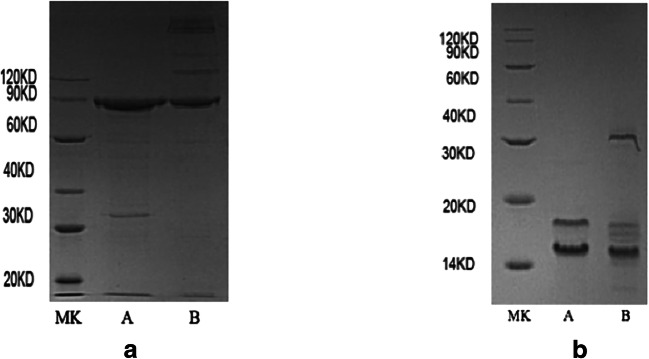
In order to establish a high-throughput drug screening assay, recombinant Ppt2p and Acp1p were successfully produced and purified (Fig. 1). The native Ppt2p protein was predicted to be 15.8 kDa. A vector pET43.1 that contains an N-terminal Nus-tag was added to improve solubility of the protein, so the molecular weight of recombinant Ppt2p protein was 76 kDa (Fig. 1a).
Fig. 1.
Expression and purification of recombinant C. albicans Ppt2 and Acp1. Escherichia coli BL21 DE3 cells were transformed with a pET43.1-Ppt2 (76 kDa) and b pET30-Acp1 (16 kDa), and expression of the recombinant proteins was induced with 0.1 mM IPTG. Recombinant proteins were purified using a chelating SFF (Ni) column. Fractions were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Lane A = reduced protein; lane B = non-reduced protein; MK = molecular weight markers
The recombinant protein Acp1p came out with two bands (Fig. 1b). A similar result also occurred in Aspergillus fumigatus [11]. It is possible that the upper of the two Acp1 bands is holo-ACP already pantetheinylated by the Escherichia coli PPTase during expression, and the lower band is apo-ACP.
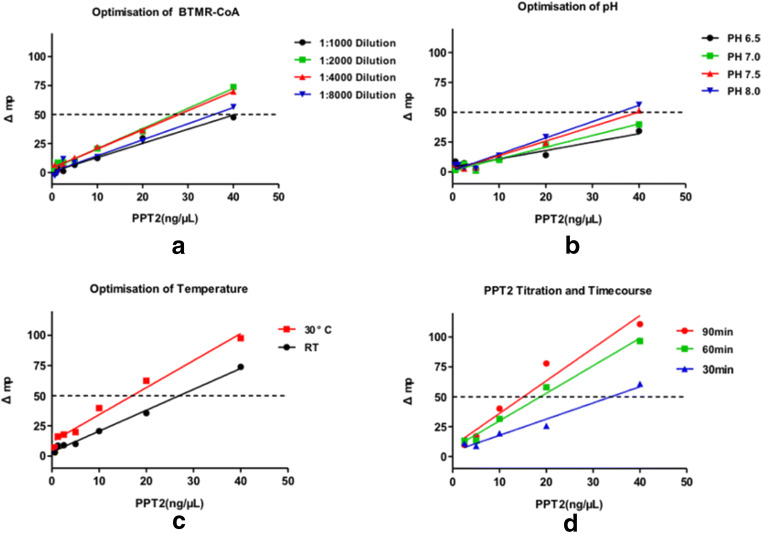
In the presence of Ppt2p, its target protein Acp1p, labelled CoA and a compound targeting Ppt2p, a decrease in FP value was observed compared with a reaction in the absence of the Ppt2p inhibitor. An addition of 20 mM EDTA stopped the reaction, allowing multiple plates to be processed simultaneously. To find out the optimal CoA-BTMR concentration, 1:1000, 1:2000, 1:4000 and 1:8000 dilutions were tested and 1:2000 dilution and 1:4000 dilution gave the best Δmp value (Fig. 2a), and we chose 1:2000 dilution for further experiments. The pH and temperature were also tested and pH 8.0 and 30 °C were optimal (Fig. 2b, c). Finally, the Ppt2 concentration and reaction duration were tested, and 40 ng/μL Ppt2p and 30 min had the optimal linearity, but Δmp was low. However, after 60 min and 90 min, the linearity and Δmp were well within the 20 ng/μL Ppt2p concentration. At a concentration of 20 ng/μL Ppt2p, Δmp was > 50. To save costs, we therefore chose 20 ng/μL as the Ppt2p concentration and a reaction duration of 60 min for the final high-throughput screening assay to identify inhibitors of C. albicans (Table 1) Ppt2 (Fig. 2d).
Fig. 2.
Optimisation of BTMR-CoA, pH, temperature, Ppt2 concentration and reaction duration. Δmp is an important indicator in the fluorescence polarization experiment. The larger Δmp value illustrates the better performance of the experiment. It is generally considered that Δmp > 50 is an accurate standard for experiments. a Reactions with 1:1000, 1:2000, 1:4000 and 1:8000 dilutions of BTMR-CoA. b Reactions with different pH values achieved by adding NaOH. c Reactions at 30 °C and room temperature (RT). d Reactions with different concentrations of Ppt2 (from 10 to 40 ng/μL), and for different durations (30, 60 and 90 min)
Table 1.
Candida albicans strains used in this study
| Strain | Parental strain | Relevant characteristics or genotype | Reference or source |
|---|---|---|---|
| SN152 | SC5314 | ura3::imm434::URA3/ura3::imm434iro1::IRO1/iro1::imm434 his1::hisG/his1::hisG leu2/leu2 arg4/arg4 | |
| ATCC 90028 | N/A | Candida albicans ATCC strain | ATCC |
| Clinical isolates | |||
| 80 | N/A | Fluconazole-susceptible C. glabrata | – |
| 180 | N/A | Fluconazole-susceptible C. tropicalis | – |
| 13,139 | N/A | Fluconazole-resistant C. albicans | – |
| 26 | N/A | Fluconazole-resistant C. albicans | – |
| 21 | N/A | Fluconazole-resistant C. albicans | – |
| 196 | N/A | Fluconazole-susceptible C. albicans | – |
| 198 | N/A | Fluconazole-susceptible C. albicans | – |
N/A not applicable
Policresulen, procyanidine, dichlorophen, tea polyphenol and rutin are potent inhibitors of Ppt2p in FP assays
We tested 110 clinical drugs at the final concentration of 100 μM as candidate Ppt2p inhibitors using the optimized screening method described above. Of these, eight compounds achieved > 50% inhibition, with policresulen (CAS 9011-02-3/101418-00-2), procyanidine (CAS 4852-22-6), dichlorophen (CAS 97-23-4), tea polyphenol (CAS 84650-60-2) and rutin (CAS 153-18-4) achieving > 80% inhibition, and pterostilbene (CAS 537-42-8), glycyrrhetinic acid (CAS 471-53-4) and dimethyl fumarate (CAS 624-49-7) achieving between 50% and 80% inhibition. Applying the method of Zhang and colleagues, Z’ > 0.5 was obtained, indicating that the screening method is robust [15].
We then determined IC50 values for the eight compounds following duplicate serial dilution in 384-well black plates (Table 2). The result demonstrated that the procyanidine and tea polyphenol had the better IC50 values which were all less than 10 μM (Fig. S2F, G) in particular.
Table 2.
IC50 values of the eight compounds identified by the FP assay
| Compounds | Final stock (μM) | IC50 value (μM) |
|---|---|---|
| Policresulen | 100 | 14.76 ± 0.37 |
| Glycyrrhetinic acid | 300 | 99.48 ± 6.32 |
| Pterostilbene | 300 | 125.95 ± 14.65 |
| Dimethyl fumarate | 300 | 62.69 ± 1.62 |
| Dichlorophen | 100 | 40.14 ± 1.15 |
| Procyanidine | 100 | 2.31 ± 0.04 |
| Tea polyphenol | 100 | 3.74 ± 0.37 |
| Rutin | 100 | 21.21 ± 1.40 |
IC50 half maximal inhibitory concentration
Pterostilbene, procyanidine, dichlorophen and tea polyphenol gave the best MIC values in broth microdilution assays
Among the eight compounds, policresulen, rutin, glycyrrhetinic acid and dimethyl fumarate had almost no inhibitory effect on Candida spp. isolates. The MICs of these four compounds were all greater than 128 mg/L. As shown in Table 3, pterostilbene, procyanidine, dichlorophen and tea polyphenol had better MIC values via broth microdilution assay, and these four compounds exhibited the strongest inhibitory effect on C. glabrata, especially procyanidine and dichlorophen (MIC = 0.5 mg/L). By contrast, the inhibitory effect of compounds on C. tropicalis was less pronounced, and even procyanidine and dichlorophen had MICs > 100 mg/L. Among the four compounds, procyanidine appeared to be the best inhibitor of C. albicans, with MIC values < 10 mg/L except for strain 21.
Table 3.
Results of in vitro compound susceptibility testing via broth microdilution assay
| Isolates | Species | MIC (mg/L)/(μM) | ||||
|---|---|---|---|---|---|---|
| Fluconazole | Pterostilbene | Procyanidine | Dichlorophen | Tea polyphenol | ||
| 90028 | C. albicans | 0.5 (1.63) | 16 (62.43) | 8 (13.46) | 64 (237.81) | 8 (28.43) |
| 80 | C. glabrata | 1 (3.27) | 16 (62.43) | 0.5 (0.84) | 0.5 (1.86) | 4 (14.22) |
| 180 | C. tropicalis | 1 (3.27) | 32 (124.85) | > 128 (> 215.30) | 128 (475.62) | 16 (56.87) |
| 13139 | C. albicans | 128 (417.93) | 16 (62.43) | 8 (13.46) | 32 (118.91) | 16 (56.87) |
| 26 | C. albicans | 256 (835.86) | 32 (124.85) | 8 (13.46) | 32 (118.91) | 32 (113.73) |
| 21 | C. albicans | 256 (835.86) | 32 (124.85) | 32 (53.82) | 16 (59.45) | 32 (113.73) |
| 196 | C. albicans | 0.5 (1.63) | 16 (62.43) | 4 (6.73) | 8 (29.73) | 32 (113.73) |
| 198 | C. albicans | 0.5 (1.63) | 16 (62.43) | 4 (6.73) | 16 (59.45) | 16 (56.87) |
MIC minimum inhibitory concentration
Discussion
In this study, we successfully improved a novel FP-based assay for the high-throughput screening of potential Ppt2p inhibitors based on the documented paper. After screening 110 clinical drugs, eight compounds revealed the ability to inhibit Ppt2p potentially. Among the eight compounds, pterostilbene, procyanidine, dichlorophen and tea polyphenol were the most potent inhibitors with the lowest MIC values.
Among the few reports on Ppt2p, most indicate that it plays an important role in metabolism in C. albicans. This enzyme transfers the phosphopantetheinyl group of CoA to Acp1 in order to make it activate as a cofactor for the synthesis of lipoic acid. Dobb et al. (2015) demonstrated Ppt2 as a broad-spectrum novel antifungal target and proposed a raw method for further drug discovery. Building on previous research, we developed an FP-based assay for the high-throughput screening of potential Ppt2p inhibitors, and confirmed MIC values of identified candidate drugs using broth microdilution assays. The results showed that pterostilbene, procyanidine, dichlorophen and tea polyphenol had the best MIC values. Among the four compounds, MIC results of procyanidine, dichlorophen and tea polyphenol were basically consistent with their IC50 values in the FP assays. Procyanidine had the lowest IC50 value and the results of in vitro susceptibility experiments were also the best. However, the results of pterostilbene in FP assays were not satisfactory, but the in vitro susceptibility experiments of this compound had shown certain effects. We speculated that the reasons may be related to the internal environment of Candida spp. isolates. The FP assays only obtain the results from the protein level, but the internal environments of strains are relatively complex with many influence factors so the results of in vitro susceptibility experiments may appear to be contrary to the FP assays. Another noteworthy result is that the antifungal effect of these four compounds on C. glabrata is very remarkable regardless of the value of IC50. We speculated that this may be related to the haploid of C. glabrata, which is different from those of other Candida species [16]. However, there is currently no literature to prove this speculation. In addition, there have been reported that dichlorophen has the antifungal effect on fungi [17], but the specific mechanism is not clarified in the literature. So we suspect that dichlorophen acts on more than one target of Candida species, further study is needed.
It is worth mentioning that these four drugs found in the experiment have excellent inhibitory effects on FLC-resistant C. albicans. The MIC values of FLC for strains 13139, 26 and 21 were all > 128 mg/L, but the MIC values of these four drugs for resistant strains were all ≤ 32 mg/L, especially procyanidine, even reached below 10 mg/L. The currently known mechanisms of C. albicans resistance to fluconazole are mainly high expression of efflux pump proteins and mutations related to ERG11 gene [18, 19]. In our previous study, strain 13139 was found to have mutations in ERG11-related genes that caused resistance to fluconazole [20], but in the present experiment, these newly discovered drugs have superior antifungal effects against strain 13139. The result on the other hand confirms that the target for these drugs is not a known mechanism. In the present study, we suspect that the target is most likely to be the enzyme Ppt2, which requires further research to confirm the exact mechanism. These findings provide new ideas for the clinical treatment of FLC-resistant C. albicans.
In order to shorten the development cycle, reduce risks and improve the success rate of new drug development, drug repurposing has received more and more attention. Drug repurposing refers to new indications or new usages of marketed drugs. Due to the known pharmacokinetics and safety data of the marketed drugs, the development of new usages can quickly come to phase II clinical evaluation for the new clinical application as soon as possible [21]. It is reported that 70–80% FDA-approved new drugs are new dosage forms and new usages of marketed drugs [22]. With the completion of the Human Genome Project and the advancement of life sciences, human medicine has entered the era of “precise medicine”. It means that various omics studies can be carried out to reveal gene regulation mechanisms, interpret the molecular mechanisms of diseases and search for various drug targets in order to provide the biological basis for drug repurposing. Of the four potential Ppt2 inhibitors screened in this study, tea polyphenol, pterostilbene and procyanidine are established clinical drugs, which bodes well for their use as potential antifungal agents in future clinical trials. Tea polyphenol, pterostilbene and procyanidine are derived from herbal medicines [23–25], and dichlorophenol is a fungicide and algaecide. Tea polyphenol has been found to possess hypolipidemic, antioxidative and antitumor activities [23]. In the previous study, epigallocatechin gallate (main active ingredient of tea polyphenol) was reported to inhibit C. albicans by affecting biofilm formation [26]. However, another study showed that theaflavins in tea polyphenol inhibited Candida species by affecting the cell wall [27]. The exact mechanism of the inhibitory effects on Candida species is still not very clear. We suspect that it may be related to enzyme Ppt2, but this also needs further research to confirm. Pterostilbene is a natural phenolic compound found in grapes and other plants that has extremely strong antifungal activity. It has been reported that pterostilbene could treat cardiovascular diseases by altering some genes in the MAPK pathway [24]. However, no accurate mechanism for the antifungal activity of pterostilbene has been found. Procyanidine exhibits anticancer activity and induces cardiovascular protection [25, 28]. To date, almost no studies have confirmed the relationship between procyanidine and Candida species. In the present study, these four compounds were all found to have a certain inhibitory effect on clinical Candida species; thereinto, tea polyphenol and dichlorophen were previously found to have antifungal effects, but the specific mechanism remains unclear. Our experiment results can provide some new ideas for the study of the target mechanism of these four compounds, but it is still necessary to further verify whether the target of these compounds is indeed Ppt2 through gene knockout techniques and in vivo experiments.
Conclusion
In this study, we improved an FP-based assay on Dobb’s documented methods from the aspects of CoA-BTMR dilution, pH, temperature, timecourse and PPT2 titration, making it more suitable for high-throughput screening of potential Ppt2p inhibitors for use as antifungals. We then screened 110 FDA-approved drugs and measured minimum inhibitory concentration values for positive compounds in vitro using broth dilution assays. The results illustrated that 20 ng/μL PPT2, 1:2000 dilution CoA-BTMR, pH = 8.0 and 30 °C are more suitable for high-throughput screening of subsequent compounds. The FP assay showed that policresulen (CAS 9011-02-3/101418-00-2), procyanidine (CAS 4852-22-6), dichlorophen (CAS 97-23-4), tea polyphenol (CAS 84650-60-2) and rutin (CAS 153-18-4) had better inhibitory effects. Further, broth microdilution assays demonstrated that pterostilbene, procyanidine, dichlorophen and tea polyphenol gave the better MIC values in vitro. It is worth mentioning that these four drugs especially had remarkable antifungal effects against FLC-resistant C. albicans, even the MIC value of procyanidine could reach below 10 mg/L. These results will provide the potential new ideas for clinical treatment of FLC-resistant Candida species. More importantly, pterostilbene, procyanidine and tea polyphenol are all marketed drugs, which bodes well for their use as potential antifungal agents in future clinical trials.
Materials and methods
Strains and growth media
Strains used in the study are listed in Table 1. Eight Candida spp. isolates were collected from Rujin Hospital in Shanghai. Three fluconazole (FLC)-resistant C. albicans isolates, two FLC-susceptible C. albicans isolates, one FLC-susceptible C. tropicalis isolate and one FLC-susceptible C. glabrata isolate were also included. All the isolates were cultured on chromogenic medium (Chromagar, Paris, France) for identification. Candida albicans ATCC 90028 was added as the control. All the strains were stored at − 80 °C in 15% glycerol and grown on YPD medium at 30 °C. A total of 1.5% agar was added for the solid medium. Escherichia coli DH5 cells were used for the multiplying of plasmids, and the DMT chemically competent cells (TransGen Biotech, Beijing, China) were used for ligation and transformation. All the strains were cultured in Luria-Bertani (LB) broth or on LB medium which was added with 100 μg/mL ampicillin or kanamycin as required.
Protein expression and purification
The genomic DNA of C. albicans BWP17 was extracted and used as the template for amplification of PPT2 genes. PPT2F and PPT2R were used for the amplification of PPT2; and pET43.1F and pET43.1R were used for linearisation of pET43.1 (all primer sequences are included in Table S1). Amplification was implemented in a system of 50 μL including 5 unit/μL Pyrobest DNA polymerase (TaKaRa, Tokyo, Japan), 2.5 mM dNTPs, 0.2 mM primers, 100 ng genomic DNA or 0.1 ng plasmid DNA in 1× Pyrobest DNA polymerase buffer II. Purified PPT2 PCR products were cloned into the pET43.1 vector using a homologous recombination kit. Amplification of ACP1 failed; hence, the gene synthesis method was used and ACP1 was cloned into the pET30 vector using NdeI and HindIII restriction enzymes. A His-tag was also added to the C terminus for subsequent protein purification. Recombinant E. coli BL21 (DE3) cells (Miaolingbio, Beijing, China) were grown on LB medium supplemented with 100 μg/mL ampicillin (pET43.1) or 100 μg/mL kanamycin (pET30) at 37 °C. When the optical density at 600 nm (OD600) was 0.8, 0.1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) was added and cells were incubated at 18 °C overnight. Cells were then harvested by centrifugation at 10,000g for 10 min and resuspended in buffer consisting of 20 mM phosphate buffer and 500 mM NaCl (Ppt2 resuspension buffer pH = 8.0, Acp1 resuspension buffer pH = 7.4). Resuspended cells were lysed by ultrasonication and centrifuged at 12,000 rpm for 30 min, and the supernatant was applied to an AKTA prime system equipped with a chelating SFF (Ni) column for the protein purification. Protein concentration was determined by the Bradford assay.
Fluorescence polarization assay
The fluorescence polarization assay in the present study was based on the documented methods and further optimized the conditions for the better drug screening [12].
4’PPT subunit of coenzyme A (CoA) was labelled with the fluorescent dye BODIPY TMR maleimide (Life Technologies, MA, USA) as described in the previous document to monitor the transfer of the phosphopantetheine group from CoA to Acp1 [29]. The initial components were 2–8 ng/μL Ppt2, 30 ng/μL Acp1 and 1:25 CoA-BTMR with assay buffer comprising 62.5 mM TRIS-HCl and 12.5 mM MgCl2 (pH 6.75) [12]. Reagents were placed in the wells of a 384-well black plate and incubated at room temperature for 85 min and then EDTA was added to stop the reaction. FP was measured using an Envision multilabel plate reader using a 531-nm excitation filter and a 579-nm emission filter. In order to identify optimal reaction conditions, we respectively varied CoA-BTMR dilution (1:1000; 1:2000; 1:4000; 1:8000), pH (pH = 6.5; pH = 7.0; pH = 7.5; pH = 8.0), temperature (room temperature and 30 °C), Ppt2 titration (2.5 ng/μL; 5 ng/μL; 10 ng/μL; 20 ng/μL; 40 ng/μL) and duration (30 min; 60 min; 90 min) in every assay.
Drug screening assay
After optimizing the FP assay, we used it to screen 110 FDA-approved drugs from the pharmacy of Shanghai Key Laboratory of New Drug Design, School of Pharmacy, East China University of Science and Technology. Compounds in final concentrations of 100 μM were prepared using ECHO (Labcyte, CA, USA) with three replicates each (Fig. S1A). Next, CoA-BTMR, Ppt2 and Acp1 reagents were mixed (5 μL each) under optimal conditions in 384-well black plates and incubated at room temperature for 60 min. For min controls, 5 μL of reaction buffer was used instead of Ppt2. The reaction was stopped by an addition of EDTA to 20 mM, and FP values were tested. The inhibition rate of each compound was calculated from the max and min fluorescence signal values of each plate, where max is the highest value with Ppt2 and min is the lowest value without Ppt2. The inhibition rate (%) was calculated from the following equation: inhibition% = 100−(FPcompound−FPmin)/(FPmax−FPmin) × 100.
We also determined IC50 values during preliminary screening of 110 compounds with inhibition rates > 50%. The final concentrations were 300 μM when the inhibition rate was > 50%, while the final concentrations were 100 μM when the inhibition rate was > 80%. We used a Bravo automated liquid handling platform (Agilent, CA, USA) to dilute the compounds to 11 different concentrations with DMSO (Fig. S1B), and to dispense them (in duplicate) into 384-well black plates. Finally, the Graphpad prism software (http://www.graphpad.com) was used to calculate the IC50 value of each compound.
Broth microdilution assay
Broth microdilution assays were carried out based on the protocol (M27-A3) of the Clinical and Laboratory Standards Institute (CLSI) in triplicate for each compound. The compound was serially diluted twofold to acquire a range of concentrations from 128 to 0.25 mg/L. Next, 100 μL of each solution was added to a 384-well microplate. Cells of different C. albicans isolates were obtained from YPD cultures and resuspended in the saline. The cell density was modulated from 1 × 106 to 5 × 106 cells/mL with spectrophotometry. Then, 100 μL of the dilution was added to the 384-well microplate containing compounds with an inhibition rate of > 50% as determined above. The 384-well microplate was incubated at 37 °C for 48 h and then OD600 values were measured. MIC was defined as the lowest antifungal concentration which inhibited strain growth by 50% versus controls.
Electronic supplementary material
(DOCX 698 kb)
Acknowledgements
The authors thank Shanghai Key Laboratory of New Drug Design, School of Pharmacy, East China University of Science and Technology for kindly providing the clinical drugs. The authors also thank Jiangye Chen of State Key Laboratory of Molecular Biology, Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences for providing strain BWP17.
Funding information
This work was financed by grants from the Program of the National Nature Science Foundation of China (#81871706#), the Program of Shanghai Key Specialty (#ZK2012A21) and the Program of Shanghai Municipal Health and Planning Committee (#201740069) and (#201840227).
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Shuai-Shuai Ni, Email: nss1106@126.com.
Ming-Jie Xiang, Email: mjxiang123456@126.com.
References
- 1.Lionakis MS. New insights into innate immune control of systemic candidiasis. Med Mycol. 2014;52:555–564. doi: 10.1093/mmy/myu029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown GD, Denning DW, Gow NA, et al. Hidden killers: human fungal infections. Sci Transl Med. 2012;4:165rv113. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 3.Smeekens SP, van de Veerdonk FL, Kullberg BJ, Netea MG. Genetic susceptibility to Candida infections. EMBO Mol Med. 2013;5:805–813. doi: 10.1002/emmm.201201678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding X, Yan D, Sun W, Zeng Z, Su R, Su J. Epidemiology and risk factors for nosocomial non-Candida albicans candidemia in adult patients at a tertiary care hospital in North China. Med Mycol. 2015;53:684–690. doi: 10.1093/mmy/myv060. [DOI] [PubMed] [Google Scholar]
- 5.Wu JQ, Zhu LP, Ou XT, Xu B, Hu XP, Wang X, Weng XH. Epidemiology and risk factors for non-Candida albicans candidemia in non-neutropenic patients at a Chinese teaching hospital. Med Mycol. 2011;49:552–555. doi: 10.3109/13693786.2010.541948. [DOI] [PubMed] [Google Scholar]
- 6.Bayhan GI, Garipardic M, Karaman K, Akbayram S. Voriconazole-associated visual disturbances and hallucinations. Cutan Ocul Toxicol. 2016;35:80–82. doi: 10.3109/15569527.2015.1020544. [DOI] [PubMed] [Google Scholar]
- 7.Vollenbroich R, Maeder MT, Weilenmann D. Congestive heart failure related to antifungal therapy with itraconazole. Int J Cardiol. 2014;172:e170–e171. doi: 10.1016/j.ijcard.2013.12.057. [DOI] [PubMed] [Google Scholar]
- 8.Arendrup MC, Fuursted K, Gahrn-Hansen B et al (2008) Semi-national surveillance of fungaemia in Denmark 2004-2006: increasing incidence of fungaemia and numbers of isolates with reduced azole susceptibility. Clin Microbiol Infect 14:487–494 [DOI] [PubMed]
- 9.Wang H, Xiao M, Chen SC, et al. In vitro susceptibilities of yeast species to fluconazole and voriconazole as determined by the 2010 National China Hospital Invasive Fungal Surveillance Net (CHIF-NET) study. J Clin Microbiol. 2012;50:3952–3959. doi: 10.1128/JCM.01130-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crawford JM, Vagstad AL, Ehrlich KC, Udwary DW, Townsend CA. Acyl-carrier protein-phosphopantetheinyltransferase partnerships in fungal fatty acid synthases. Chembiochem. 2008;9:1559–1563. doi: 10.1002/cbic.200700659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allen G, Bromley M, Kaye SJ, Keszenman-Pereyra D, Zucchi TD, Price J, Birch M, Oliver JD, Turner G. Functional analysis of a mitochondrial phosphopantetheinyl transferase (PPTase) gene pptB in Aspergillus fumigatus. Fungal Genet Biol. 2011;48:456–464. doi: 10.1016/j.fgb.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 12.Dobb KS, Kaye SJ, Beckmann N, Thain JL, Stateva L, Birch M, Oliver JD. Characterisation of the Candida albicans phosphopantetheinyl transferase Ppt2 as a potential antifungal drug target. PLoS One. 2015;10:e0143770. doi: 10.1371/journal.pone.0143770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ashburn TT, Thor KB. Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov. 2004;3:673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 14.Chong CR, Sullivan DJ., Jr New uses for old drugs. Nature. 2007;448:645–646. doi: 10.1038/448645a. [DOI] [PubMed] [Google Scholar]
- 15.Zhang JH, Chung TDY, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 16.Glockner A, Cornely OA. Candida glabrata-unique features and challenges in the clinical management of invasive infections. Mycoses. 2015;58:445–450. doi: 10.1111/myc.12348. [DOI] [PubMed] [Google Scholar]
- 17.Phuengkham H, Teeranachaideekul V, Chulasiri M, Nasongkla N. Preparation and optimization of chlorophene-loaded nanospheres as controlled release antimicrobial delivery systems. Pharm Dev Technol. 2016;21:8–13. doi: 10.3109/10837450.2014.959180. [DOI] [PubMed] [Google Scholar]
- 18.St Georgiev V. Membrane transporters and antifungal drug resistance. Curr Drug Targets. 2000;1:261–284. doi: 10.2174/1389450003349209. [DOI] [PubMed] [Google Scholar]
- 19.Sanglard D, Ischer F, Koymans L, et al. Amino acid substitutions in the cytochrome P-450 lanosterol 14alpha-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob Agents Chemother. 1998;42:241–253. doi: 10.1128/AAC.42.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiang MJ, Liu JY, Ni PH, Wang S, Shi C, Wei B, Ni YX, Ge HL. Erg11 mutations associated with azole resistance in clinical isolates of Candida albicans. FEMS Yeast Res. 2013;13:386–393. doi: 10.1111/1567-1364.12042. [DOI] [PubMed] [Google Scholar]
- 21.Boucher HW, Talbot GH, Benjamin DK, Bradley J, Guidos RJ, Jones RN, Murray BE, Bonomo RA, Gilbert D, for the Infectious Diseases Society of America Infectious Diseases Society of A: 10 x ‘20 progress-development of new drugs active against gram-negative bacilli: an update from the Infectious Diseases Society of America. Clin Infect Dis. 2013;56:1685–1694. doi: 10.1093/cid/cit152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thangamani S, Mohammad H, Younis W, Seleem M. Drug repurposing for the treatment of staphylococcal infections. Curr Pharm Des. 2015;21:2089–2100. doi: 10.2174/1381612821666150310104416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yin H, Deng Y, Wang H, Liu W, Zhuang X, Chu W. Tea polyphenols as an antivirulence compound disrupt quorum-sensing regulated pathogenicity of Pseudomonas aeruginosa. Sci Rep. 2015;5:16158. doi: 10.1038/srep16158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin HC, Hsieh MJ, Peng CH, Yang SF, Huang CN. Pterostilbene inhibits vascular smooth muscle cells migration and matrix metalloproteinase-2 through modulation of MAPK pathway. J Food Sci. 2015;80:H2331–H2335. doi: 10.1111/1750-3841.13002. [DOI] [PubMed] [Google Scholar]
- 25.Huynh HT, Teel RW. Selective induction of apoptosis in human mammary cancer cells (MCF-7) by pycnogenol. Anticancer Res. 2000;20:2417–2420. [PubMed] [Google Scholar]
- 26.Evensen NA, Braun PC. The effects of tea polyphenols on Candida albicans: inhibition of biofilm formation and proteasome inactivation. Can J Microbiol. 2009;55:1033–1039. doi: 10.1139/W09-058. [DOI] [PubMed] [Google Scholar]
- 27.Sitheeque MA, Panagoda GJ, Yau J, et al. Antifungal activity of black tea polyphenols (catechins and theaflavins) against Candida species. Chemotherapy. 2009;55:189–196. doi: 10.1159/000216836. [DOI] [PubMed] [Google Scholar]
- 28.Sano A, Uchida R, Saito M, et al. Beneficial effects of grape seed extract on malondialdehyde-modified LDL. J Nutr Sci Vitaminol (Tokyo) 2007;53:174–182. doi: 10.3177/jnsv.53.174. [DOI] [PubMed] [Google Scholar]
- 29.La Clair JJ, Foley TL, Schegg TR, et al. Manipulation of carrier proteins in antibiotic biosynthesis. Chem Biol. 2004;11:195–201. doi: 10.1016/j.chembiol.2004.02.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 698 kb)