Abstract
This study aimed to evaluate virulence factors and genetic markers of antimicrobial resistance in 400 Staphylococcus aureus strains isolated from bovine mastitis in four Brazilian states, as well as to assess the association between these characteristics and field information. Virulence factors and drug resistance genes were identified by PCR screening. Biofilm-forming and hemolytic phenotype were detected using Congo red Tryptic Soy Broth and defibrinated sheep blood agar, respectively. Of all isolates, 83.5% were biofilm-forming and 98.5% strains exhibited biofilm gene icaAD, and a significant association between phenotype and genotype for biofilm was observed (P = 0.0005). Hemolysin genes were observed in 82.85% (hla+hlb+), 16.5% (hla+) and 0.75% (hlb+) isolates, whereas the hemolytic phenotype exhibited was complete and incomplete hemolysis in 64.25%, complete in 28.25%, incomplete in 4.75%, and negative in 2.75% of the strains. Virulence factors genes luk, seb, sec, sed, and tst were observed in 3.5%, 0.5%, 1%, 0.25%, and 0.74% isolates, respectively. The gene blaZ was detected in 82.03% of penicillin-resistant isolates, whereas tetK and aac(6′)-Ie–aph(2′)-Ia were observed in 33.87% and 45.15% of the tetracycline and aminoglycosides-resistant isolates, respectively. Fluoroquinolone resistance gene mepA was detected for the first time in S. aureus from bovine mastitis. Resistance genes tetM (3.22%), tetL (1.61%), ermA (14.29%), ermB (14.29%), ermC (33.3%), ermT (9.52%), ermY (4.76%), msrA (9.52%), and mphC (9.52%) were also detected among resistant isolates. No association between virulence factors or antimicrobial-resistant genes and year of isolation, geographic origin, or antimicrobial resistance profile was observed. Our results showed that S. aureus strains isolated from bovine mastitis in the four Brazilian states sampled are mainly biofilm-forming and hemolytic, whereas virulence genes associated with enterotoxins, luk and tst, were less frequently observed. Moreover, a wide variety of resistance genes that confer resistance to almost all classes of antimicrobial agents approved for use in animals and humans were found. Overall, the data point to a great pathogenic potential of S. aureus associated with bovine mastitis and to the non-negligible risks to public health of staphylococcal infections from animal origin.
Electronic supplementary material
The online version of this article (10.1007/s42770-020-00363-5) contains supplementary material, which is available to authorized users.
Keywords: Staphylococci, Intramammary infection, Hemolysins, Biofilm, mepA
Introduction
Bovine mastitis is one of the most common diseases that affects the world dairy production, being responsible for decreasing quantity and quality of milk produced [1]. Staphylococcus aureus is one of the main pathogens isolated from bovine mastitis in different parts of the world where dairy farming is expressive [1]. Indeed, studies conducted in countries from Europe, Africa, and America have reported a high prevalence of S. aureus in dairy cows reaching 41% in France (1995–2012), 47.2% in herds from Italy (2012–2013) [2], 74% in Ethiopia (2014–2015) [3], and Canada (2003–2005) [4]. In Brazil, several studies have showed the importance of S. aureus in the epidemiology of mastitis in cattle [5–8]. Although it can be involved in clinical mastitis (especially after calving), the infection is usually subclinical, causing no visible changes in milk or udder [1].
The S. aureus’ ability to cause infections are related to the expression of various virulence factors, which are structures, products, or mechanisms frequently acquired by mobile genetic elements [9–11]. It is well documented that S. aureus strains from bovine mastitis can produce a wide variety of extracellular toxins, such as enterotoxins, encoded mainly by sea, seb, sec, sed, and see genes, toxic shock syndrome toxin 1 (tst), Panton–Valentine leukocidin (PVL) (luk), alpha, and beta hemolysins (hla and hlb), among others [10, 12, 13]. The ability to produce toxins by S. aureus strains from animal origin is not only of animal health but also of public health concern, since some of these toxins, such as enterotoxins, hemolysins and toxic shock syndrome toxin 1, in addition to favoring the infection, are also thermostable and remain active even after the thermal treatments used in milk [14]. Furthermore, the production of extracellular polymeric substances (EPS), mainly exopolysaccharide, appears to play a crucial role in the infection, adhesion, and colonization of the mammary glandular epithelium [15]. Thus, formation of biofilm also ensures the successful colonization and maintenance of S. aureus in the host tissues.
In addition to being an important factor for the colonization, biofilm formation by S. aureus strains isolated from bovine mastitis, which expresses the icaA and icaD genes, may be associated with antimicrobial resistance [15]. The mechanisms responsible for drug resistance include physical and chemical diffusion barriers formed by the exopolysaccharide matrix, which makes the penetration of antimicrobials difficult, besides creating microenvironments that antagonize the drug [5]. Antimicrobial resistance is a major problem in animal and public health, and an alarming increase in drug resistance among S. aureus strains has been reported worldwide [16–18]. Antimicrobial resistance genes (ARG) commonly reported in Staphylococcus spp. isolated from cattle are mainly mecA and blaZ (β-lactam resistance) [18, 19]; tetK, tetL, and tetM, (tetracycline resistance) [20]; ermA, ermB, ermC, ermT, ermY, msrA, and mphC [macrolide, lincosamide, streptogramin B (MLSB), and macrolide phosphotransferase resistance] [17, 21]; aac(6′)-Ie–aph(2′)-Ia [aminoglycoside modifying enzyme (AME)] [17]; and mepA, grlA/grlB, and gyrA/gyrB (fluoroquinolone resistance) [22]. Staphylococci carrying such resistance genes play a key role in the spread of resistance as they can easily exchange their resistance genes by horizontal transfer with staphylococci or other Gram-positive bacteria from animal or human origin, which increases the risk of antimicrobial resistance transmission from animals to humans [16, 22].
A further understanding of the potential for damage of S. aureus isolates from milk is of great importance considering its zoonotic potential, since this agent is considered as one of the main pathogens causing food poisoning [23] and the principal agent responsible for the contagious mastitis worldwide [1]. From animal and also public health’s points of view, it is essential to determine which virulence factors and, chiefly, resistance genes are carried by S. aureus isolates from a certain region. Therefore, the aims of this study were to evaluate (i) virulence factors and (ii) genetic markers of drug resistance of S. aureus strains isolated from bovine mastitis in four Brazilian states (Minas Gerais, São Paulo, Rio de Janeiro, and Goiás), as well as (iii) the temporal and spatial association of these characteristics.
Material and methods
Bacterial strains and culture conditions
A total of 400 S. aureus strains isolated from cows with mastitis were used in the present study. These are representative S. aureus strains from the Collection of Microorganisms of Agribusiness Interest from Empresa Brasileira de Pesquisa Agropecuária (Embrapa) Gado de Leite, isolated between 1994 and 2016 from different Brazilian states. Embrapa is a Brazilian reference center for bovine mastitis research, and its collection comprises S. aureus strains from great temporal and geographic distributions, covering the main milk-producing regions in Brazil. The isolates were randomly sampled from the 1168 S. aureus strains of the Embrapa’s collection to obtain about 33% of all S. aureus in the collection and a minimum of 30% of strains from each state (supplementary Table S1). In addition, a minimum of 20% of isolates was sampled per year, with the exception of 5 years (2005, 2006, 2007, 2008, and 2013) of the 22 years sampled (1994 to 2016), for which some strains did not grow on thawing. The distribution of the isolates sampled per year and state is shown in the Fig. 1a. Of the 400 strains, 31 were isolated from bulk tank milk samples, whereas the remaining (369) were obtained from individual cow’s milk samples (combined sample from all quarters or from individual mammary quarters). The bacteriological analysis was carried out by culturing 10 μL of milk on 5% defibrinated sheep blood agar at 37 °C for 24 to 48 h. Colonies were identified by Gram test, morphology, size, pigmentation, and biochemical tests [24].
Fig. 1.
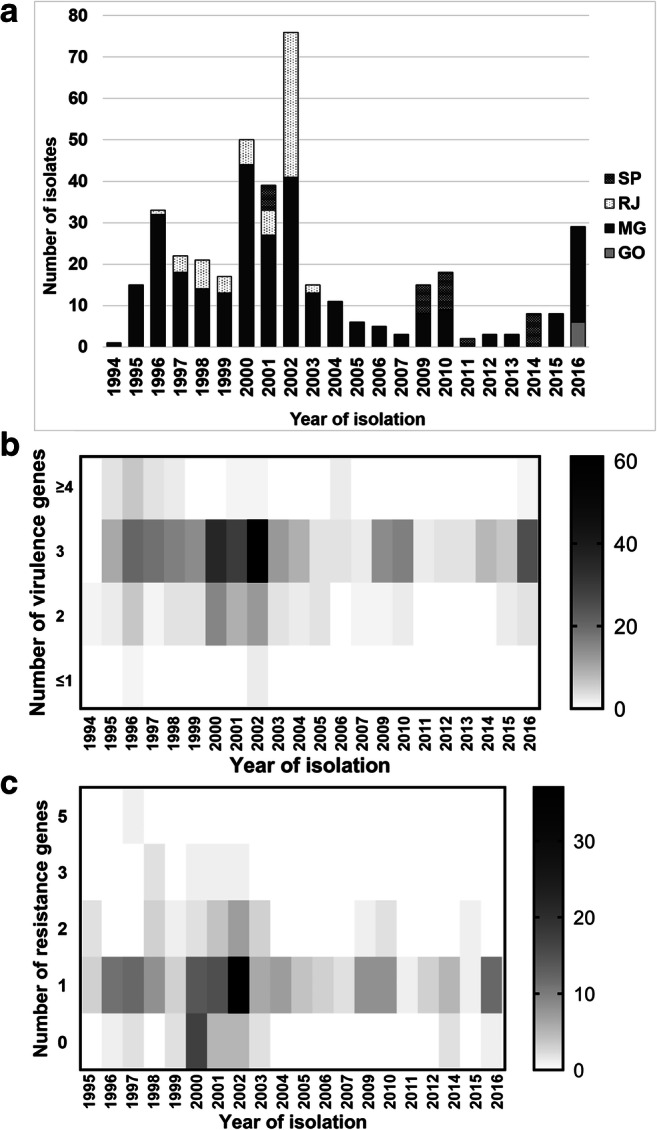
Distribution, virulence, and antimicrobial-resistant profile of Staphylococcus aureus isolated from bovine mastitis in Brazil, 1994–2016. a Distribution of S. aureus according to the year of isolation and Brazilian state (SP São Paulo, RJ Rio de Janeiro, MG Minas Gerais, and GO Goiás). b Distribution of the number of virulence genes among S. aureus isolated from bovine mastitis in São Paulo, Rio de Janeiro, Minas Gerais, and Goiás states, Brazil, 1994–2016, according to the year of isolation. c Distribution of the number of antimicrobial resistance genes among S. aureus isolated from bovine mastitis in São Paulo, Rio de Janeiro, Minas Gerais, and Goiás states, Brazil, 1994–2016, according to the year of isolation
The antimicrobial susceptibility profile of all studied strains was carried out previously by Aizawa et al. [25] using agar disk diffusion method and the following antimicrobial disks (Sensifar®, Brazil): cefoxitin (30 μg), oxacillin (1 μg), ampicillin (10 μg), enrofloxacin (5 μg), ciprofloxacin (5 μg), cephalothin (30 μg), ceftiofur (30 μg), amoxicillin + clavulanic acid (20/10 μg), erythromycin (15 μg), neomycin (30 μg), gentamicin (10 μg), tetracycline (30 μg), sulfamethoxazole + trimethoprim (1.25/23.75 μg), and penicillin-novobiocin (10UI/30 μg). Only isolates that exhibited a resistance phenotype to any of the antimicrobials previously tested were evaluated for the presence of drug resistance genes, according to the profile observed. The search for genes associated with antimicrobial resistance was restricted to strains that showed resistant phenotype to allow more robust inferences about the pathogenic potential of S. aureus strains assessed. The search for genes associated with virulence was carried out in all isolates.
The isolates were cultured on Brain Heart Infusion (BHI) agar (Merck, Germany) plates, at 37 °C, for 24 h, in aerobic conditions. All strains were stored in BHI broth (Merck, Germany) supplemented with 20% glycerol (Sigma-Aldrich, USA) at − 80 °C.
DNA extraction
After growth on BHI plates, all strains were suspended in phosphate-buffered saline (PBS) (0.01 M, pH 7.4, all reagents from Merck, Germany) and centrifuged, and the pellet was submitted to genomic DNA extraction with guanidium thiocyanate (Merck, Germany), according to Pitcher et al. [26]. The quantity and quality of DNA extracted were assessed by spectrophotometry using the NanoVue™ spectrophotometer (GE Healthcare, USA). DNA samples were kept at − 20 °C until the analysis.
Identification of S. aureus
In addition to phenotypic identification [24], all strains were also tested for the presence of conserved thermonuclease gene (nuc) by PCR (Table 1) to be confirmed as S. aureus [27]. Visualization of the amplified PCR products was performed in 1.0% agarose gel in tris-borate-EDTA buffer (TBE) (89 mM Tris Base, 89 mM boric acid, and 2 mM EDTA; pH 8.0; all from Sigma-Aldrich, USA) and stained with ethidium bromide (0.5 mg/mL) (Ludwig Biotecnologia Ltda, Brazil). Following electrophoresis, the gels were visualized under ultraviolet light and photographed (L-PIX EX, Loccus Biotechnology, Brazil). The molecular weight marker 100 bp DNA ladder (Kasvi, Brazil) was used in all electrophoresis.
Table 1.
Genes investigated in Staphylococcus aureus isolated from bovine mastitis in this study by PCR
| Target | Gene | Sequence (5′ to 3′) | Amplicon size (bp)a | Positive control (ATCC)b | Reference | Modifications |
|---|---|---|---|---|---|---|
| Thermostable nuclease (S. aureus species-specific) | nuc |
F AGTTCAGCAAATGCATCACA R TAGCCAAGCCTTGACGAACT |
400 | 25,923 | [27] | Annealing at 56 °C for 30 s and extension at 72 °C for 30 s |
| Staphylococcal enterotoxin A | sea |
F GGTTATCAATGTGCGGGTGG R CGGCACTTTTTTCTCTTCGG |
102 | 13,565 | [28] | 2.5 mM MgCl2 |
| Staphylococcal enterotoxin B | seb |
F GTATGGTGGTGTAACTGAGC R CCAAATAGTGACGAGTTAGG |
164 | 14,458 | [28] | |
| Staphylococcal enterotoxin C | sec |
F AGATGAAGTAGTTGATGTGTATGG R CACACTTTTAGAATCAACCG |
451 | 19,095 | [28] | |
| Staphylococcal enterotoxin D | sed |
F CCAATAATAGGAGAAAATAAAAG R ATTGGTATTTTTTTTCGTTC |
278 | 23,235 | [28] | |
| Staphylococcal enterotoxin E | see |
F AGGTTTTTTCACAGGTCATCC R CTTTTTTTTCTTCGGTCAATC |
209 | 27,644 | [28] | |
| Resistance to methicillin | femA |
F AAAAAAGCACATAACAAGCG R GATAAAGAAGAAACCAGCAG |
132 | 25,923 | [28] | Annealing at 57 °C for 1 min and extension at 72 °C for 1 min |
| Toxic shock syndrome toxin 1 | tst |
F ACCCCTGTTCCCTTATCATC R TTTTCAGTATTTGTAACGCC |
326 | 33,586 | [28] | |
| Panton–Valentine leukocidin (PVL) | luk |
F ATCATTAGGTAAAATGTCTGGACATGATCCA R GCATCAASTGTATTGGATAGCAAAAGC |
433 | 25,923 | [29] | Annealing at 62 °C for 1 min and extension at 72 °C for 1 min |
| Alpha-hemolysin | hla |
F CTGATTACTATCCAAGAAATTCGATTG R CTTTCCAGCCTACTTTTTTATCAGT |
209 | 8096 | [10] | Unchanged |
| Beta-hemolysin | hlb |
F GTGCACTTACTGACAATAGTGC R GTTGATGAGTAGCTACCTTCAGT |
309 | 13,565 | [10] | |
| Biofilm | icaAD |
F CCTAACTAACGAAAGGTAGG R TTAGCGTTGGGTATTCCCTC |
1266 | 51,651 | [11] |
aBase pairs (bp)
bAmerican Type Culture Collection (ATCC)
Phenotypic detection of biofilm and hemolysin production
For phenotypic identification of biofilm-forming strains, four colonies of each strain were inoculated in trypticase broth (TSB) (Merck, Germany) supplemented with Congo Red (0.8 g/L) and sucrose (36 g/L) (Sigma-Aldrich, USA), and incubated for 48 h at 37 °C, as described by Lee et al. [30]. Staphylococcus aureus ATCC 51651 and S. chromogenes, isolated from bovine mastitis belonging to the Collection of Microorganisms from Laboratório de Bacteriologia, Departamento de Medicina Veterinária, Universidade Federal de Lavras, were used as positive and negative controls in all assays, respectively (supplementary Fig. S1A).
Strains were tested for hemolysis production on 5% (v/v) defibrinated sheep blood agar (Merck, Germany) cultured at 37 °C for 24 h [24]. The hemolytic phenomenon was then observed, and zones of complete hemolysis were considered due to alpha-hemolysin, whereas darkening zones surrounding the colonies (incomplete hemolysis) were regarded as beta-hemolysin zone (supplementary Fig. S1B). Staphylococcus aureus ATCC 25923 (complete hemolytic phenotype) and S. hyicus (non-hemolytic), isolated from bovine mastitis belonging to the Collection of Microorganisms from Laboratório de Bacteriologia, Departamento de Medicina Veterinária, Universidade Federal de Lavras, were used as control for comparative analyses.
Detection of virulence genes
Detection of the biofilm genes icaAD [11]; enterotoxins sea, seb, sec, sed, and see [28]; hemolysins hla and hlb [10]; toxic shock syndrome toxin (TSST-1) tst and femA [28]; and Panton–Valentine leukocidin (PVL) luk [29] was performed by PCR screening using primers and conditions stated in Table 1. Positive controls are also described in Table 1. All reagents of the PCR mix without template DNA were routinely used in each assay, as negative control. Agarose gel electrophoresis for PCR products was performed as described in Section Identification of S.aureus.
Detection of antimicrobial resistance genes
Strains described as resistant, according to the phenotype previously observed [25] were screened for the presence of resistant genes. Positive controls, primers, and PCR conditions used in all PCR assays for detection of resistance genes are summarized in Table 2. Positive controls are also described in Table 2. All reagents of the PCR mix without template DNA were routinely used in each assay, as negative control. Agarose gel electrophoresis for PCR products was performed as described in Section Identification of S.aureus.
Table 2.
Antimicrobial resistance genes investigated in resistant Staphylococcus aureus isolated from bovine mastitis in this study by PCR
| Target | Gene | Primer sequence (5′ to 3′) | Amplicon size (bp)a | Positive controlb | Reference |
|---|---|---|---|---|---|
| β-Lactam resistance | blaZ |
F CAGTTCACATGCCAAAGAG R TACACTCTTGGCGGTTTC |
772 | 60 | [31] |
| Macrolide resistance—rRNA erm methylase | erm(A) |
F TCTAAAAAGCATGTAAAAGAA R CTTCGATAGTTTATTAATATTAG |
645 | 75, 76, 78 | [32] |
| Macrolide resistance—rRNA erm methylase | erm(B) |
F GAAAAGTACTCAACCAAATA R AGTAACGGTACTTAAATTGTTTA |
639 | 76, 60 | [32] |
| Macrolide resistance—rRNA erm methylase | erm(C) |
F TCAAAACATAATATAGATAAA R GCTAATATTGTTTAAATCGTCAAT |
642 | 184, 398 | [32] |
| Macrolide/lincosamide/streptogramin B (MLSB) resistance | erm(T) |
F CCGCCATTGAAATAGATCCT R TTCTGTAGCTGTGCTTTCAAAAA |
200 | 161 | [33] |
| Macrolide resistance—rRNA erm methylase | erm(Y) |
F AGGCCCCTTTTAAAGACGAAGGCA R GGCGCGATTGTTCATTTTAAGGCCC |
320 | 60 | [33] |
| Macrolide resistance—efflux pump | msr(A) |
F GGCACAATAAGAGTGTTTAAAGG R AAGTTATATCATGAATAGATTGTCCTGTT |
940 | 60, 352 | [9] |
| Macrolide resistance—macrolide phosphotransferase | mph(C) |
F ATGACTCGACATAATGAAAT R CTACTCTTTCATACCTAACTC |
900 | 60, 352 | [31] |
| Tetracycline resistance (efflux pump) | tet(L) |
F CATTTGGTCTTATTGGATCG R ATTACACTTCCGATTTCGG |
456 | 240 | [34] |
| Tetracycline resistance (efflux pump) | tet(K) |
F TTAGGTGAAGGGTTAGGTCC R GCAAACTCATTCCAGAAGCA |
697 | 184, 82 | [34] |
| Tetracycline resistance (ribosomal protection) | tet(M) |
F GTTAAATAGTGTTCTTGGAG R CTAAGATATGGCTCTAACAA |
657 | 75, 78 | [34] |
| Aminoglycoside resistance—aminoglycoside-modifying enzyme (AME) | aac(6′ -Ie–aph(2′)-Ia |
F CAGAGCCTTGGGAAGATGAAG R CCTCGTGTAATTCATGTTCTGGC |
348 | 137, 386 | [35] |
| Fluoroquinolone resistance (efflux pump) | mepA |
F ATGTTGCTGCTGCTCTGTTC R TCAACTGTCAAACGATCACG |
718 | ATCCc 33,591 | [36] |
| Fluoroquinolone resistance (topoisomerase IV mutation) | grlA |
F TGCCAGATGTTCGTGATGGT R TGGAATGAAAGAAACTGTCTC |
339 | ATCC 33591 | [37] |
| Fluoroquinolone resistance (DNA gyrase mutation) | gyrA |
F TCGTGCATTGCCAGATGTTCG R TCGAGCAGGTAAGACTGACGG |
394 | ATCC 33591 | [37] |
aBase pairs (bp)
bStrains used as positive controls were from the collection of the Laboratório de Bacteriologia, Departamento de Medicina Veterinária, Universidade Federal de Lavras [60]
cAmerican Type Culture Collection (ATCC)
Multidrug resistance was defined as resistance to three or more antimicrobial class. The antimicrobial groups were defined according to Clinical and Laboratory Standards Institute (CLSI) M100 manual (28th ed.) [38].
Screening for mutations in gyrA and grlA genes
For ciprofloxacin- and enrofloxacin-resistant strains, PCR amplification of quinolone resistance–determining regions (QRDRs) gyrA and grlA was carried out as described above. PCR-amplified DNA was separated by agarose gel electrophoresis (described in Section Identification of S.aureus) to verify the efficiency of amplification, purified using a PCR purification kit (Invitek, USA), and sequenced using Big Dye™ 3.1 (Applied Biosystems, USA) on an ABI-3500 automatic sequencer (Applied Biosystems, USA). The sequences obtained were submitted to quality evaluation by the Phred software (reliability index > 20) [39], grouped in a consensus with the CAP3 (Sequence Assembly Program) software [40]. Amino acid changes were identified by comparison with published wild-type sequences of GrlA encoded by the grlA gene [41] and GyrA encoded by the gyrA gene [42]. The comparisons were performed with aid of the software BioEdit 7.2 [43] using the sequences of S. aureus strains MRSA252 (ATCC BAA-1720™) and RN4220 (derived from NCTC8325-4) for identification of mutations potentially associated with antimicrobial resistance.
Statistical analyses
Prevalence was obtained in cross tabulations and expressed as percentage. Associations between the variables were carried out by univariate analysis using chi-square or Fisher’s exact tests. In all cases, a P value ≤ 0.05 was defined as significant. All statistical analyses were performed using GraphPad Prism 8.1 (GraphPad Software, USA).
Results
Identification of S. aureus isolates and prevalence of biofilm production ability and biofilm-associated genes
All 400 isolates were confirmed as S. aureus by PCR amplification of gene nuc. Prevalence of biofilm formation was 83.5% (334/400) among the tested S. aureus isolates. PCR analysis for detection of the icaAD biofilm gene revealed that 98.5% (394/400) isolates harbored icaAD gene (Table 3). Interestingly, 83.25% (333/400) of the isolates that phenotypically were biofilm producers also exhibited icaAD gene; however, 15.25% (61/394) did not produce biofilm but harbored these genes. A significant association between the phenotype and genotype for biofilm production was observed (P = 0.0005). In contrast, no association between the biofilm phenotype or genotype and the other variables tested, such as year of isolation, geographic origin, and antimicrobial resistance profile, was observed.
Table 3.
Prevalence of virulence factor genes in Staphylococcus aureus strains isolated from bovine mastitis in São Paulo, Rio de Janeiro, Minas Gerais, and Goiás states, Brazil, 1994–2016, assessed by PCR
| Gene | No. of isolates | Prevalence (%) |
|---|---|---|
| Enterotoxins | ||
| sea+ | 0/400 | 0 |
| seb+ | 2/400 | 0.5 |
| sec+ | 4/400 | 1.0 |
| sed+ | 1/400 | 0.25 |
| see+ | 0/400 | 0 |
| Toxic shock syndrome toxin – 1 (TSST) | ||
| tst+ | 3/400 | 0.74 |
| Hemolysin | ||
| hla+hlb− | 66/400 | 16.5 |
| hla−hlb+ | 3/400 | 0.75 |
| hla+hlb+ | 329/400 | 82.85 |
| hla−hlb− | 2/400 | 0.5 |
| Panton–Valentine leukocidin (PVL) | ||
| luk+ | 14/400 | 3.5 |
| Biofilm | ||
| icaAD+ | 394/400 | 98.5 |
Prevalence of virulence genes and hemolytic phenotype
Table 3 summarizes the frequency of virulence genes investigated among the S. aureus strains isolated from bovine mastitis. After the icaAD gene, the most common in the studied isolates were those coding for alpha and beta hemolysins. Double hemolytic phenotype (complete and incomplete) was exhibited by 64.25% (257/400) of the strains, while 28.25% (113/400) showed only complete hemolysis, 4.75% (19/400) only incomplete hemolysis, and 2.75% (11/400) were non-hemolytic. A significant association between the phenotype and genotype for hemolysin production was observed (P < 0.05).
In order to analyze the frequency of virulence genes according to the year of isolation, years were grouped based on the cumulative distribution in percentiles according to the number of isolates (25%, 50%, and 75%) (1994–1998, 1999–2001, 2002–2004, and 2005–2016), and the strains were also classified according to the number of virulence genes found into classes as follows ≤1, 2, 3, and ≥4 genes (Fig. 1). Most of the isolates showed at least three of the virulence genes tested [77.0% (308/400)], being icaAD and hla the most prevalent among the isolates. Distribution of S. aureus strains according to the number of virulence genes exhibited and the year of isolation is shown in the Fig. 1b). No pattern was observed for the presence of virulence genes over the years among the isolates tested. In addition, no association between virulence factor genes or hemolytic phenotype and year of isolation, geographic origin or antimicrobial resistance profile was observed.
Prevalence of antimicrobial resistance genes
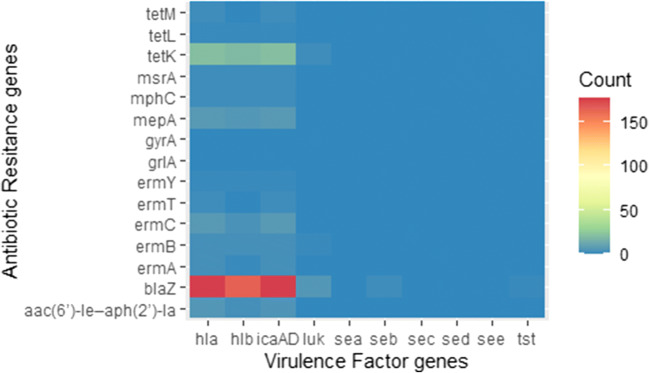
Prevalence of ARG among resistant S. aureus isolated from bovine mastitis in the four sampled Brazilian states (Minas Gerais, São Paulo, Rio de Janeiro, and Goiás) is shown in Table 4. The most frequently found genes among resistant S. aureus isolates (only strains resistant to a given class were tested for the corresponding resistance gene) were mepA (fluoroquinolone resistance), blaZ (β-lactam resistance), aac(6′)-Ie–aph(2′)-Ia (aminoglycoside resistance), tetK (tetracycline resistance), and ermC (macrolide resistance) (Table 4). Although some mutations were observed in the comparison of gyrA and grlA sequences of quinolone-resistant strains and the strain RN4220 (GenBank: AY661734.1), none of the changes has resulted in amino acid exchange and could not be associated with resistance to quinolones (supplementary Fig. S1). Thirty-two tested isolates exhibited two (n = 26), three (n = 5), and five (n = 1) ARG simultaneously (supplementary Table S2 and S3). No association between ARG and year of isolation, geographic origin, or antimicrobial resistance profile was observed.
Table 4.
Prevalence of antimicrobial-resistant genes among resistant Staphylococcus aureus isolates from bovine mastitis in São Paulo, Rio de Janeiro, Minas Gerais, and Goiás states, Brazil, 1994–2016, assessed by PCR
| Antimicrobial class | Gene | Number of resistant strainsa | Number of strains showing ARGa (%) |
|---|---|---|---|
| Penicillin | blaZ | 217 | 178 (82.03) |
| Tetracyclines | tetK | 62 | 21 (33.87) |
| tetL | 62 | 1 (1.61) | |
| tetM | 62 | 2 (3.22) | |
| Macrolides | ermA | 21 | 3 (14.29) |
| ermB | 21 | 3 (14.29) | |
| ermC | 21 | 7 (33.30) | |
| ermT | 21 | 2 (9.52) | |
| ermY | 21 | 1 (4.76) | |
| msrA | 21 | 2 (9.52) | |
| mphC | 21 | 2 (9.52) | |
| Aminoglycosides | aac(6′)-Ie-aph(2′)-Ia | 13 | 6 (45.15) |
| Quinolones | mepA | 7 | 7 (100) |
aPreviously determined by [25]
bAntimicrobial resistance genes
Association between virulence and antimicrobial resistance
Combinations for the presence of virulence genes and ARG were observed, albeit not common [2.5% (10/400)] (supplementary Table S3 and Fig. 2). Furthermore, no pattern was observed for the presence of ARG over the years among the isolates tested (Fig. 1c).
Fig. 2.
Profile of virulence genes (icaAD, hla, hlb, luk, sea, seb, sec, sed, see, and tst) and antimicrobial resistance genes [blaZ, tetK, tetl, tetM, ermA, ermB, ermC, ermT, ermY, mrsA, mphC, mepA, and aac(6′)-Ie–aph(2′)-Ia] among S. aureus isolated from bovine mastitis in São Paulo, Rio de Janeiro, Minas Gerais, and Goiás states, Brazil, 1994–2016, assessed by PCR
Discussion
In the present study, we investigated some of the major virulence factors and genetic markers of antimicrobial resistance in S. aureus strains isolated from bovine mastitis. Our results showed that Brazilian staphylococci from animal origin have a great potential to cause severe infections, since they exhibited mainly a hemolytic and biofilm forming profile, in addition to a wide variety of resistance genes. Strains were tested for the biofilm production, and the presence of icaAD gene was also investigated, as this gene is commonly detected in biofilm-forming S. aureus strains isolated from bovine mastitis. A high frequency (83.25%) of biofilm-forming strains that also harbored the icaAD gene was observed, as detected by others in studies with S. aureus isolates from bovine mastitis [44–46]. As expected, a significant association was observed between the phenotype and genotype for biofilm-forming ability. These results suggest that icaAD is a crucial gene for biofilm formation in S. aureus. However, the presence of icaAD gene in non-biofilm-forming strains (15.25%) could be explained considering that biofilm formation is regulated and influenced by the quorum sensing system [47], which can result in non-expression of the operon. On the other hand, expression of the biofilm phenotype by strains that did not carry icaAD genes, observed in low frequency, may be due to the existence of other ica-independent biofilm formation mechanisms like cell wall–anchored proteins: staphylococcal surface protein SasX and SasG, clumping factors A and B, serine-aspartate repeat protein SdrC, staphylococcal protein A, and fibronectin-binding proteins A and B [15, 48]. The ability to produce biofilm is important for bacterial pathogenesis since it allows the bacteria to form microbial communities, better able to survive the aggressions of the host and the environment, especially those from the immune system, antimicrobials, disinfectants, or even the lack of nutrients [48]. The high frequency in which the biofilm-forming phenotype/genotype was found among the S. aureus mastitis isolates tested indicates that this is a particularly important characteristic for the bovine mammary gland colonization, probably having a role in the spread of the pathogen and long-term/persistent infections.
As observed for biofilm formation, the majority of S. aureus isolates also exhibited phenotype and genotype for hemolysin production, which were also significantly associated, clearly showing the pathogenic potential of these isolates. Indeed, our findings for the presence of hemolysin genes (hla and hlb) and hemolytic phenotype revealed that a large proportion (82.85% and 64.25%) can produce alpha and beta hemolysins, which has also been reported by others [49, 50]. Therefore, the community risk associated with infections acquired through contact or consumption of animal products considering the production of alpha and beta hemolysins by S. aureus from mastitis are even higher compared to the other toxins investigated in the present study, since both, but especially beta hemolysin, have stability against inactivation at high temperatures (thermostable below 90 °C for 30 min) [49, 51]. Moreover, the interaction between alpha and beta hemolysins increase the adherence to bovine mammary epithelial cells and the proliferation of S. aureus [52], which have implications also for the pathogenesis of bovine mastitis. In fact, as well as to the biofilm-forming ability, the results of the present study on the presence of genes encoding hemolysins also suggest that these toxins may be associated with the successful infection and great adaptation of S. aureus to the bovine mammary gland. Moreover, it has been suggested that hemolysins have a role in the formation of the S. aureus biofilm, suggesting a synergistic action of these virulence mechanisms [51].
In addition to the ability to form biofilm and produce hemolysis, S. aureus can also produce a wide variety of enterotoxins, frequently involved in foodborne disease outbreaks [23]. The majority (95%) of S. aureus food poisoning cases are caused by enterotoxins sea, seb, sec, sed, and see [53]; however, in this study enterotoxins genes were observed in low frequency among the S. aureus strains (Table 33). Similarly, studies conducted in Turkey and Brazil also found low frequency or absence of the sec gene in S. aureus isolated from cattle [8, 12]. With regard to the seb and sed genes, the low frequency was also corroborated by other findings, since the frequency of these genes seems to be low in Staphylococcus spp. or were not observed [54, 55]. Likewise, in the present study, neither sea nor see genes were detected, similar to the results found by Yang et al. [49] that did not observe any sea-positive S. aureus (n = 39) strains among isolates from bovine clinical mastitis in China. These findings point to a low risk of food poisoning associated with enterotoxins encoded by sea, seb, sec, sed, and see genes among S. aureus from cattle in the four Brazilian states sampled. However, it is worth to note that these staphylococcal enterotoxins keep their biological and immunological activities even following pasteurization, food processing, and exposure to gastrointestinal proteases [23], which indicates non-negligible public health risk, although with the low frequency observed. In addition, some enterotoxins have also been associated with an important role in the pathogenesis of bovine mastitis, since they can induce bovine mammary epithelial cell (bMEC) apoptosis [54].
Likewise, the tst gene responsible for the production of TSST-1 was also detected in the present study in low frequency (0.74%) in S. aureus isolates from bovine mastitis, similar to what was observed in China (2.6%) [49]. Nonetheless, it is interesting to note that the two tst positive strains identified also exhibited the sec gene. A comparable relationship between the presence of these two genes has been reported in the literature [56]. Both toxins can exhibit various biological activities and act as superantigens for cells of the bovine immune system, contributing to pathological mechanisms of bovine mastitis [51]. Moreover, considering that Staphylococcal enterotoxins (SEs) and TSST-1 can keep their biological and immunological activities after pasteurization [14], detection of strains able to produce both toxins in cow’s milk samples also represents an important threat to public health. Although found in low frequency, the expression of these toxins could lead to episodes of food poisoning even in milk products subjected to heat treatment.
The luk gene was also exhibited by only a few isolates, which was not surprising, since in other studies the PVL gene was rarely detected in S. aureus strains isolated from bovine mastitis [13, 57]. However, as stated before for other virulence factors observed, despite of the low prevalence, PVL-positive strains from bovine milk could be a significant risk to the community, due to the pore-forming characteristic of PVL toxin, its association with necrosis of skin and soft tissues and with community-acquired methicillin-resistant S. aureus (MRSA) [13].
The present study also analyzed the distribution of ARG among resistant S. aureus isolated from bovine mastitis in the Brazilian states sampled. The blaZ gene, screened among β-lactam–resistant strains (ampicillin, amoxicillin + clavulanic acid, penicillin-novobiocin, ceftiofur), was the main genetic marker of resistance for this antimicrobial class. The assessment of these ARG helps to determine strategies for treatment, control and prevention of dissemination of resistance between animals and humans, especially MRSA (mecA-positive strains), since the resistance to β-lactams are principally conferred by mecA and blaZ genes, and their presence implies resistance to almost all β-lactams agents [19]. Recently, the mecC gene has also emerged as an important mechanism of resistance to this antimicrobial group among S. aureus from mastitis [18]. Nonetheless, in contrast to blaZ, all strains were previously demonstrated negative for mecA and mecC genes [25]. Additionally, in a proportion of the isolates that exhibited resistance to β-lactams [39/217 (18%)], blaZ was also detected. Thereby, further studies are needed to identify the genetic determinant of resistance for these isolates that were phenotypically β-lactam resistant.
For tetracycline-resistant strains, by far the most frequent gene found was tetK (n = 21), followed by tetM (n = 2) and tetL (n = 1), which was also frequently observed in a study conducted by Martini et al. [20] in S. aureus isolated from bovine mastitis in Minas Gerais state, Brazil. The detection of these genes in S. aureus from animal origin is of high concern to public health, since these genes are transferred by mobile genetic elements, which may facilitate the spread of several resistance genes and consequently can lead to treatment failure in both veterinary and human medicine [16].
The investigation of the genetic determinants of resistance of other antimicrobial classes widely used for the treatment of staphylococcal infections, as macrolides, lincosamide, and streptogramin B (MLSB), revealed that most of the isolates resistant to macrolides (erythromycin) carried the gene ermC, as well as observed by Lina et al. [9] in S. aureus isolates. On the other hand, low frequency of ermA and ermB genes was observed in the present study, as it has also been reported elsewhere [17], showing that these genes are not commonly detected in S. aureus from bovine mastitis. Similarly, the other genes related to macrolides resistance (ermT, msrA, and mphC) were found in lower frequency, suggesting that these mechanisms are probably less important in the resistance to this antimicrobial class in S. aureus from animal origin. Moreover, two isolates were detected harboring msrA and mphC, of which one also carried the ermY, blaZ, and ermB genes (supplementary Tables S2 and S3). Interestingly, mphC often occurs linked to msrA, and the presence of mphC gene alone confers only low-level resistance to macrolides [21].
Although aminoglycosides are widely used for mastitis treatment in cattle [1], few studies have been focused in the identification of mechanism of resistance against this antimicrobial class in S. aureus. In the present study, the aminoglycoside resistance gene aac(6′)-Ie-aph(2′)-Ia was detected in high proportion (45.15%) of aminoglycoside-resistant S. aureus, suggesting that this gene is an important mechanism of resistance to this antimicrobial group and could be associated with mastitis treatment failure in cattle. Indeed, a similar study has also reported a high prevalence of aac(6′)-Ie-aph(2′)-Ia in S. aureus isolated from mastitis [17].
In general, two important mechanisms are responsible for resistance to fluoroquinolones among S. aureus strains. The first one is attributed to mutations occurring in the QRDR of grlA/grlB (topoisomerase IV) and gyrA/gyrB (DNA gyrase), whereas the second is mediated by drug efflux [41, 42]. Several efflux pumps have been described in S. aureus, including those encoded by norA, norB, norC, mdeA, mepA, sepA, and sdrM genes [58]. In this study, it was identified for the first time that the fluoroquinolone resistance gene mepA in all isolates of S. aureus from bovine mastitis were phenotypically resistant to ciprofloxacin or enrofloxacin. The importance of mepA detection among bovine mastitis S. aureus goes beyond resistance to fluoroquinolones, as this gene also confers resistance to a wide range of compounds, including various dyes and biocides [59], such as iodine, quaternary ammonium and chlorhexidine, widely used in post-milking teat dipping. On the other hand, mutations in QRDR associated with resistance were not observed in quinolone-resistant strains, reinforcing that mepA-encoded efflux pump was responsible for the observed phenotype.
Albeit virulence factors and the ability to resist to antimicrobial drugs in S. aureus have been demonstrated to be closely associated, since both contribute to successfully host colonization and dissemination into a population and thereby are usually synergistically regulated [51], no significative association was observed between the virulence and antimicrobial resistance genes assessed. This result can be partly explained by the low frequency observed for some genes encoding virulence factors and ARG or even due to the non-probability sampling used. However, the findings of the present study call attention to the adoption of a One Health approach to the growing challenges in human and animal health, such as staphylococcal infections, mainly related to drug resistance, one of the key priorities of this initiative. Furthermore, although no livestock-associated MRSA (LA-MRSA) has been identified among the mastitis isolates investigated, which have been mainly implicated in severe infections and deaths in the humans [16], the resistance genes observed, all located in mobile elements, point to the risk of antimicrobial resistance transfer of these genes for other Staphylococcus spp. or even for other genera of medical importance. Additionally, despite the direct comparison of the results of the present study with others also conducted in Brazil is not possible, due to the difference in the distribution of the isolates (geographical and temporal) and different methodologies used to assess virulence factors and resistance genes; other studies performed in S. aureus from different Brazilian states also point to the importance of biofilm production and presence of enterotoxins and antimicrobial resistance genes (blaZ) for the pathogenesis of mastitis, besides the potential food poisoning risk associated with dairy products [5–7].
Conclusions
Our results showed that S. aureus strains isolated from bovine mastitis in the four Brazilian states sampled carried mainly biofilm-forming and hemolytic genes, whereas virulence genes associated with enterotoxins, PVL and TSST-1 were less frequently observed. Moreover, a wide variety of resistance genes that confer resistance to almost all classes of antimicrobial agents approved for use in animals and in human population were found. Overall, the data point to a great pathogenic potential of S. aureus associated with bovine mastitis and to the non-negligible risks to public health of staphylococcal infections from animal origin.
Electronic supplementary material
(PNG 4518 kb)
(PDF 57 kb)
(DOCX 30 kb)
Acknowledgments
VCP is grateful to Capes and OAS (Organization of American States) for her fellowship. MBH and APL are thankful to CNPq for their fellowships.
Availability of data and material
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials.
Code availability
Not applicable
Funding information
This study was supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação de Amparo à Pesquisa de Minas Gerais (Fapemig), Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP 2015/10332-6), and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior.
Compliance with ethical standards
Conflicts of interest/competing interests
The authors declare that they have no conflict of interest.
Ethics approval
Not applicable.
Consent to participate
Not applicable
Consent for publication
All authors gave the consent for publication.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ruegg PL. A 100-year review: mastitis detection, management, and prevention. J Dairy Sci. 2017;100(12):10381–10397. doi: 10.3168/jds.2017-13023. [DOI] [PubMed] [Google Scholar]
- 2.Cortimiglia C, Luini M, Bianchini V, Marzagalli L, Vezzoli F, Avisani D, Bertoletti M, Ianzano A, Franco A, Battisti A. Prevalence of Staphylococcus aureus and of methicillin-resistant S. aureus clonal complexes in bulk tank milk from dairy cattle herds in Lombardy Region (Northern Italy) Epidemiol Infect. 2016;144(14):3046–3051. doi: 10.1017/S0950268816001576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abebe R, Hatiya H, Abera M, Megersa B, Asmare K. Bovine mastitis: prevalence, risk factors and isolation of Staphylococcus aureus in dairy herds at Hawassa milk shed, South Ethiopia. BMC Vet Res. 2016;12(1):270. doi: 10.1186/s12917-016-0905-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olde Riekerink RG, Barkema HW, Scholl DT, Poole DE, Kelton DF. Management practices associated with the bulk-milk prevalence of Staphylococcus aureus in Canadian dairy farms. Prev Vet Med. 2010;97(1):20–28. doi: 10.1016/j.prevetmed.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Marques VF, Motta CC, Soares BD, Melo DA, Coelho SM, Coelho ID, Barbosa HS, Souza MM. Biofilm production and beta-lactamic resistance in Brazilian Staphylococcus aureus isolates from bovine mastitis. Braz J Microbiol. 2017;48(1):118–124. doi: 10.1016/j.bjm.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silveira-Filho VM, Luz IS, Campos AP, Silva WM, Barros MP, Medeiros ES, Freitas MF, Mota RA, Sena MJ, Leal-Balbino TC. Antibiotic resistance and molecular analysis of Staphylococcus aureus isolated from cow's milk and dairy products in northeast Brazil. J Food Protect. 2014;77(4):583–591. doi: 10.4315/0362-028x.jfp-13-343. [DOI] [PubMed] [Google Scholar]
- 7.Araújo RMP, Peixoto RM, Peixoto LJS, Gouveia GV, Costa MM. Virulence factors in Staphylococcus aureus and quality of raw milk from dairy cows in a semiarid region of northeastern Brazil. Acta Sci Vet. 2017;45(1491):10.22456/1679–10.9216.80637. [Google Scholar]
- 8.Rall V, Miranda E, Castilho I, Camargo C, Langoni H, Guimarães F, Júnior JA, Júnior AF. Diversity of Staphylococcus species and prevalence of enterotoxin genes isolated from milk of healthy cows and cows with subclinical mastitis. J Dairy Sci. 2014;97(2):829–837. doi: 10.3168/jds.2013-7226. [DOI] [PubMed] [Google Scholar]
- 9.Lina G, Quaglia A, Reverdy ME, Leclercq R, Vandenesch F, Etienne J. Distribution of genes encoding resistance to macrolides, lincosamides, and streptogramins among staphylococci. Antimicrob Agents Chemother. 1999;43(5):1062–1066. doi: 10.1128/AAC.43.5.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jarraud S, Mougel C, Thioulouse J, Lina G, Meugnier H, Forey F, Nesme X, Etienne J, Vandenesch F. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect Immun. 2002;70(2):631–641. doi: 10.1128/IAI.70.2.631-641.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun F, Wang Q, Xia P. PCR analysis of clinically isolated Staphylococcus aureus biofilm associated gene. Acta Acad Med Militaris Tertiae. 2003;31(15):1147–1149. [Google Scholar]
- 12.Boynukara B, Gulhan T, Alisarli M, Gurturk K, Solmaz H. Classical enterotoxigenic characteristics of Staphylococcus aureus strains isolated from bovine subclinical mastitis in Van, Turkey. Int J Food Microbiol. 2008;125(2):209–211. doi: 10.1016/j.ijfoodmicro.2008.03.024. [DOI] [PubMed] [Google Scholar]
- 13.Shrivastava N, Sharma V, Shrivastav A, Nayak A, Rai AK. Prevalence and characterization of Panton-Valentine leukocidin-positive Staphylococcus aureus in bovine milk in Jabalpur district of Madhya Pradesh, India. Vet World. 2018;11(3):316. doi: 10.14202/vetworld.2018.316-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tam K, Torres VJ (2019) Staphylococcus aureus secreted toxins and extracellular enzymes. Microbiol Spectrum 7(2). 10.1128/microbiolspec.GPP3-0039-2018 [DOI] [PMC free article] [PubMed]
- 15.Cucarella C, Tormo MÁ, Úbeda C, Trotonda MP, Monzón M, Peris C, Amorena B, Lasa Í, Penadés JR. Role of biofilm-associated protein bap in the pathogenesis of bovine Staphylococcus aureus. Infect Immun. 2004;72(4):2177–2185. doi: 10.1128/IAI.72.4.2177-2185.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinross P, Petersen A, Skov R, Van Hauwermeiren E, Pantosti A, Laurent F, Voss A, Kluytmans J, Struelens MJ, Heuer O, Monnet DL (2017) Livestock-associated meticillin-resistant Staphylococcus aureus (MRSA) among human MRSA isolates, European Union/European economic area countries, 2013. Euro Surveill 22(44). 10.2807/1560-7917.es.2017.22.44.16-00696 [DOI] [PMC free article] [PubMed]
- 17.Qu Y, Zhao H, Nobrega DB, Cobo ER, Han B, Zhao Z, Li S, Li M, Barkema HW, Gao J. Molecular epidemiology and distribution of antimicrobial resistance genes of Staphylococcus species isolated from Chinese dairy cows with clinical mastitis. J Dairy Sci. 2019;102(2):1571–1583. doi: 10.3168/jds.2018-15136. [DOI] [PubMed] [Google Scholar]
- 18.García-Álvarez L, Holden MT, Lindsay H, Webb CR, Brown DF, Curran MD, Walpole E, Brooks K, Pickard DJ, Teale C. Methicillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect Dis. 2011;11(8):595–603. doi: 10.1016/S1473-3099(11)70126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Becker K, Larsen AR, Skov RL, Paterson GK, Holmes MA, Sabat AJ, Friedrich AW, Köck R, Peters G, Kriegeskorte A. Evaluation of a modular multiplex-PCR methicillin-resistant Staphylococcus aureus detection assay adapted for mecC detection. J Clin Microbiol. 2013;51(6):1917–1919. doi: 10.1128/JCM.00075-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martini CL, Lange CC, Brito MA, Ribeiro JB, Mendonca LC, Vaz EK. Characterisation of penicillin and tetracycline resistance in Staphylococcus aureus isolated from bovine milk samples in Minas Gerais, Brazil. J Dairy Res. 2017;84(2):202–205. doi: 10.1017/S0022029917000061. [DOI] [PubMed] [Google Scholar]
- 21.Lüthje P, Schwarz S. Antimicrobial resistance of coagulase-negative staphylococci from bovine subclinical mastitis with particular reference to macrolide–lincosamide resistance phenotypes and genotypes. J Antimicrob Chemother. 2006;57(5):966–969. doi: 10.1093/jac/dkl061. [DOI] [PubMed] [Google Scholar]
- 22.Silva NC, Guimarães FF, Manzi MP, Júnior AF, Gómez-Sanz E, Gómez P, Langoni H, Rall VL, Torres C. Methicillin-resistant Staphylococcus aureus of lineage ST398 as cause of mastitis in cows. Lett Appl Microbiol. 2014;59(6):665–669. doi: 10.1111/lam.12329. [DOI] [PubMed] [Google Scholar]
- 23.Cretenet M, Even S, Le Loir Y. Unveiling Staphylococcus aureus enterotoxin production in dairy products: a review of recent advances to face new challenges. J Dairy SciTechnol. 2011;91(2):127–150. doi: 10.1007/s13594-011-0014-9. [DOI] [Google Scholar]
- 24.Markey B, Leonard F, Archambault M, Cullinane A, Maguire D (2013) Clinical veterinary microbiology E-book. Elsevier Health Sciences
- 25.Aizawa J, Souza-Filho AF, Guimarães AS, Vasconcelos CG, Brito MAV, Sellera FP, Cortez A, Heinemann MB. Retrospective multicenter study reveals absence of MRSA-associated bovine mastitis in Brazil (1994 to 2016) J Infect Dev Countr. 2019;13(06):581–583. doi: 10.3855/jidc.11406. [DOI] [PubMed] [Google Scholar]
- 26.Pitcher DG, Saunders NA, Owen RJ. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol. 1989;8(4):151–156. doi: 10.1111/j.1472-765X.1989.tb00262.x. [DOI] [Google Scholar]
- 27.Cremonesi P, Luzzana M, Brasca M, Morandi S, Lodi R, Vimercati C, Agnellini D, Caramenti G, Moroni P, Castiglioni B. Development of a multiplex PCR assay for the identification of Staphylococcus aureus enterotoxigenic strains isolated from milk and dairy products. Mol Cell Probes. 2005;19(5):299–305. doi: 10.1016/j.mcp.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 28.Mehrotra M, Wang G, Johnson WM. Multiplex PCR for detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin 1, and methicillin resistance. J Clin Microbiol. 2000;38(3):1032–1035. doi: 10.1128/JCM.38.3.1032-1035.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lina G, Piémont Y, Godail-Gamot F, Bes M, Peter M-O, Gauduchon V, Vandenesch F, Etienne J. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999;29(5):1128–1132. doi: 10.1086/313461. [DOI] [PubMed] [Google Scholar]
- 30.Lee J-S, Bae Y-M, Han A, Lee S-Y. Development of Congo red broth method for the detection of biofilm-forming or slime-producing Staphylococcus sp. LWT Food Sci Technol. 2016;73:707–714. doi: 10.1016/j.lwt.2016.03.023. [DOI] [Google Scholar]
- 31.Schnellmann C, Gerber V, Rossano A, Jaquier V, Panchaud Y, Doherr MG, Thomann A, Straub R, Perreten V. Presence of new mecA and mph(C) variants conferring antibiotic resistance in Staphylococcus spp. isolated from the skin of horses before and after clinic admission. J Clin Microbiol. 2006;44(12):4444–4454. doi: 10.1128/jcm.00868-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sutcliffe J, Grebe T, Tait-Kamradt A, Wondrack L. Detection of erythromycin-resistant determinants by PCR. Antimicrob Agents Chemother. 1996;40(11):2562–2566. doi: 10.1128/AAC.40.11.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gomez-Sanz E, Torres C, Lozano C, Fernandez-Perez R, Aspiroz C, Ruiz-Larrea F, Zarazaga M. Detection, molecular characterization, and clonal diversity of methicillin-resistant Staphylococcus aureus CC398 and CC97 in Spanish slaughter pigs of different age groups. Foodborne Pathog Dis. 2010;7(10):1269–1277. doi: 10.1089/fpd.2010.0610. [DOI] [PubMed] [Google Scholar]
- 34.Aarestrup FM, Agersø Y, Ahrens P, Jørgensen JCØ, Madsen M, Jensen LB. Antimicrobial susceptibility and presence of resistance genes in staphylococci from poultry. Vet Microbiol. 2000;74(4):353–364. doi: 10.1016/S0378-1135(00)00197-8. [DOI] [PubMed] [Google Scholar]
- 35.Vakulenko SB, Donabedian SM, Voskresenskiy AM, Zervos MJ, Lerner SA, Chow JW. Multiplex PCR for detection of aminoglycoside resistance genes in enterococci. Antimicrob Agents Chemother. 2003;47(4):1423–1426. doi: 10.1128/AAC.47.4.1423-1426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Couto I, Costa SS, Viveiros M, Martins M, Amaral L. Efflux-mediated response of Staphylococcus aureus exposed to ethidium bromide. J Antimicrob Chemother. 2008;62(3):504–513. doi: 10.1093/jac/dkn217. [DOI] [PubMed] [Google Scholar]
- 37.Pan X-S, Hamlyn PJ, Talens-Visconti R, Alovero FL, Manzo RH, Fisher LM. Small-colony mutants of Staphylococcus aureus allow selection of gyrase-mediated resistance to dual-target fluoroquinolones. Antimicrob Agents Chemother. 2002;46(8):2498–2506. doi: 10.1128/AAC.46.8.2498-2506.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.CLSI . CLSI supplement M100. 27. Wayne: Clinical and Laboratory Standards Institute; 2017. [Google Scholar]
- 39.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I Accuracy assessment. Genome Res. 1998;8(3):175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 40.Huang X, Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9(9):868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamagishi J, Kojima T, Oyamada Y, Fujimoto K, Hattori H, Nakamura S, Inoue M. Alterations in the DNA topoisomerase IV grlA gene responsible for quinolone resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1996;40(5):1157–1163. doi: 10.1128/AAC.40.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ito H, Yoshida H, Bogaki-Shonai M, Niga T, Hattori H, Nakamura S. Quinolone resistance mutations in the DNA gyrase gyrA and gyrB genes of Staphylococcus aureus. Antimicrob Agents Chemother. 1994;38(9):2014–2023. doi: 10.1128/AAC.38.9.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 44.Castelani L, Pilon LE, Martins T, Pozzi CR, Arcaro JRP. Investigation of biofilm production and ica A and ica D genes in Staphylococcus aureus isolated from heifers and cows with mastitis. J Anim Sci. 2015;86(3):340–344. doi: 10.1111/asj.12284. [DOI] [PubMed] [Google Scholar]
- 45.Li L, Yang H, Liu D, He H, Wang C, Zhong J, Gao T, Zeng Y. Analysis of biofilm formation and associated gene detection in Staphylococcus isolates from bovine mastitis. Afr J Biotechnol. 2012;11(8):2113–2118. doi: 10.5897/AJB11.081. [DOI] [Google Scholar]
- 46.Vasudevan P, Nair MKM, Annamalai T, Venkitanarayanan KS. Phenotypic and genotypic characterization of bovine mastitis isolates of Staphylococcus aureus for biofilm formation. Vet Microbiol. 2003;92(1):179–185. doi: 10.1016/S0378-1135(02)00360-7. [DOI] [PubMed] [Google Scholar]
- 47.Tang JN, Zhou R, Wang HN, Zeng ZG. Accessory gene regulator in Staphylococcus biofilm formation and infection. J Cent South Univ. 2008;33(11):1066–1070. [PubMed] [Google Scholar]
- 48.Figueiredo AMS, Ferreira FA, Beltrame CO, Cortes MF. The role of biofilms in persistent infections and factors involved in ica-independent biofilm development and gene regulation in Staphylococcus aureus. Crit Rev Microbiol. 2017;43(5):602–620. doi: 10.1080/1040841X.2017.1282941. [DOI] [PubMed] [Google Scholar]
- 49.Yang FL, Li XS, Liang XW, Zhang XF, Qin GS, Yang BZ. Detection of virulence-associated genes in Staphylococcus aureus isolated from bovine clinical mastitis milk samples in Guangxi. Trop Anim Health Prod. 2012;44(8):1821–1826. doi: 10.1007/s11250-012-0143-z. [DOI] [PubMed] [Google Scholar]
- 50.Silva ER, Boechat JUD, Martins JCD, Ferreira WPB, Siqueira AP, da Silva N. Hemolysin production by Staphylococcus aureus species isolated from mastitic goat milk in Brazilian dairy herds. Small Ruminant Res. 2005;56(1–3):271–275. doi: 10.1016/j.smallrumres.2004.04.011. [DOI] [Google Scholar]
- 51.Pérez V, Costa G, Guimarães A, Heinemann M, Lage A, Dorneles E (2020) Relationship between virulence factors and antimicrobial resistance in Staphylococcus aureus from bovine mastitis. J Glob Antimicrob Res 22:792–802. 10.1016/j.jgar.2020.06.010 [DOI] [PubMed]
- 52.Cifrian E, Guidry A, Bramley A, Norcross N, Bastida-Corcuera F, Marquardt W. Effect of staphylococcal β toxin on the cytotoxicity, proliferation and adherence of Staphylococcus aureus to bovine mammary epithelial cells. Vet Microbiol. 1996;48(3–4):187–198. doi: 10.1016/0378-1135(95)00159-X. [DOI] [PubMed] [Google Scholar]
- 53.Hennekinne J-A, Ostyn A, Guillier F, Herbin S, Prufer A-L, Dragacci S. How should staphylococcal food poisoning outbreaks be characterized? Toxins. 2010;2(8):2106–2116. doi: 10.3390/toxins2082106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Y, Chen W, Ali T, Alkasir R, Yin J, Liu G, Han B. Staphylococcal enterotoxin H induced apoptosis of bovine mammary epithelial cells in vitro. Toxins. 2014;6(12):3552–3567. doi: 10.3390/toxins6123552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ruaro A, Andrighetto C, Torriani S, Lombardi A. Biodiversity and characterization of indigenous coagulase-negative staphylococci isolated from raw milk and cheese of North Italy. Food Microbiol. 2013;34(1):106–111. doi: 10.1016/j.fm.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 56.Fitzgerald J, Hartigan P, Meaney W, Smyth C. Molecular population and virulence factor analysis of Staphylococcus aureus from bovine intramammary infection. J Applied Microbiol. 2000;88(6):1028–1037. doi: 10.1046/j.1365-2672.2000.01071.x. [DOI] [PubMed] [Google Scholar]
- 57.Fluit A. Livestock-associated Staphylococcus aureus. Clin Microbiol Infect. 2012;18(8):735–744. doi: 10.1111/j.1469-0691.2012.03846.x. [DOI] [PubMed] [Google Scholar]
- 58.Poole K. Efflux pumps as antimicrobial resistance mechanisms. Ann Med. 2007;39(3):162–176. doi: 10.1080/07853890701195262. [DOI] [PubMed] [Google Scholar]
- 59.Correia S, Poeta P, Hébraud M, Capelo JL, Igrejas G. Mechanisms of quinolone action and resistance: Where do we stand? J Med Microbiol. 2017;66(5):551–559. doi: 10.1099/jmm.0.000475. [DOI] [PubMed] [Google Scholar]
- 60.Silva JR, Castro GAC, Gonçalves MS, Custódio DAC, Mian GF, Costa GM. In vitro antimicrobial susceptibility and genetic resistance determinants of Streptococcus agalactiae isolated from mastitic cows in Brazilian dairy herds. Semina: Ciênc Agrár. 2017;38(4):2581–2594. doi: 10.5433/1679-0359.2017v38n4Supl1p2581. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PNG 4518 kb)
(PDF 57 kb)
(DOCX 30 kb)
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials.