Abstract
A retrospective study of the epidemiology of vancomycin-resistant enterococci (VRE) in a regional hospital of central Italy in 2001–2018 demonstrated an increased VRE prevalence since 2016. A total of 113 VRE isolates, 89 E. faecium (VREfm) and 24 E. faecalis (VREfs), were collected in the study period. All strains showed high-level resistance to vancomycin; 107 also showed teicoplanin resistance. Altogether, 84 VREfm and 20 VREfs carried vanA, whereas 5 VREfm and 1 VREfs carried vanB. MLST analysis documented that 89 VREfm isolates mainly belonged to ST78, ST80, and ST117. Most strains were isolated from 2001 to 2007, ST78 being the predominant clone. VREfm re-emerged in 2016 with a prevalence of the ST80 lineage. Most VREfs were isolated from 2001 to 2006; although they belonged to 7 different STs, there was a prevalence of ST88 and ST6. Notably, ST88 was sporadically recovered throughout the study period. The increasing rate of VREfm isolation from 2016 to 2018 may be related to the influx of new successful clones and to the renewed and widespread use of vancomycin. Improved infection control measures in hospital wards should be adopted to limit the spread of new epidemic VRE strains.
Keywords: Enterococcus faecium, Enterococcus faecalis, Vancomycin resistance, vanA gene, vanB gene, MLST
Introduction
Among human gut commensals, enterococci have a special ability to colonize healthy carriers and hospital patients, to adapt to adverse environmental conditions, and to evolve and transmit antimicrobial resistance determinants. Enterococci are opportunistic pathogens commonly associated to urinary tract infections, sepsis, and infective endocarditis [1], Enterococcus faecalis and Enterococcus faecium being the most clinically relevant species. Whereas E. faecalis is the more virulent, E. faecium rapidly acquires multidrug resistance determinants, a feature that has led to the marked increase of this pathogen as a cause of human infections [1, 2]. In recent decades, the gradual evolution of hospital-associated lineages of E. faecium has made it a major nosocomial pathogen worldwide [1].
Vancomycin was largely used in the 1980s as the drug of choice to treat Clostridioides difficile enterocolitis as well as severe hospital-acquired infections caused by aminoglycoside- and ampicillin-resistant enterococci and methicillin-resistant Staphylococcus aureus (MRSA) [3]. Its heavy clinical use induced a steady increase in vancomycin-resistant enterococci (VRE) until the early 2000s. In Europe, their emergence was also ascribed to the administration of the glycopeptide avoparcin as a growth promoter in farm animals, which in the EU was banned in 1997 (Commission Directive 97/6 EC) [4]. The failure of antibiotic treatments against VRE is a serious and growing problem that is associated to increased mortality and rising hospital costs [2]. Moreover, patients infected with or colonized by VRE constitute a reservoir of resistant hospital clones [5, 6]. In hospital patients, VRE bacteremia is often due to the predominance of VRE in the gastrointestinal microbiota after vancomycin treatment [1].
In Italy, VRE recovery declined from 2003 to 2013 as a consequence of the reduced use of glycopeptides in humans and animals [7]. A decline was also detected in 2013–2016 in some European countries and in North America, likely due to regional and national recommendations to use vancomycin more sparingly and to efforts aimed at preventing the spread of MRSA and VRE infections (https://www.ecdc.europa.eu/sites/default/files/documents/AMR-surveillance-Europe-2016) [8].
Recently, VRE incidence has been showing an upward trend in several European countries, including Italy (www.ecdc.europa.eu/en/publications-data/surveillance-antimicrobial-resistance-europe-2017) [9–12]. Although nine vancomycin resistance gene clusters have been described to date, vanA and vanB (both transferable) are still those most commonly found. The VanA phenotype is characterized by high-level resistance to both vancomycin and teicoplanin, whereas the VanB phenotype is associated to resistance to vancomycin alone [13].
Vancomycin resistance has also been associated to hospital-adapted lineages of E. faecium (such as those belonging to Sequence Types ST17, ST18 and ST78) and E. faecalis (ST6). Notably, vancomycin resistance is much more common in E. faecium [8]. Horizontal propagation of mobile genetic elements plays a greater role than clonal diffusion in resistance spread [14].
A comprehensive picture of VRE epidemiology in Italy is not available. A retrospective study was conducted to characterize the VRE strains isolated at the Marche regional hospital in Ancona (central Italy) from 2001 to 2018.
Material and methods
Bacterial strains
We investigated 113 VRE, 89 E. faecium (VREfm), and 24 E. faecalis (VREfs), isolated from clinical specimens (mostly urine and blood) collected in 2001–2018 in different hospital wards of a regional teaching hospital in central Italy with almost 1000 beds. Only one isolate per patient was included in the study. Isolates were cultured on brain-heart infusion agar and VRE screening plates containing 6 mg/L vancomycin (both from Oxoid, Basingstoke, UK). Strains were identified by matrix-assisted laser desorption/ionization time-of flight mass spectrometry (MALDI-TOF/MS) (Bruker Daltonik GmbH, Bremen, Germany).
Antimicrobial susceptibility testing
Antibiotic susceptibility patterns were demonstrated by the Vitek-2 system (bioMèrieux, Marcy-l’Etoile, France). Vancomycin and teicoplanin minimal inhibitory concentrations (MICs) were determined by the agar microdilution method [15]; the results were interpreted according to the EUCAST MIC breakpoints (www.eucast.org). E. faecalis ATCC 29212 was used for quality control.
vanA and vanB gene detection by polymerase chain reaction (PCR)
Two primer pairs were used to detect the vanA (FW 5’-GGGAAAACGACAATTGC-3′/ RV 5′-GTACAATGCGGCCGTTA–3′) and vanB genes (FW 5′- ATGGGAAGCCGATAGTC-3′/RV 5′- GATTTCGTTCCTCGACC-3′) [16].
Bacterial DNA was obtained by resuspending some colonies collected from a Slanetz Bartley agar plate in 200 μl sterile distilled water and by boiling them for 10 min in a water bath. Then, 5 μl of suspension was added in a final volume of 25 μl of mastermix containing 0.2 μM of each primer for vanA and vanB, 500 mM dNTP mix, 7 mM MgCl2, and 2 U Dream Taq DNA polymerase (ThermoFisher Scientific, Waltham, MA, USA). PCR conditions were as follows: 94 °C for 3 min; 30 cycles of 94 °C for 1 min, 54 °C for 1 min, and 72 °C for 1 min; and 72 °C for 5 min. PCR was performed in a GeneAmp PCR System 9700 (Applied Biosystems System 9700 GeneAmp PCR Thermal Cycler). PCR products were resolved by electrophoresis on 1.5% agarose gel [16]. E. faecium BM4147 (vanA) and E. faecalis ATCC 51299 (vanB) were the positive controls.
Typing assays
Multilocus sequence typing (MLST) was performed as recommended in the MLST database (www.pubmlst.org).
Isolates with identical STs were considered as members of a single lineage, those that differed in only one locus were considered as single-locus variants (SLV), and those that differed in two loci were considered as double-locus variants (DLV). Using eBURST analysis, STs sharing 5 or 6 of the 7 loci were clustered into a clonal complex (CC). The goeBURST 1.2.1 algorithm was used to cluster the STs (http://www.phyloviz.net/).
Pulsed-field gel electrophoresis (PFGE) typing was performed as described previously [17]. Briefly, genomic DNA extracted from cells embedded in agarose plugs was digested with SmaI endonuclease (New England Biolabs, Beverly, MA), and the resulting fragments were separated by PFGE. DNA patterns were analyzed with BioNumerics software version 7.0 (Applied Maths Scientific Software Development, Sint-Martens-Latem, Belgium). The dendrogram was built by applying the Dice similarity coefficient, with 1.5% optimization and 2.0% tolerance. Clustering was obtained using the unweighted pair group method with arithmetic mean. Strains showing a pattern similarity > 80% were considered closely related and grouped into clusters.
Results and discussion
We investigated 113 VRE, 89 VREfm, and 24 VREfs, isolated in 2001–2018 in a regional hospital in central Italy for their vancomycin and teicoplanin resistance genotype and phenotype and clonal relatedness. All isolates showed high-level resistance to vancomycin (MIC range, 32; > 256 mg/L) and 107 also to teicoplanin (MIC range, 4; > 256 mg/L). The remaining 6 strains (3 VREfm and 3 VREfs) showed teicoplanin MICs < 0.5 mg/L.
PCR screening demonstrated vanA in 92% of isolates (84 VREfm and 20 VREfs) and vanB in 5.3% (5 VREfm and 1 VREfs). The remaining 3 (2.7%) isolates (all VREfs) were negative for both determinants. Although 9 van operons related to vancomycin resistance have been described, the vanA and vanB gene clusters are still prevalent among clinical VRE isolates, with varying frequencies in different countries [13]. In recent surveys, a higher proportion of VREfm and VREfs showing the VanA phenotype has been described in Europe and North America, whereas in Australia vanB-positive VREfm strains were involved in more than 50% of bacteremias caused by VRE in 2015 [8, 18].
In this study, vanA was the most frequent vancomycin resistance determinant among VREfm and VREfs, in line with previous Italian studies [7, 19].
MLST analysis assigned the 89 VREfm isolates to 9 STs (Fig. 1a). The most common were ST78 (54%), ST80 (15%), and ST117 (10%) and were followed by ST1590 and ST1115 (n = 4), ST17 (n = 3), ST18 and ST1666 (n = 2), and ST145, ST202, and ST1626 (n = 1). Interestingly, more that 60% of the VREfm isolates belonged to ST17, ST18, and ST78, which are well-known members of CC17, one of the most widespread nosocomial clones of E. faecium. The vanA strains were distributed in all STs, whereas vanB isolates were found only in ST18, ST117, and ST80. eBURST analysis grouped all STs in a single CC, since each ST was a SLV (ST17, ST18, ST117, ST145, ST1115, ST1590, ST1626, and ST1666) or a DLV (ST80 and ST202) of the putative founder ST78.
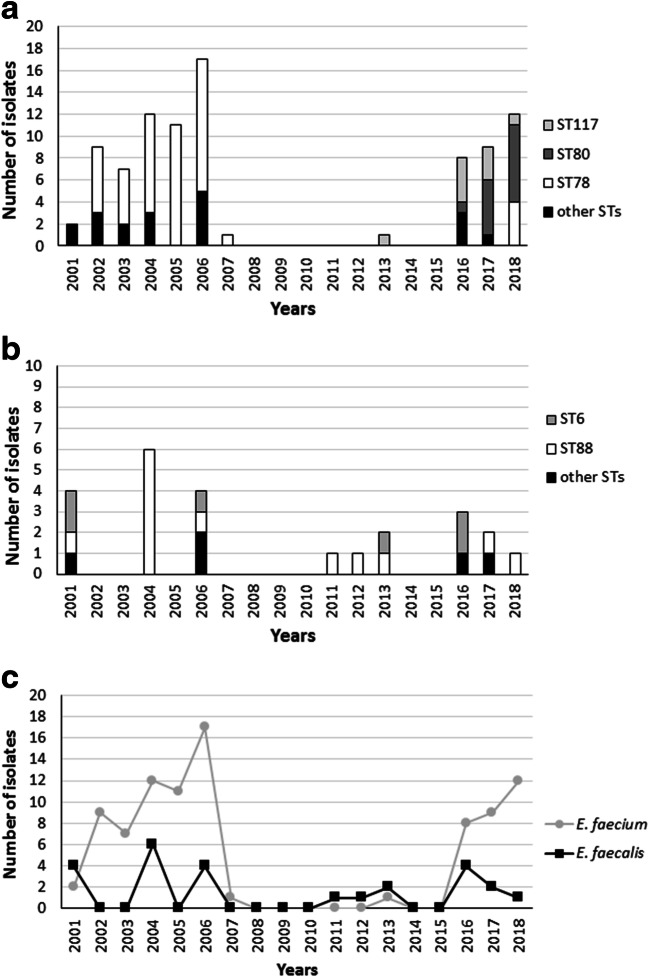
Fig. 1.
ST distribution of VREfm isolates from 2001 to 2018 (a). ST distribution of VREfs isolates from 2001 to 2018 (b). Total distribution of VRE isolates from 2001 to 2018 (c)
The analysis of VREfm distribution revealed that most (n = 59) were isolated from 2001 to 2007, the largest number (n = 17) being isolated in 2006, and that ST78 (accounting for 74% of the isolates) was predominant throughout this period. No VREfm was recovered in 2008–2015 with the exception of a single ST117 strain isolated in 2013. VREfm re-emerged in 2016 (n = 8) and was also isolated in 2017 (n = 9) and 2018 (n = 12). The most common STs were ST117 in 2016 and ST80 in 2017 and 2018. Although ST78 was again recovered in 2018, ST80 is currently the dominant lineage in the hospital (Fig. 1c and Fig. 2a). The replacement of ST78 by ST117 and ST80 may be related to a greater adaptability of these new VRE clones to the clinical setting.
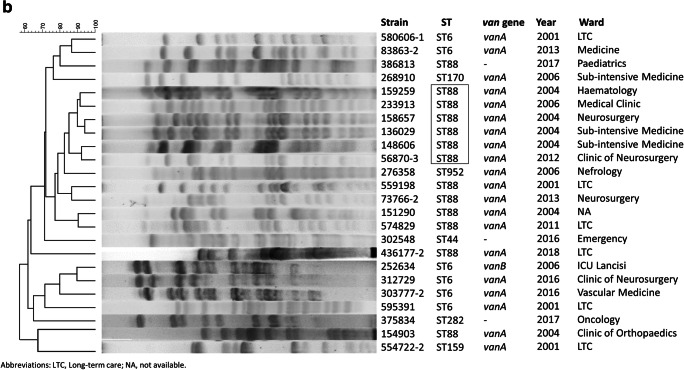
Fig. 2.
SmaI-PFGE pattern and dendrogram of VREfm (a) and VREfs isolates (b). The main pulsotypes are highlighted in a square box
The ST78 strains circulating until 2006 clustered in two main, closely related pulsotypes and were collected in different wards, whereas a new ST78 clonal group with a different pulsotype (similarity < 80%) was established in 2018. The strains isolated in 2017–2018 belonging to ST117 and ST80 show no clonal relationship, as documented by their pulsotypes (Fig. 2a).
The 24 VREfs belong to 7 STs (Fig. 1b), which according to eBURST analysis were unrelated. The most represented was ST88 (n = 13) followed by ST6 (n = 6) and by ST159, ST170, ST952, ST44, and ST282. The vanA VREfs (n = 20) were distributed among all the STs, whereas the single vanB VREfs belonged to ST6. The 79% of VREfs belonged to ST88 and ST6 which include nosocomial multidrug-resistant enterococcal strains [20]. Moreover, ST6 (CC2) has frequently been reported as a cause of invasive infection [20]. Although a limited number of VREfs were recovered during the study period, most (58%) were isolated in 2001–2006; notably, the dominant clone (ST88) continued to be sporadically recovered throughout the study period. Altogether, the VREfs are genetically diverse, except for a small ST88 cluster that encompassed isolates recovered from different wards and sample types in 2004–2006 and in 2012 (Fig. 2b). The overall distribution of VREfm and VREfs in the study period is shown in Fig. 1c.
Although the prevalence of specific VREfm and VREfs clones was observed in the hospital in 2001–2018, PFGE analysis disclosed an overall genetic heterogeneity among strains within the same lineage. This finding suggests that the diffusion of vanA resistance could be due to the dissemination of Tn1546-like elements rather than to clonal diffusion of a single strain [14].
This study reports an increasing incidence of VRE infections in an Italian hospital from 2016 to 2018 that is in line with reports from other European countries [9–12]. It is also in line with the literature showing that VREfm has overtaken VREfs as agents of hospital-acquired infections [21].
As in Denmark [22], the increase in VRE infections in our hospital could be related to the influx of new successful VRE lineages such as ST80. The increasing tolerance of VRE to the biocides used in hospital settings [21] and the renewed widespread use of vancomycin to treat severe infections caused by multidrug-resistant enterococci and MRSA may also be contributing factors. Furthermore, the use of broad-spectrum antibiotics (particularly carbapenems, third-generation cephalosporins) or antibiotic combinations (such as piperacillin/tazobactam) against Gram-negative bacteria in the hospital environment may favor the selection of intestinal VRE, thus increasing the risk of VRE infections [8].
An in-depth knowledge of the genotype of endemic nosocomial VRE and of the relatedness of the different isolates is critical to limit the fast dissemination of old and new VRE clones in clinical settings. The re-emergence of endemic VRE in European (and Italian) hospitals emphasizes the need for stringent control measures to reduce the risk of dissemination of resistant clones, such as the isolation of infected/colonized patients, more accurate disinfection procedures, and improved antimicrobial treatments.
Funding information
This work was supported by internal funding and by Progetto Strategico di Ateneo 2017–Polytechnic University of Marche “In the hunt of new antibiotics: active compounds from both chemical synthesis and natural sources.”
Compliance with ethical standards
Conflict of interest
The authors declare that there are no conflicts of interest.
Ethics approval
Ethical approval was waived by the institutional ethics committee due to the retrospective nature of the study and to the fact that all procedures were part of routine care.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Arias CA, Murray BE. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol. 2012;10:266–278. doi: 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cattoir V, Giard JC. Antibiotic resistance in Enterococcus faecium clinical isolates. Expert Rev Anti-Infect Ther. 2014;12:239–248. doi: 10.1586/14787210.2014.870886. [DOI] [PubMed] [Google Scholar]
- 3.Levine DP. Vancomycin: a history. Clin Infect Dis. 2006;42(Suppl 1):S5–S12. doi: 10.1086/491709. [DOI] [PubMed] [Google Scholar]
- 4.Torres C, Alonso CA, Ruiz-Ripa L, León-Sampedro R, Del Campo R, Coque TM (2018) Antimicrobial resistance in Enterococcus spp of animal origin. Microbiol Spectr 6(4) [DOI] [PubMed]
- 5.Willems RJ, Top J, van Santen M, Robinson DA, Coque TM, Baquero F, Grundmann H, Bonten MJ. Global spread of vancomycin-resistant Enterococcus faecium from distinct nosocomial genetic complex. Emerg Infect Dis. 2005;11:821–828. doi: 10.3201/1106.041204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deshpande LM, Fritsche TR, Moet GJ, Biedenbach DJ, Jones RN. Antimicrobial resistance and molecular epidemiology of vancomycin-resistant enterococci from North America and Europe: a report from the SENTRY antimicrobial surveillance program. Diagn Microbiol Infect Dis. 2007;58:163–170. doi: 10.1016/j.diagmicrobio.2006.12.022. [DOI] [PubMed] [Google Scholar]
- 7.Del Grosso M, Kärki T, D'Ancona FP, Pantosti A. Decrease of vancomycin resistance in Enterococcus faecium isolates from bloodstream infections in Italy from 2003 to 2013. Antimicrob Agents Chemother. 2015;59:3690–3691. doi: 10.1128/AAC.00513-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pfaller MA, Cormican M, Flamm RK, Mendes RE, Jones RN. Temporal and geographic variation in antimicrobial susceptibility and resistance patterns of enterococci: results from the SENTRY antimicrobial surveillance program, 1997–2016. Open Forum Infect Dis. 2019;6:S54–S62. doi: 10.1093/ofid/ofy344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Remschmidt C, Schröder C, Behnke M, Gastmeier P, Geffers C, Kramer TS. Continuous increase of vancomycin resistance in enterococci causing nosocomial infections in Germany − 10 years of surveillance. Antimicrob Resist Infect Control. 2018;7:54. doi: 10.1186/s13756-018-0353-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Markwart R, Willrich N, Haller S, Noll I, Koppe U, Werner G, Eckmanns T, Reuss A. The rise in vancomycin-resistant Enterococcus faecium in Germany: data from the German antimicrobial resistance surveillance (ARS) Antimicrob Resist Infect Control. 2019;8:147. doi: 10.1186/s13756-019-0594-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buetti N, Wassilew N, Rion V, Senn L, Gardiol C, Widmer A, Marschall J, for Swissnoso Emergence of vancomycin-resistant enterococci in Switzerland: a nation-wide survey. Antimicrob Resist Infect Control. 2019;8:16. doi: 10.1186/s13756-019-0466-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hammerum AM, Justesen US, Pinholt M, Roer L, Kaya H, Worning P, Nygaard S, Kemp M, Clausen ME, Nielsen KL, Samulioniené J, Kjærsgaard M, Østergaard C, Coia J, Søndergaard TS, Gaini S, Schønning K, Westh H, Hasman H, Holzknecht BJ. Surveillance of vancomycin-resistant enterococci reveals shift in dominating clones and national spread of a vancomycin-variable vanA Enterococcus faecium ST1421-CT1134 clone, Denmark, 2015 to March 2019. Euro Surveill. 2019;24(34):1900503. doi: 10.2807/1560-7917.ES.2019.24.34.1900503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahmed MO, Baptiste KE. Vancomycin-resistant enterococci: a review of antimicrobial resistance mechanisms and perspectives of human and animal health. Microb Drug Resist. 2018;24:590–606. doi: 10.1089/mdr.2017.0147. [DOI] [PubMed] [Google Scholar]
- 14.Freitas AR, Tedim AP, Francia MV, Jensen LB, Novais C, Peixe L, Sánchez-Valenzuela A, Sundsfjord A, Hegstad K, Werner G, Sadowy E, Hammerum AM, Garcia-Migura L, Willems RJ, Baquero F, Coque TM. Multilevel population genetic analysis of vanA and vanB Enterococcus faecium causing nosocomial outbreaks in 27 countries (1986-2012) J Antimicrob Chemother. 2016;71:3351–3366. doi: 10.1093/jac/dkw312. [DOI] [PubMed] [Google Scholar]
- 15.CLSI, editor. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 11th ed. CLSI standard M07. Clinical and Laboratory Standard Institute: Wayne; 2018. [Google Scholar]
- 16.Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995;33:24–27. doi: 10.1128/JCM.33.1.24-27.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brenciani A, Morroni G, Pollini S, Tiberi E, Mingoia M, Varaldo PE, Rossolini GM, Giovanetti E. Characterization of novel conjugative multiresistance plasmids carrying cfr from linezolid-resistant Staphylococcus epidermidis clinical isolates from Italy. J Antimicrob Chemother. 2016;71:307–313. doi: 10.1093/jac/dkv341. [DOI] [PubMed] [Google Scholar]
- 18.Coombs GW, Daley DA, Lee YT, Pang S, Bell JM, Turnidge JD, Australian Group on Antimicrobial Resistance Australian Group on Antimicrobial Resistance (AGAR) Australian Enterococcal Sepsis Outcome Programme (AESOP) Annual Report 2015. Commun Dis Intell. 2018;42:S2209–S6051. [PubMed] [Google Scholar]
- 19.Bressan R, Knezevich A, Monticelli J, Campanile F, Busetti M, Santagati M, Dolzani L, Milan A, Bongiorno D, Di Santolo M, Tonin EA, Stefani S, Luzzati R, Lagatolla C. Spread of vancomycin-resistant Enterococcus faecium isolates despite validated infection control measures in an italian hospital: antibiotic resistance and genotypic characterization of the endemic strain. Microb Drug Resist. 2018;24:1148–1155. doi: 10.1089/mdr.2017.0314. [DOI] [PubMed] [Google Scholar]
- 20.Kuch A, Willems RJ, Werner G, Coque TM, Hammerum AM, Sundsfjord A, Klare I, Ruiz-Garbajosa P, Simonsen GS, van Luit-Asbroek M, Hryniewicz W, Sadowy E. Insight into antimicrobial susceptibility and population structure of contemporary human Enterococcus faecalis isolates from Europe. J Antimicrob Chemother. 2012;67:551–558. doi: 10.1093/jac/dkr544. [DOI] [PubMed] [Google Scholar]
- 21.Alotaibi SMI, Ayibiekea A, Pedersen AF, Jakobsen L, Pinholt M, Gumpert H, Hammerum AM, Westh H, Ingmer H. Susceptibility of vancomycin-resistant and – sensitive Enterococcus faecium obtained from Danish hospitals to benzalkonium chloride, chlorhexidine and hydrogen peroxide biocides. J Med Microbiol. 2017;66:1744–1751. doi: 10.1099/jmm.0.000642. [DOI] [PubMed] [Google Scholar]
- 22.Pinholt M, Bayliss SC, Gumpert H, Worning P, Jensen VVS, Pedersen M, Feil EJ, Westh H. WGS of 1058 Enterococcus faecium from Copenhagen, Denmark, reveals rapid clonal expansion of vancomycin-resistant clone ST80 combined with widespread dissemination of a vanA-containing plasmid and acquisition of a heterogeneous accessory genome. J Antimicrob Chemother. 2019;74:1776–1785. doi: 10.1093/jac/dkz118. [DOI] [PubMed] [Google Scholar]