Abstract
Detection of bovine viral diarrhea virus (BVDV) in aborted fetus samples is often difficult due to tissue autolysis and inappropriate sampling. Studies assessing different methods for BVDV identification in fetal specimens are scarce. The present study evaluated the agreement between different diagnostic techniques to detect BVDV infections in specimens from a large number of bovine aborted fetuses and neonatal deaths over a period of 22 years. Additionally, genetic, serological, and pathological analyses were conducted in order to characterize BVDV strains of fetal origin. Samples from 95 selected cases from 1997 to 2018 were analyzed by antigen-capture ELISA (AgELISA), nested RT-PCR (RT-nPCR), and real-time RT-PCR (RT-qPCR). In addition, amplification and sequencing of the 5′UTR region were performed for phylogenetic purposes. Virus neutralization tests against the BVDV-1a, BVDV-1b, and BVDV-2b subtypes were conducted on 60 fetal fluids of the selected cases. Furthermore, the frequency and severity of histopathological lesions were evaluated in BVDV-positive cases. This study demonstrated that RT-nPCR and RT-qPCR were more suitable than AgELISA for BVDV detection in fetal specimens. However, the agreement between the two RT-PCR methods was moderate. The BVDV-1b subtype was more frequently detected than the BVDV-1a and BVDV-2b subtypes. Neutralizing antibodies to any of the three subtypes evaluated were present in 94% of the fetal fluids. Microscopically, half of the BVDV-positive cases showed a mild non-suppurative inflammatory response. These results emphasize the need to consider different methods for a diagnostic approach of BVDV associated to reproductive losses.
Electronic supplementary material
The online version of this article (10.1007/s42770-020-00296-z) contains supplementary material, which is available to authorized users.
Keywords: BVDV, Abortion, Diagnosis, Phylogenetic analysis, Pathology
Introduction
Bovine viral diarrhea virus (BVDV) belongs to the genus Pestivirus, family Flaviviridae. This virus has a worldwide distribution and causes infections in ruminants. The viral genome consists of a single-stranded, positive sense RNA molecule of approximately 12.3 kb in length. Based on genetic and antigenic characteristics, BVDV can be divided into two species, BVDV-1 and BVDV-2 [1]. According to the recently proposed taxonomy of the genus Pestivirus [2], these two species correspond to Pestivirus A and Pestivirus B, respectively. To follow the terminology consistent with previous reports, the previous names were used in the study. Also, according to the sequence analysis of the 5′unstranslated region (5′UTR), BVDV-1 has been further classified into at least 21 subtypes and BVDV-2 has been classified into 4 subtypes [3]. Each BVDV species has two biotypes, non-cytopathic (ncp) and cytopathic (cp), according to their cytopathogenicity on cell cultures [1].
Reproductive losses due to fetal infections by BVDV represent a great economic impact for the cattle industry [4]. Outcomes of BVDV fetal infections depend on the gestational age when they occur. Infections during the first trimester can cause embryonic death, mummification, abortion, and the generation of persistently infected (PI) calves. Also, BVDV infections between 80 and 150 days of gestation may result in congenital defects, while infections during late gestation, between 125 and 285 days, can lead to abortion or the birth of weak calves [4]. When infection occurs after 150–180 days of gestation, the fetus is able to mount an immune response, clear the virus, and be born seropositive [5].
Diagnostic methods commonly used for BVDV detection include virus isolation (VI), immunohistochemistry, antigen-capture ELISA (AgELISA), nucleic acid detection, and serological tests [6, 7]. Regarding BVDV diagnosis on reproductive disorders, the accuracy of these methods may be affected by several factors such as inappropriate sampling and autolysis [8, 9]. Although VI has been considered as the “gold standard” method for BVDV diagnosis in different clinical presentations [6, 7], the use of molecular techniques has become more common and widely accepted because of its high sensitivity [10]. VI only detects infectious virus, whereas RT-PCR methods do not require viable virus to get a positive result. Thus, it is important to consider that detection of BVDV RNA does not imply that infective virus is present [7, 11].
Fetal lesions following BVDV experimental infections have been investigated [12, 13], but unfortunately, histopathological descriptions of BVDV naturally infected and aborted fetuses or dead neonatal calves are scarce [14, 15].
BVDV detection in cases of reproductive disorders represents a clear evidence of virus circulation, and it is important to be considered when control programs are proposed [16]. Although VI is the “gold standard” for virus detection, there is uncertainty about the selection of diagnostic methods to detect BVDV in samples from aborted fetuses and neonatal deaths. Additionally, evaluation or validation of different assays for BVDV identification in those types of samples is limited [8, 16, 17]. Moreover, there is no published information on the comparison of diagnostic methods and performance of phylogenetic, serologic, and pathologic studies of BVDV in specimens from cases of bovine abortion.
Therefore, this study was initially conducted to compare the ability of diagnostic methods to detect BVDV in stored samples from aborted fetuses and neonates over a 22-year period. In addition, the phylogenetic grouping of BVDV strains, the levels of neutralizing antibodies in fetal fluids, and the lesions observed in BVDV-positive cases were investigated for a comprehensive understanding of reproductive disorders associated with this virus.
Materials and methods
Cases, necropsy, and selection criteria
Ninety-five archived cases of bovine aborted fetuses and stillborn calves from 67 beef and 14 dairy farms were selected for this study. These cases were submitted for postmortem examination (between 1997 and 2018) to the Specialized Veterinary Diagnostic Service (SVDS) at INTA Balcarce, Argentina. In each case, fetal age was estimated by the crown-rump length or was provided by veterinary practitioners. Full necropsy and sampling procedures were performed as described previously for routine diagnosis of reproductive pathogens including Brucella abortus, Campylobacter fetus, Leptospira spp., Tritrichomonas foetus, Neospora caninum, BVDV, and bovine herpesvirus [18, 19].
In this study, the selection criteria of the 95 cases included previous BVDV isolation on Madin-Darby bovine kidney (MDBK) cells (16 cases); presence of microscopic lesions compatible with BVDV infection (48 cases); BVDV isolation and presence of microscopic lesions (12 cases); presence of congenital malformations and compatible microscopic lesions (18 cases); BVDV isolation, presence of microscopic lesions, and congenital malformations (1 case).
BVDV isolation was attempted from spleen tissue samples according to standard procedures [20]. When spleen was not available, other samples such as lung, brain, lymph node, serum, or fetal fluid were used. Overall, 95 samples (for antigen and molecular detection) and 60 fetal fluids (of the 95 selected cases, for neutralizing antibody detection) from thoracic and abdominal cavities (hereafter referred to as fetal fluids) were stored at − 80 °C. Diagnostic methods (AgELISA, nested RT-PCR, real-time RT-PCR) and virus neutralization (VN) test are described below in detail and were performed during 2018 and 2019.
AgELISA
To detect the non-structural protein 3 (NS3) of BVDV, a commercial AgELISA kit (BIO K 337-Monoscreen, Bio-X Diagnostics, Belgium) was used following the manufacturer’s instructions. The optical density (OD) values were measured using a plate reader at 450 nm. The net OD value for each sample was calculated by subtracting the raw OD value of the negative control well from the raw OD value of the sample well. The same procedure was carried out for the positive control (antigen) well. The value for each sample was calculated using a formula [% value = (delta OD sample/delta OD positive)*100]. Positive or negative status was considered if the sample value was greater or lower than 7% compared to the positive control, respectively, as indicated by the manufacturer.
RNA purification
Total RNA from 100 mg of tissue or 100 μl of fluid (serum or fetal fluid) was extracted by using TRIzol® Reagent (Cat. No. 15596, Invitrogen, USA) and following the manufacturer’s instructions. RNA was diluted in 40 μl of RNase-free water and stored at − 80 °C until analyzed.
Nested RT-PCR
A nested RT-PCR (RT-nPCR) protocol [21] with some modifications [22] was performed for the detection and genotyping of BVDV in archived samples. Amplification of specific fragments of the NS5B gene enables the differentiation of BVDV-1 (369 pb) and BVDV-2 (615 pb) [21].
Real-time RT-PCR (RT-qPCR)
Reverse transcription (RT) was carried out using universal pan-pestivirus primers 324/326 [23], which target a fragment of the 5′UTR region. For complementary DNA (cDNA) synthesis, 4 μl of RNA sample, 0.3 μl of each primer (20 μM), and 0.4 μl of DMSO were denatured at 95 °C for 5 min. Thereafter, 2 μl of RT buffer, 1 μl of deoxynucleotide mix (10 mM each), 0.12 μl (24 U) of M-MLV, and 0.88 μl of RNase-free water were incubated at 37 °C for 60 min and 70 °C for 5 min.
The PCRs contained 0.6 μl (5 μM) of each primer (Pesti-qF/Pesti-qR) [24], 5 μl of 1X PCR Master Mix (FastStart Universal SYBR Green Master Rox, Roche, Germany), 1.8 μl of RNase-free water, and 2 μl of cDNA template in a final volume of 10 μl. Amplification and detection of the specific product (160 bp) were carried out in duplicate on an ABI 7500 cycler (Applied Biosystems, CA, USA) and using these conditions: 50 °C for 2 min, 95 °C for 10 min, 40 cycles of denaturation at 95 °C for 15 s, and annealing/extension at 60 °C for 1 min. After amplification, melt analysis was performed. Samples that gave both a typical amplification curve and a melt temperature value between 80.0 and 82.5 °C were considered positive. Quantification cycle (Cq) values were also recorded.
Phylogenetic analyses
The RNA from BVDV-positive samples (mainly spleen), identified by at least one of three diagnostic methods and/or previous VI, was included for this purpose. A fragment of the 5′UTR (288 bp) was amplified by RT-PCR using pan-pestivirus primers 324/326 [23]. Briefly, phylogenetic analyses were performed with the neighbor-joining method, Kimura-2 parameter genetic distance model, and bootstrapping of 1000 replicates as described [22]. Because the 5′UTR could not be amplified from the sample of one case (13–434), the NS5B gene fragment was analyzed.
Nucleotide sequences were deposited in the GenBank database (accession numbers MK684367 to MK684395). Also, three sequences of strains included in the analyses have been previously reported: 98–204 (JX848359), 08–724 (JX679693) [25], and 10–636 (MH294527) [22].
VN test of fetal fluids
Sixty fetal fluids of the 95 selected cases were available to perform a VN test according to international guidelines [11]. Cell culture-adapted and cp BVDV strains from Argentina (Laboratory of Veterinary Virology, SVDS, INTA Balcarce) were used for this test, which included BVDV-1a (13–558; accession number MK558700), BVDV-1b (00–693; MK558701), and BVDV-2b (14–663; MK558702). These subtypes are the most frequent in Argentina [25].
Fetal fluids were centrifuged at 7500g for 5 min, and the supernatants were inactivated at 56 °C for 30 min. Twofold serial dilutions (1:4 to 1:32) of the supernatants in minimal essential medium (MEM) were performed. Dilutions of fetal fluids were mixed with 100 TCID50 of each virus strain in 96-well plates and incubated for 60 min at 37 °C in 5% CO2. Then, 2 × 104 MDBK cells suspended in MEM plus 10% fetal bovine serum (MEM + 10% FBS) were added to each well, and the plates were incubated for 72 h at 37 °C in 5% CO2. Wells with MEM + 10% FBS were used as negative controls, and back titrations of the working virus strains were conducted on each plate. Samples that showed neutralization at a dilution of 1:32 were retested in dilutions from 1:4 to 1:512.
Histopathology
Heart, lung, and brain tissue samples were selected from BVDV-positive cases for histopathological evaluation. The severity of microscopic lesions was classified using a semi-quantitative score, which was based on the proportion of tissue section that showed microscopic lesions: 0 = absence of lesion; 1 = mild, 25% of tissue affected; 2 = moderate, 25–50% of tissue affected; and 3 = severe, > 50% of tissue affected [26].
Data analyses
The proportions of fetuses from each age category and BVDV subtypes were analyzed by the chi-square test for homogeneity of proportions (Proc FREQ, SAS Studio v3.6, SAS Institute Inc. Cary, NC, USA). The proportion of positive cases for each diagnostic technique was analyzed by the Q Cochran and McNemar test (Proc FREQ, SAS). When necessary, multiple pairwise comparisons were adjusted using the Bonferroni method. Agreement between techniques was estimated according to Cicchetti and Feinstein [27] and Gwet’s Agreement Coefficient (AC1 coefficient) [28] (“rel” package v1.3.3, R v3.5.1, R Core Team 2018, Vienna, Austria). Ninety-five percent confidence intervals (CI95%) were calculated using the likelihood ratio method. Interpretation of the AC1 coefficients was performed according to McHugh [29].
VN titers for each virus strain were converted to a log2 according to the Spearman-Kärber method [30] and expressed as geometric mean titers. The statistical analyses were performed with Friedman test (Proc FREQ, SAS). Neutralizing antibody (NAb) titers equal to or greater than 2 (dilution 1:4) were considered positive. In order to determine if the serologic responses of the selected cases were specific for BVDV-1a, BVDV-1b, or BVDV-2b subtypes, NAb titers of the positive samples were compared using the following formula: RsubtypeX = (3 × titer against subtype X) / (titer against subtype X + titer against subtype Y + titer against subtype Z) [31]. If the ratio (R) value of one fetal fluid sample for a certain virus strain (X) was > 0.2 with respect to the R values of the other virus strains (Y and Z), this indicated that the sample had higher NAb titers for that virus strain (X). Conversely, if the R value was < 0.2, the sample was considered equivocal, without a predominant NAb titer [31].
Statistical analysis for the proportions of tissues with histologic lesions was performed with the Q Cochran test, whereas the score of severity was analyzed by Kruskal-Wallis test for non-parametric data. For all statistical tests conducted, a 5% significance level was used.
Results
Detection of BVDV
Samples from 37 out of 95 selected cases (39%) were positive for BVDV by at least one or more diagnostic methods (AgELISA, RT-nPCR, or RT-qPCR). In spite of a previous positive BVDV isolation result, samples from three cases (97–097, 98–096, and 09–688) were negative by all three methods (Online Resource 1). Nevertheless, the BVDV isolates and information on these cases (age and lesions) were included for phylogenetic and statistical analyses. Thus, 40 BVDV-positive cases were considered for this study (Table 1).
Table 1.
Summary of BVDV detection in 40 cases of bovine abortion and neonatal death using AgELISA and RT-PCR-based methods
| Selection criteria | Number of cases | Year | Age | AgELISAa positive | RT-nPCRb positive | RT-qPCRc | Subtype | NAb responsed | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1° | 2° | 3° | Neo | Pos | Cq, range | 1a | 1b | 2b | Pos | Neg | n.d. | |||||
| Microscopic lesions | 10 | 2010–2018 | 0 | 2 | 6 | 2 | 0 | 5 | 8 | 30.0–37.4 | 1 | 4 | 0 | 2 | 1 | n. |
| Microscopic lesions; congenital malformations | 2 | 2014–2015 | 0 | 0 | 1 | 1 | 0 | 2 | 2 | 21.2–33.0 | 0 | 2 | 0 | 1 | 0 | 7 |
| VI + | 16 | 1997–2018 | 2 | 0 | 10 | 1 | 6 | 11 | 12 | 19.1–36.4 | 2 | 10 | 1 | 4 | 0 | 1 |
| VI +; microscopic lesions | 11 | 2004–2017 | 1 | 3 | 7 | 0 | 2 | 7 | 10 | 21.2–32.5 | 3 | 6 | 2 | 7 | 0 | 12 |
| VI +; microscopic lesions; congenital malformations | 1 | 2013 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 24.3 | 1 | 0 | 0 | 0 | 0 | 4 |
| Total | 40 | 1997–2018 | 3 | 5 | 24 | 5 | 8 | 26 | 33 | 19.1–37.4 | 7 | 22 | 3 | 14 | 1 | 4 |
Age: data not available in three cases; Subtype: data not available in eight cases
1°, 2°, and 3° trimester of gestation; Neo neonate; Cq quantification cycle value; VI+ positive BVDV isolation; n.d. not done
aAntigen-capture ELISA, BIO K 337-Monoscreen Ag ELISA BVDV, Bio-X Diagnostics, Belgium
bNested RT-PCR (Gilbert et al., 1999) [21]
cReal-time RT-PCR (Mari et al., 2016) [24]
dNeutralizing antibodies detected by virus neutralization test
Bovine viral diarrhea virus was more frequently detected in fetuses during the third trimester of gestation (n = 24; 64.9%, CI95% = 48.9–78.9) compared to the first trimester (n = 3; 8.1%, CI95% = 2.1–19.7), second trimester (n = 5; 13.5%, CI95% = 5.1–26.8), and neonates (n = 5; 13.5%, CI95% = 5.1–26.8) (p < 0.01) (Table 1).
Comparative performance of diagnostic methods
The proportions of BVDV-positive samples detected by RT-nPCR (n = 26; 27.4%, CI95% = 19.1–36.9) and RT-qPCR (n = 33; 34.7%, CI95% = 25.7–44.6) were not statistically different (p > 0.42), whereas positive samples detected by AgELISA (n = 8; 8.4%, CI95% = 3.9–15.1) were significantly lower than those detected by any of the two RT-PCR-based methods (p < 0.01).
Agreement among the three diagnostic methods is presented in Table 2. A high percentage of negative agreement was observed among all of them. Positive agreement between AgELISA and each RT-PCR method was poor, whereas this value was moderate between the two RT-PCR methods. According to the AC1 coefficient, there was moderate agreement between AgELISA and RT-nPCR or between the two RT-PCR methods.
Table 2.
Agreement among AgELISA and RT-PCR-based methods for BVDV detection in bovine aborted fetuses and neonatal deaths
| Techniques | N | Percentage agreementd | Gwet’s agreement coefficiente | |||
|---|---|---|---|---|---|---|
| Negative | Positive | AC1 | CI95% | Concordancef | ||
| AgELISAa vs. RT-nPCRb | 95 | 87.2 | 41.2 | 0.70 | 0.56–0.84 | Weak to strong |
| AgELISA vs. RT-qPCRc | 95 | 81.9 | 34.1 | 0.57 | 0.40–0.74 | Moderate to weak |
| RT-nPCR vs. RT-qPCR | 95 | 88.5 | 74.6 | 0.72 | 0.58–0.86 | Weak to strong |
aAntigen-capture ELISA, BIO K 337-Monoscreen Ag ELISA BVDV, Bio-X Diagnostics, Belgium
bNested RT-PCR (Gilbert et al., 1999) [21]
cReal-time RT-PCR (Mari et al., 2016) [24]
dEstimated according to Cicchetti and Feinstein (1990) [27]
eEstimated according to Gwet (2008) [28]
fInterpretation according to McHugh (2012) [29]
RT-PCR-based methods were discordant in 15 cases, 11 cases negative by RT-nPCR were positive by RT-qPCR, whereas 4 cases positive by RT-nPCR were negative by RT-qPCR (Online Resource 1). Cq values of the RT-qPCR ranged from 19.1 to 36.4 (mean 25.9) in samples with previous BVDV isolation, whereas Cq values ranged from 21.2 to 37.4 (mean 33.7) in samples previously negative by VI.
Phylogenetic analyses
Of the 40 BVDV-positive cases, 31 5′UTR sequences (25 from original samples and 6 from cell culture supernatants) and one NS5B gene fragment sequence (original sample) were used for phylogenetic grouping. BVDV strains could be classified as BVDV-1a, BVDV-1b, and BVDV-2b based on this analysis (Fig. 1a, b). Statistically, BVDV-1b (n = 22, 68.7%, CI95% = 51.7–82.9) was more frequently detected than BVDV-1a (n = 7, 21.9%, CI95% = 10.1–38.0; p = 0.02) and BVDV-2b (n = 3, 9.4%, CI95% = 2.4–22.5; p < 0.01), whereas there was no significant difference between the proportions of BVDV-1a and BVDV-2b (p = 0.62). Unfortunately, BVDV genotyping could not be performed in eight cases.
Fig. 1.
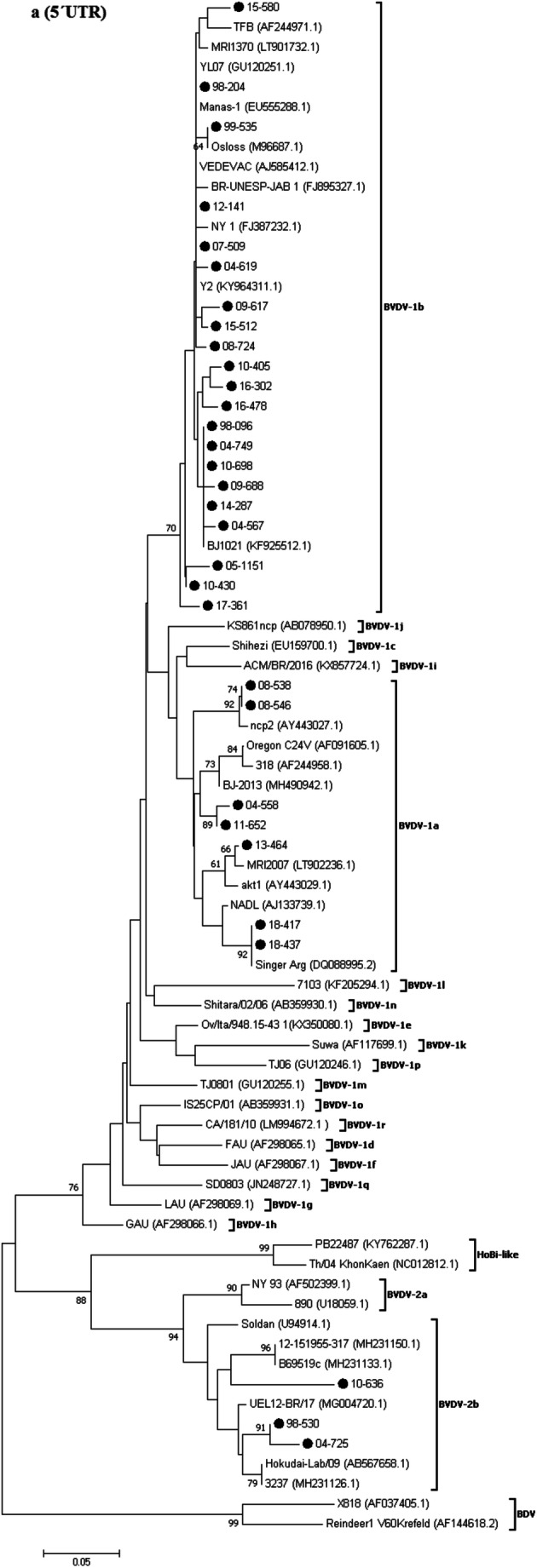
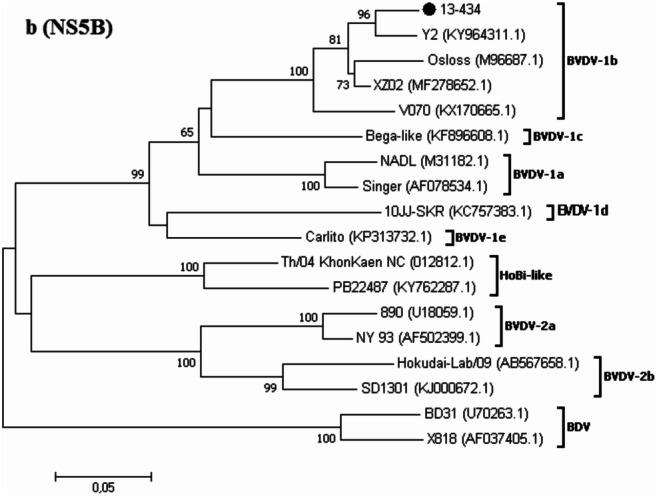
Phylogenetic trees based on the partial nucleotide sequences of the genomic regions 5′UTR (a) and NS5B (b). Genetic distances were calculated using the Kimura-2 correction parameter, and phylogenetic trees were constructed using the neighbor-joining method. Bootstrap values (1000 replicates) above 60% are shown. BVDV strains studied in the current study are highlighted by dots. The GenBank accession numbers of each BVDV reference strain are indicated in brackets. Nucleotide sequences from border disease virus (BDV) were included for out-group rooting
VN test of fetal fluids
Of the 60 fetal fluids processed, 51 could be evaluated to detect NAb response and 9 were excluded because of their toxicity to cultured cells. A total of 48 fluids tested positive for NAb titers against BVDV (94%, 48/51), and 3 fetal fluids tested negative (6%, 3/51). Of the fluids positive for NAb, 14 belonged to BVDV-positive cases (Table 1) and 34 belonged to cases negative to BVDV by any of the diagnostic techniques, including VI. In contrast, of the 3 fluids negative for NAb, 1 case was positive for BVDV and showed microscopic lesions (13–434; Online Resource 1), whereas the other 2 cases were negative for BVDV and presented microscopic lesions.
The 14 fetal fluids positive for NAb from BVDV-positive cases were classified as follows: presence of microscopic lesions (9 cases); congenital malformations and microscopic lesions (1 case); and no microscopic lesions (3 cases). In one case, necropsy was not performed. On the other hand, the 34 fetal fluids positive for NAb but negative for BVDV were classified as follows: presence of microscopic lesions (24 cases); congenital malformations and microscopic lesions (7 cases); and no microscopic lesions (3 cases).
Overall, the geometric mean NAb titers were higher for BVDV-1b (3.66, CI95% = 3.16–4.17) and BVDV-2b (3.71, CI95% = 3.30–4.12) than for BVDV-1a (2.96, CI95% = 2.42–3.51) (p < 0.05). The distribution of the predominant NAb titers against each of the three BVDV subtypes is shown in Fig. 2.
Fig. 2.
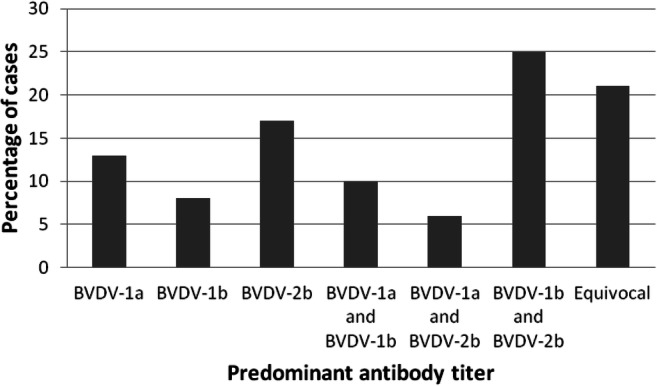
Distribution of the predominant neutralizing antibody responses directed to each BVDV subtype in 48 fetal fluid samples from selected cases. BVDV-1a: predominant titer against BVDV-1a; BVDV-1b: predominant titer against BVDV-1b; BVDV-2b: predominant titer against BVDV-2b; BVDV-1a and BVDV-1b: predominant titers against BVDV-1a and BVDV-1b; BVDV-1a and BVDV-2b: predominant titers against BVDV-1a and BVDV-2b; BVDV-1b and BVDV-2b: predominant titers against BVDV-1b and BVDV-2b; Equivocal: no predominant titer
Gross pathology and microbiology of BVDV-positive cases
Necropsy was performed in 32 out of the 40 BVDV-positive cases. The most relevant pathological findings included corneal opacity (9%, 3/32), hydrothorax (19%, 6/32), hydropericardium (6%, 2/32), ascites (22%, 7/32), subcutaneous edema (22%, 7/32), and congestion of the meninges, lungs, and kidneys (19%, 6/32). Three of them also revealed congenital malformations including hydrocephalus (13–464, 15–580); microencephaly and cerebellar hypoplasia (15–580); doming of the head, brachygnathism, and nasal deviation (14–287); and alopecia and hypotrichosis (15–580). Nevertheless, these three BVDV-positive cases only accounted for a low proportion (16%, 3/19) of all the congenital malformations (20%, 19/95) observed on the 95 selected cases.
Concomitant infection of BVDV with Neospora canimun was identified in three fetuses, whereas Leptospira spp. were detected in one fetus. The rest of the BVDV-positive cases were negative to the other important reproductive pathogens.
Histopathology of BVDV-positive cases
Microscopic lesions in the 32 BVDV-positive cases showed no significant differences neither in frequency (p = 0.61) nor in severity (p = 0.81) (Table 3). Based on the mean severity score, the proportion of tissues that presented microscopic lesions was less than 25%. The main histopathological findings observed in the heart, lungs, and brain are described (Table 3). The most frequent microscopic lesion was a non-suppurative inflammatory response, predominantly of lymphocytes and macrophages.
Table 3.
Frequency, severity, and main microscopic lesions in organs from 32 BVDV-positive cases of bovine abortion and neonatal death confirmed using AgELISA and RT-PCR-based methods
| Tissue | % cases with lesion*, a | CI95% | Mean score of severity*, b | CI95% |
| Heart | 55.6 | 37.0–73.2 | 0.77 | 0.44–1.11 |
| Lung | 51.9 | 33.5–69.9 | 0.63 | 0.35–0.90 |
| Brain | 44.4 | 26.9–63.1 | 0.70 | 0.35–1.08 |
| Lesion | Number of cases with lesions | |||
| Heart | ||||
| Non-suppurative myocarditis | 5 | |||
| Non-suppurative pericarditis | 6 | |||
| Non-suppurative pericarditis and myocarditis | 6 | |||
| Myocardial necrosis | 3 | |||
| Myocardial and epicardial hemorrhage | 1 | |||
| Lungs | ||||
| Interstitial non-suppurative pneumonia | 13 | |||
| Suppurative bronchopneumonia | 1 | |||
| Interlobular hemorrhage | 1 | |||
| Brain | ||||
| Non-suppurative encephalitis | 3 | |||
| Non-suppurative meningitis | 7 | |||
| Non-suppurative meningoencephalitis | 2 | |||
| Neuronal degeneration and necrosis | 1 | |||
| Midbrain hemorrhage | 1 | |||
| Cerebellar dysplasia | 2 | |||
| Cortical dysgenesis | 1 | |||
*p > 0.05
aCochran Q test
bKruskal-Wallis test
Discussion
Establishing BVDV as the etiological agent of abortion or neonatal death is often a difficult task. The poor state of preserved specimens due to autolysis usually has negative impact on diagnostic test performance [16]. To assess a relationship between viral detection and abortion, it is necessary to associate such finding with compatible fetal pathology, antibody response, and herd history, while ruling out other potential reproductive pathogens [17, 32]. Thus, a multi-pronged approach should be applied in order to confirm BVDV as responsible for reproductive losses. To the authors’ knowledge, there are no previous studies that address several diagnostic techniques in a substantial number of spontaneous BVDV reproductive cases over a long period of time.
The BVDV-positive cases evaluated in this study ranged from fetuses in the first trimester of gestation to 11-day-old neonates. However, most of them were fetuses from the third trimester of gestation. This observation may be explained by the fact that small fetuses are difficult to find under field conditions [18]. Accordingly, this finding cannot be entirely attributable to a higher incidence of BVDV infection during late gestation. Although most BVDV abortions occur in early gestation, this virus should not be ruled out as a cause of abortion in late-term fetuses [4, 33].
This study evaluated different diagnostic methods for identifying BVDV in samples collected over a 22-year period. It is possible, therefore, that the integrity of some analytes (viral RNA and antigens) could have been adversely affected by storage time. The high percentage of negative agreement (over 80%) among the three methods may reflect similar specificity for the tests. On the other hand, the moderate and weak agreement (AC1 = 0.70 and 0.57) and the poor positive agreement (41.2 and 34.1%) between AgELISA and RT-PCR methods, under the conditions of this study, are attributable to the low proportion of positive cases detected by AgELISA (8.4%). These results may be caused by protein degradation due to autolysis or long-term preservation. The method requires the presence of intact viral protein in the samples [16]. Thus, AgELISA appears to be of limited value for archived or autolytic samples such as fetal tissues. In the present study, spleen and occasionally lymph node, brain, or lung tissues were used for AgELISA as indicated by the manufacturer, but fetal fluids [17] or skin [16, 17] seems to be more suitable for this purpose.
The AC1 coefficient and the percentage of positive agreement between the RT-PCR methods were moderate in this retrospective study. This finding is related to the discrepancy observed in 15 cases. Although the proportions of positive cases detected by both molecular methods were not statistically different, 11 more cases were detected by RT-qPCR than by RT-nPCR. However, four other cases (all classified as BVDV-1b) were positive by the RT-nPCR but negative by the RT-qPCR. The reasons for this disagreement are not known. One possibility is that primers used in both methods are based on different gene targets, the non-coding region 5′UTR and the coding region for the non-structural protein NS5B. Since both gene regions have been considered highly conserved among pestiviruses [34, 35], differences in the ability of the two primer pairs to recognize the BVDV subtypes are not expected. Another possible explanation of the discordant results between the molecular methods is the smaller size of the amplicon from the RT-qPCR (160 bp) compared to the RT-nPCR (1124 bp for the first step and 604 or 360 bp for the second step). Fragmentation of RNA due to postmortem tissue degradation, long storage time [36], or the presence of environmental contaminants in the samples [9] can hamper the amplification of products over 400 bp with respect to shorter PCR products (70–250 bp) [36]. In the present work, less than half of the RNA samples showed an OD 260/280 ratio greater than the recommended value of 1.8 (data not shown). In the present study, it is therefore possible that poor RNA quality could have impaired BVDV detection by the RT-nPCR, which amplified a longer product in comparison to the RT-qPCR. Further research is needed to probe if amplicon size has the same effect on RT-PCR efficiency in autolytic or fetal specimens as previously described for formalin-fixed tissues [37].
Samples with previous BVDV isolation on cell culture had lower Cq values than samples with negative VI, suggesting a high viral load in those specimens positive by VI. However, 3 cases selected because of previous positive VI were negative in both RT-PCR methods. Moreover, they were also negative to the RT-PCR using the pan-pestivirus primers 324/326 (data not shown). Thus, other factors, such as viral RNA degradation during storage or the presence of inhibitors in the samples, cannot be excluded. In this regard, Ridpath et al. [9] have shown that contaminants present in fetal samples archived for 36 months may significantly reduce the detection of BVDV by molecular techniques. Nevertheless, both RT-PCR methods used in this study detected the virus in samples stored at − 80 °C for up to 21 years, so this finding shows that viral RNA could be stable for such a long period of time. Thus, molecular techniques should be valuable diagnostic tools to evaluate autolytic samples or archived tissues for retrospective studies.
The present study describes the genetic variability of several field strains of BVDV associated with reproductive disease. BVDV-1b was more frequent than BVDV-1a or BVDV-2b, as previously reported in Argentina for other disease syndromes [25]. Between 1984 and 1999, BVDV-1a was the most prevalent subtype identified in Argentina [38]. Since commercial vaccines used in Argentina are generally based on inactivated BVDV-1a and occasionally contain inactivated BVDV-2 [25], the selection pressure induced by the vaccination could have led to the emergence of BVDV-1b as the predominant subtype between 1998 and 2018, as described herein and by Pecora et al. [25]. Although the data in this study show that the genetic diversity of BVDV in Argentina has not increased since 1984, there is still a need to continue the surveillance of the BVDV genetic diversity.
Multiple BVDV strains should be used in the VN test to assess antibody status [6]. To the authors’ knowledge, this study compares for the first time the levels of NAb titers on fetal fluids from aborted fetuses and neonatal calves against three BVDV subtypes (BVDV-1a, BVDV-1b, and BVDV-2b). These subtypes were previously described in Argentinean cattle herds [20, 25, 38]. The presence of NAb titers for BVDV in 94% of the fetal fluids confirms the high circulation of this virus in Argentinean cattle [39] and shows the potential risk that BVDV represents to the reproductive performance of bovine herds. This remarkably high percentage of fetal fluids with NAb could be due to sample selection. Firstly, all the abortion and neonate cases evaluated were either confirmed or highly suspected of BVDV infection. Secondly, most fetuses corresponded to the 3° trimester of gestation or were neonates, a stage in which the animal is immunocompetent [5]. The presence of specific antibodies in fetal fluids is indicative of viral exposure, but the specific virus may not necessarily be the cause of abortion or death [40, 41]. Interpretation of NAb titers should be done within the context of individual animal and herd clinical history.
Gross pathological findings observed during the postmortem examinations were consistent with those described previously for BVDV natural infections [14, 32]. Also, half of the cases herein evaluated had mild histologic lesions in the heart, lungs, and brain, which were characterized by a non-suppurative inflammatory response. These findings agree with Kirkbride [14] and Murray [15] who reported that most BVDV-aborted fetuses had mild or no microscopic lesions.
Regarding cases with congenital malformations, BVDV was only identified in 16% (3/19). Thus, other possible etiologies could have been implicated such as inherited disorders, toxins, and other infectious agents. Nevertheless, NAb titers against BVDV were detected in 8/10 (80%) of the fetal fluids from these cases with malformations. Since antibody presence in fetal fluids is indicative of viral infection [40, 41], BVDV could be implicated in more cases of malformations than those confirmed in this work.
Bovine viral diarrhea virus is frequently detected in fetuses that were aborted due to other causes [14]. In the current study, Leptospira spp. and Neospora caninum were also identified in one and three BVDV-positive cases, respectively. Since BVDV has an immunosuppressive effect, the exposure of a pregnant cow may increase its susceptibility to other reproductive pathogens [32, 41]. Although other common reproductive agents circulating in Argentina [18, 19] were not identified in the rest of the BVDV-positive cases, the possibility of undetected infectious agents cannot be excluded.
Direct or indirect evidence of viral presence does not necessarily confirm that BVDV is responsible for abortion [32]. In 21 cases of this study, virus detection combined with compatible fetal lesions and the presence of NAb titers confirmed BVDV as the cause of abortion or neonatal death. However, in the other cases, the direct role of BVDV could not be established because of the absence of microscopic lesions or co-infection with other reproductive agents.
Conclusions
In summary, RT-nPCR and RT-qPCR proved to be more suitable than AgELISA to detect BVDV in specimens from fetal and neonatal deaths, even with samples stored for up to 21 years. The AC1 coefficient revealed a moderate agreement between both molecular methods. Therefore, the choice of any of these techniques may depend on different factors such as sample autolysis, amplification product size, and lab infrastructure. Even with these potential limitations, it is recommended that molecular techniques be incorporated into diagnostic protocols for BVDV reproductive losses. The detection of BVDV-1a, BVDV-1b, and BVDV-2b confirms the circulation of these subtypes as reported in previous investigations and reinforces the role of this virus as a potential risk for bovine reproductive health. The use of multiple diagnostic methods for accurate and reliable detection of BVDV in fetal tissues may be needed. The information provided herein could be useful to improve the diagnostic approach for BVDV-related fetal and neonatal deaths.
Electronic supplementary material
(PDF 393 kb)
Acknowledgments
We thank technicians and colleagues at the Specialized Veterinary Diagnostic Service of INTA Balcarce, Argentina.
Repositories GenBank accession numbers
MK684367 to MK684395.
Funding
This research was supported by the Agencia Nacional de Promoción Científica y Tecnológica, Grant PICT Start Up 2014-0308 and INTA Specific Project PNSA 1115053.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Maximiliano J. Spetter, Enrique L. Louge Uriarte and Erika A. González Altamiranda contributed equally to this work.
References
- 1.Ridpath JF, Bolin SR, Dubovi EJ. Segregation of bovine viral diarrhea virus into genotypes. Virology. 1994;205:66–74. doi: 10.1006/viro.1994.1620. [DOI] [PubMed] [Google Scholar]
- 2.Smith DB, Meyers G, Bukh J, Gould EA, Monath T, Muerhoff AS, Pletnev A, Rico-Hesse R, Stapleton J, Simmonds P, Becher P. Proposed revision to the taxonomy of the genus Pestivirus, family Flaviviridae. J Gen Virol. 2017;98:2106–2112. doi: 10.1099/jgv.0.000873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yeşilbağ K, Alpay G, Becher P. Variability and global distribution of subtypes of bovine viral diarrhea virus. Viruses. 2017;9:128. doi: 10.3390/v9060128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grooms DL. Reproductive consequences of infection with bovine viral diarrhea virus. Vet Clin Food Anim. 2004;20:5–19. doi: 10.1016/j.cvfa.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 5.Hansen TR, Smirnova NP, Van Campen H, Shoemaker ML, Ptitsyn AA, Bielefeldt-Ohmann H. Maternal and fetal response to fetal persistent infection with bovine viral diarrhoea virus. Am J Reprod Immunol. 2010;64:295–306. doi: 10.1111/j.1600-0897.2010.00904.x. [DOI] [PubMed] [Google Scholar]
- 6.Saliki JT, Dubovi EJ. Laboratory diagnosis of bovine viral diarrhea virus infections. Vet Clin Food Anim Pract. 2004;20:69–83. doi: 10.1016/j.cvfa.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 7.Dubovi EJ. Laboratory diagnosis of bovine viral diarrhea virus. Biologicals. 2013;41:8–13. doi: 10.1016/j.biologicals.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Ellis JA, Martin K, Norman GR, Haines DM. Comparison of detection methods for bovine viral diarrhea virus in bovine abortions and neonatal death. J Vet Diagn Investig. 1995;7:433–436. doi: 10.1177/104063879500700402. [DOI] [PubMed] [Google Scholar]
- 9.Ridpath JF, Neill JD, Chiang YW, Waldbillig J. Stability of bovine viral diarrhea virus 1 nucleic acid in fetal bovine samples stored under different conditions. J Vet Diagn Investig. 2014;26:6–9. doi: 10.1177/1040638713512315. [DOI] [PubMed] [Google Scholar]
- 10.Lanyon SR, Hill FI, Reichel MP, Brownlie J. Bovine viral diarrhoea: pathogenesis and diagnosis. Vet J. 2014;199:201–209. doi: 10.1016/j.tvjl.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 11.OIE (2018) World Organization for Animal Health. Manual of diagnostic tests and vaccines for terrestrial animals. Retrieved from https://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/3.04.07_BVD.pdf. Accessed 29 September 2019
- 12.Bielefeldt-Ohmann H, Tolnay AE, Reisenhauer CE, Hansen TR, Smirnova N, Van Campen H. Transplacental infection with non-cytopathic bovine viral diarrhoea virus types 1b and 2: viral spread and molecular neuropathology. J Comp Pathol. 2008;138:72–85. doi: 10.1016/j.jcpa.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 13.Casaro APE, Kendric JW, Kennedy PC. Response of the bovine fetus to bovine viral diarrhea-mucosal disease virus. Am J Vet Res. 1977;32:1543–1562. [PubMed] [Google Scholar]
- 14.Kirkbride CA. Viral agents and associated lesions detected in a 10-year study of bovine abortions and stillbirths. J Vet Diagn Investig. 1992;4:374–379. doi: 10.1177/104063879200400402. [DOI] [PubMed] [Google Scholar]
- 15.Murray RD. Lesions in aborted bovine fetuses and placenta associated with bovine viral diarrhoea virus infection. Arch Virol. 1991;3:217–224. doi: 10.1007/978-3-7091-9153-8_26. [DOI] [PubMed] [Google Scholar]
- 16.Ridpath JF, Chiang YW, Waldbillig J, Neill JD. Stability of bovine viral diarrhea virus antigen in ear punch samples collected from bovine fetuses. J Vet Diagn Investig. 2009;21:346–349. doi: 10.1177/104063870902100307. [DOI] [PubMed] [Google Scholar]
- 17.Graham DA, Beggs N, Mawhinney K, Calvert V, Cunningham B, Rowan-Layberry L, McLaren I. Comparative evaluation of diagnostic techniques for bovine viral diarrhoea virus in aborted and stillborn fetuses. Vet Rec. 2009;164:56–58. doi: 10.1136/vr.164.2.56. [DOI] [PubMed] [Google Scholar]
- 18.Campero CM, Moore DP, Odeón AC, Cipolla AL, Odriozola E. Aetiology of bovine abortion in Argentina. Vet Res Commun. 2003;27:359–369. doi: 10.1023/A:1024754003432. [DOI] [PubMed] [Google Scholar]
- 19.Morrell EL, Campero CM, Cantón GJ, Odeón AC, Moore DP, Odriozola E, Paolicchi F, Fiorentino MA. Current trends in bovine abortion in Argentina. Pesq Vet Bras. 2019;39:12–19. doi: 10.1590/1678-5150-pvb-5668. [DOI] [Google Scholar]
- 20.Odeón AC, Risatti G, Kaiser GG, Leunda MR, Odriozola E, Campero CM, Donis RO. Bovine viral diarrhea virus genomic associations in mucosal disease, enteritis and generalized dermatitis outbreaks in Argentina. Vet Microbiol. 2003;96:133–144. doi: 10.1016/S0378-1135(03)00210-4. [DOI] [PubMed] [Google Scholar]
- 21.Gilbert SA, Burton KM, Prins SE, Deregt D. Typing of bovine viral diarrhea viruses directly from blood of persistently infected cattle by multiplex PCR. J Clin Microbiol. 1999;37:2020–2023. doi: 10.1128/JCM.37.6.2020-2023.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spetter MJ, Louge Uriarte EL, González Altamiranda EA, Leunda ML, Pereyra SB, Verna AE, Odeón AC. Dual natural infection with bovine viral diarrhea virus −1 and −2 in a stillborn calf: tissue distribution and molecular characterization. Open Vet J. 2018;8:493–497. doi: 10.4314/ovj.v8i4.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vilcek S, Herring AJ, Herring JA, Nettleton PF, Lowings JP, Paton DJ. Pestiviruses isolated from pigs, cattle and sheep can be allocated into at least three genogroups using polymerase chain reaction and restriction endonuclease analysis. Arch Virol. 1994;136:309–323. doi: 10.1007/BF01321060. [DOI] [PubMed] [Google Scholar]
- 24.Mari V, Losurdo M, Lucente MS, Lorusso E, Elia G, Martella V, Patruno G, Buonavoglia D, Decaro N. Multiplex real-time RT-PCR assay for bovine viral diarrhea virus type 1, type 2 and HoBi-like pestivirus. J Virol Methods. 2016;229:1–7. doi: 10.1016/j.jviromet.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pecora A, Malacari DA, Ridpath JF, Aguirreburualde MP, Combessies G, Odeón AC, Romera SA, Golemba MD, Wigdorovitz A. First finding of genetic and antigenic diversity in 1b-BVDV isolates from Argentina. Res Vet Sci. 2014;96:204–212. doi: 10.1016/j.rvsc.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 26.Perez SE, Bretschneider G, Leunda MR, Osorio FA, Flores EF, Odeón AC. Primary infection, latency, and reactivation of bovine herpesvirus type 5 in the bovine nervous system. Vet Pathol. 2002;39:437–444. doi: 10.1354/vp.39-4-437. [DOI] [PubMed] [Google Scholar]
- 27.Cicchetti DV, Feinstein AR. High agreement but low kappa: II. Resolving the paradoxes. J Clin Epidemiol. 1990;43:551–558. doi: 10.1016/0895-4356(90)90159-M. [DOI] [PubMed] [Google Scholar]
- 28.Gwet KL. Computing inter-rater reliability and its variance in the presence of high agreement. Br J Math Stat Psychol. 2008;61:29–48. doi: 10.1348/000711006X126600. [DOI] [PubMed] [Google Scholar]
- 29.McHugh ML. Interrater reliability: the kappa statistic. Biochem Med. 2012;22:276–282. doi: 10.11613/BM.2012.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lennette EH. General principles underlying laboratory diagnosis of viral and rickettsial infections. In: Lennette EH, Schmidt NJ, editors. Schmidt’s diagnostic procedures for viral and rickettsial infections. 4. New York: American Public Health Association; 1969. pp. 1–65. [Google Scholar]
- 31.Pecora A, Perez Aguirreburualde MS, Malacari DA, Zabal O, Sala JM, Konrad JL, Caspe SG, Bauermann F, Ridpath J, Dus Santos MJ. Serologic evidence of HoBi-like virus circulation in Argentinean water buffalo. J Vet Diagn Investig. 2017;29:926–929. doi: 10.1177/1040638717720246. [DOI] [PubMed] [Google Scholar]
- 32.Anderson ML. Disorders of cattle. In: Njaa BL, editor. Kirkbride’s diagnosis of abortion and neonatal loss in animals. 4. Oxford: Wiley-Blackwell; 2012. pp. 13–48. [Google Scholar]
- 33.Ridpath JF. Bovine viral diarrhea virus: global status. Vet Clin Food Anim Pract. 2010;26:105–121. doi: 10.1016/j.cvfa.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 34.Newcomer BW, Neill JD, Marley MS, Ridpath JF, Givens MD. Mutations induced in the NS5B gene of bovine viral diarrhea virus by antiviral treatment convey resistance to the compound. Virus Res. 2013;174:95–100. doi: 10.1016/j.virusres.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 35.Schweizer M, Peterhans E. Pestiviruses. Annu Rev Anim Biosci. 2014;2:141–163. doi: 10.1146/annurev-animal-022513-114209. [DOI] [PubMed] [Google Scholar]
- 36.Fleige S, Pfaffl MW. RNA integrity and the effect on the real-time qRT-PCR performance. Mol Asp Med. 2006;27:126–139. doi: 10.1016/j.mam.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 37.Gruber AD, Moennig V, Hewicker-Trautwein M, Trautwein G. Effect of formalin fixation and long-term storage on the detectability of bovine viral-diarrhoea-virus (BVDV) RNA in archival brain tissue using polymerase chain reaction. J Vet Sci. 1994;41:654–661. doi: 10.1111/j.1439-0450.1994.tb00276.x. [DOI] [PubMed] [Google Scholar]
- 38.Jones LR, Zandomeni R, Weber EL. Genetic typing of bovine viral diarrhea virus isolates from Argentina. Vet Microbiol. 2001;81:367–375. doi: 10.1016/S0378-1135(01)00367-4. [DOI] [PubMed] [Google Scholar]
- 39.Odeón AC, Späth EJA, Paloma EJ, Leunda MR, Fernández Sainz IJ, Pérez SE, Kaiser GG, Draghi MG, Cetrá BM, Cano A. Prevalencia de anticuerpos al virus de diarrea viral bovina, herpesvirus bovino y virus sincicial respiratorio bovino en Argentina. Rev Med Vet. 2001;82:216–220. [Google Scholar]
- 40.Hyndman L, Vilcek S, Conner J, Nettleton PF. A novel nested reverse transcription PCR detects bovine viral diarrhoea virus in fluids from aborted bovine fetuses. J Virol Methods. 1998;71:69–76. doi: 10.1016/S0166-0934(97)00206-1. [DOI] [PubMed] [Google Scholar]
- 41.Kirkbride CA, Johnson MW. Serologic examination of aborted ovine and bovine fetal fluids for the diagnosis of border disease, bluetongue, bovine viral diarrhea, and leptospiral infections. J Vet Diagn Investig. 1989;1:132–138. doi: 10.1177/104063878900100208. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 393 kb)


