Abstract
The clade A members of serine/threonine protein phosphatase 2Cs (PP2Cs) play crucial roles in plant growth, development, and stress response via the ABA signaling pathway. But little is known about other PP2C clades in plants. Our previous study showed that maize the ZmPP2C26, a clade B member of ZmPP2Cs, negatively regulated drought tolerance in transgenic Arabidopsis. However, the upstream regulatory mechanism of ZmPP2C26 remains unclear. In the present study, the expression of ZmPP2C26 gene in maize was analyzed by quantitative real time PCR (qRT-PCR). The results showed that the expression of ZmPP2C26 in shoot and root was both significantly inhibited by drought stress. Subsequently, a 2175 bp promoter of ZmPP2C26 was isolated from maize genome (P2175). To validate whether the promoter possess some key cis-element and negatively drive ZmPP2C26 expression in drought stress, three 5´-deletion fragments of 1505, 1084 and 215 bp was amplified from P2175 and were fused to β-glucuronidase (GUS) and luciferase gene (LUC) to produce promoter::GUS and promoter::LUC constructs, and transformed into tobacco, respectively. Transient expression assays indicated that all promoters could drive GUS and LUC expression. The GUS and LUC activity were both significantly inhibited by PEG-6000 treatment. Notably, the − 1084 to − 215 bp promoter possess one MBS element and inhibits the expression of GUS and LUC under drought stress. Meanwhile, we found that the 215 bp length is enough to drive ZmPP2C26 expression. These findings will provide insights into understanding the transcription-regulatory mechanism of ZmPP2C26 negatively regulating drought tolerance.
Keywords: Maize, Serine/threonine protein phosphatase 2C, Promoter, Drought stress
Introduction
Protein phosphorylation and dephosphorylation is a dynamic balance process and widespread in prokaryotes and eukaryotes. Plants can select phosphorylation or dephosphorylation of substrate molecules to realize the cascade of signals and enable plants to respond to any stimuli (Luan et al. 2003; Ma et al. 2009; Schweighofer et al. 2004). The protein phosphatases (PPs) catalyze the dephosphorylation and play crucial roles in plant growth and development. According to the difference of homology and substrate specificity, PPs are divided into eight families, including PP1, PP2A, PP2B, PP2C, PP4, PP5, PP6 and PP7. Among these PPs, PP2C, a kind of multifunctional monomer enzyme, belongs to serine/threonine PPs and specifically dephosphorylates the phosphorylated serine/threonine residues of proteins in vivo. PP2Cs have been implicated as a negative regulator of protein kinase cascades, which are activated by environmental stress in eukaryotes (Park et al. 2009; Rodriguez 1998).
Till date, there are 83, 80 and 130 PP2C members identified by bioinformatic analyses in Arabidopsis, rice and maize, respectively (Antoni et al. 2012; Wang et al. 2018). Likewise, PP2Cs are divided into 13 clades of A-K. The Clade A members of PP2Cs interact with PYR/PYL/RCAR ABA receptors, then act on SnRK2 in ABA signaling pathway, to regulate plant growth, development and stress response, such as seed size, dormancy, germination, and drought resistance (Bhaskara et al. 2012; Han et al. 2018; He et al. 2019; Komatsu et al. 2013; Lu et al. 2017; Nishimura et al. 2018; Park et al.2009; Xiang et al. 2017). However, the functions of clade B members are rarely reported. In Arabidopsis, AP2C1, a clade B PP2C, interacts with CBL-interacting protein kinase 9 (CIPK9) in the cytoplasm to dephosphorylate CIPK9 and thus negatively regulate plant tolerance to low-K+ stress (Singh et al. 2018). Additionally, other clade B PP2Cs regulate seed germination, stomatal development, and defense response via mitogen-activated protein kinase (MAPK) signaling pathway in Arabidopsis (Brock et al. 2010; Julijia et al. 2010; Meskiene et al. 1998; Schweighofer et al. 2007; Shubchynskyy et al. 2017; Sidonskaya et al. 2016; Umbrasaite et al. 2010). In maize, some clade A PP2C members have also been characterized and confirmed to increase seed germination and regulate stress response (He et al. 2019; Hu et al. 2010; Xiang et al. 2017). However, the function of other clade PP2C members of maize remains obscure.
In our previous study, maize ZmPP2C26 was identified as clade B members of ZmPP2Cs and found to negatively regulate drought tolerance in transgenic Arabidopsis (Wang et al. 2018; Zhang et al. 2018). However, the upstream regulatory mechanism remains unknown. In this study, hence, the expression of ZmPP2C26 in maize was analyzed by qRT-PCR. Subsequently, the ZmPP2C26 promoter was isolated, truncated at 5′-end, fused with GUS and l LUC gene, which were used for promoter driven-activity evaluation to provide insights into exploring the potential upstream factors acting on ZmPP2C26 under drought stress.
Materials and methods
Plant materials and growth condition
The seeds of maize inbred lines 87-1 were germinated in a petri dish. Subsequently, the seedlings were transplanted into a plastic mesh grid for hydroponic culture at 28 °C under a photoperiod of 14 h light/10 h dark and used for PEG treatment.
Tobacco (Nicotiana benthamiana) seeds were evenly sown in the flowerpot with nutrient soil and allowed to germinate and grow at 25 °C for 2 weeks. The seedlings were then transplanted into some small pots, and grown in a tissue culture chamber under a 16 h light/8 h night for 6 weeks at 25 °C and used for Agrobacterium transformation.
RNA extraction and qRT-PCR ananlysis
At the three-leaf stage, maize seedings were divided into three groups and treated with 16% (w/v) PEG-6000 solution. At 0 (control), 3, 6, 9, 12, 24 and 48 h of treatment, the shoot and root were sampled and immediately ground in liquid nitrogen for RNA extraction with three replicates, respectively. Total RNA was extracted using RNAiso plus kit (TaKaRa, Japan), quantified using NanoDrop™ OneC (ThermoScientific, USA), and reverse transcribed into cDNA using the PrimeScript™ reagent kit (TaKaRa, Japan) according to the manufacturer’s instruction. The specific primer pairs qPf/qPr and qGf/qGr (Table1) were designed using PrimerBlast (https://www.ncbi.nlm.nih.gov/tools/primer-blast/), synthesized at Sangon biotech (Shanghai, China), used to amplify 358 bp fragment of ZmPP2C26 gene, and 171 bp fragment of ZmGAPDH gene that was used as internal reference, respectively. The cDNA samples were used as templates for qRT-PCR by using ChamQ Universal SYBR qPCR Master Mix (Vazyme, Nanjing) in CFX96™ Real Time System (Bio-Rad, USA). As described by Sun et al. (2020), the two-step temperature cycle was performed as follows: 95 °C for 30 s; 40 cycles of 95 °C for 10 s, 60 °C for 20 s; at the end of the last cycle, the temperature was increased to 95 °C at 0.5 °C/s, so that the melting curve could be calculated and used to differentiate specific and non-specific amplicons. The 2−ΔΔCT method of the CFX Manger™ software version 2.0 (Bio-Rad, USA) was used to normalize the deferential gene expression between ZmPP2C26 and ZmGAPDH gene.
Table 1.
The primers used in the study
| Primer name | Sequence (5′-3′) | Description |
|---|---|---|
| qPf | GGGAGGACGAGAAGGAAAGG | qRT-PCR of ZmPP2C26 gene |
| qPr | AACTGCACGATCACGACACT | |
| qGf | TGAATGGCAAGCTCACTGGT | qRT-PCR of ZmGAPDH gene |
| qGr | TGAATGGCAAGCTCACTGGT | |
| PF0 | TCACTTTACCATTTTTATGCGGGA | Forward primer for P2175 amplification |
| PR | CGGGGCTAGGGTTTTTTTTTCTTTC | Reverse primer for P2175 amplification |
| gus-PF0 | acgacggccagtgccaagcttTCACTTTACCATTTTTATGCGGGA | Forward primers for PCR of r promoter::GUS construct |
| gus-PF1 | acgacggccagtgccaagcttGAGCAAATCGACTCCATCCTT | |
| gus-PF2 | acgacggccagtgccaagcttAGCGTCGCGTCTATCCTTCTC | |
| gus-PF3 | acgacggccagtgccaagcttCTACGAAACCGCAAAGTCCAT | |
| gus-PR | tgtgattgtgatgtatctagaCGGGGCTAGGGTTTTTTTTTCTTTC | Reverse primer for PCR of promoter::GUS construct |
| luc- PF0 | ctatagggcgaattgggtacc TCACTTTACCATTTTTATGCGGGA | Forward primers for PCR of promoter::LUC construct |
| luc- PF1 | ctatagggcgaattgggtaccGAGCAAATCGACTCCATCCTT | |
| luc- PF2 | ctatagggcgaattgggtaccAGCGTCGCGTCTATCCTTCTC | |
| luc- PF3 | ctatagggcgaattgggtaccCTACGAAACCGCAAAGTCCAT | |
| luc- PR | caggaattcgatatcaagcttCGGGGCTAGGGTTTTTTTTT | Reverse primer for PCR of promoter::LUC construct |
Isolation and sequence analysis of ZmPP2C26 promoter
The sequence of 2500 bp 5′ flanking region to start codon (ATG) of ZmPP2C26 gene (GenBank accession. KJ855114.1) was retrieved from MaizeGDB and used as reference to design primers to amplify ZmPP2C26 promoter from genomic DNA of maize inbred line 87-1. The specific primers PF0/PR (Table 1) was designed and used to amplify ZmPP2C26 promoter. The PCR product was subcloned into the pMD19-T Vector (TaKaRa, Japan) and confirmed by sequencing. Finally, the 2175 bp fragment was obtained, considered as the full-length promoter and used for cis-acting element analysis by using online programs PlantCARE (https://www.plantcare.co.uk/, Lescot et al. 2002).
Construction of promoter::GUS and promoter::LUC Plasmids
To validate the promoter driven-activity and explore the potential key segments of 2175 bp promoter, three 5′ deleted fragments of ZmPP2C26 promoter in different size (− 1505, − 1084, and − 215 bp to − 1 bp; the “A” in “ATG” of ZmPP2C26 was designated as + 1,) were amplified from 2175 bp promoter using the specific primers (Table 1) that were designed according to the position of cis-acting elements via CE Design V1.04 software. To generate promoter::GUS constructs, the PCR products were inserted into the Xba I/Hind III site of the pRI201-GUS plasmid to replace 35S promoter using ClonExpress II One Step Cloning Kit (Vazyme, Nanjing). To generate promoter::LUC constructs, the PCR products were inserted into the Kpn I/Hind III site of pGreenII0800-LUC plasmid to drive LUC gene. In the pGreenII0800-LUC plasmid, there is a Renilla luciferase gene (REN) and 35S promoter, which was used as reference for LUC activity test.
The reconstructed plasmids were transformed into Agrobacterium tumefaciens GV3101 strain by freeze and thaw method, and used for tobacco transformation. The pRI201-GUS and pGreenII0800-LUC vector both containing 35S promoter was used as positive control for GUS and LUC activity analysis, respectively.
Tobacco transformation and stress treatment
The leaves of 7-weeks-old tobacco seedlings were used for Agrobacterium infection according the methods described by Yu et al. (2019). The GV3101 strains with recombinant plasmids were cultured overnight at 28 °C until OD600 reached at 0.5–0.7 using LB liquid medium containing 50 mg/L rifampicin and 50 mg/L kanamycin. The Agro-cells were centrifugated for 5 min at 8000 r/min and resuspended in the transformation buffer containing 10 mM MES, pH 5.6, 10 mM MgCl2, and 100 μM acetosyringone, and used for infiltration of tobacco leaf abaxial surfaces. After Agro-infiltration, the plants were cultured 36–48 h at 25 °C. For drought stress, the infiltrated leaves were incubated in the liquid 1/2 MS medium supplemented with 20% (w/v) PEG-6000 for 2 h and 4 h. The infiltrated leaves and wild type leaves incubated in the liquid 1/2 MS medium without PEG were used as control. The leaves infiltrated by Agro-cells were sampled and used for further study.
Measurement of GUS activity
Histochemical GUS staining was performed by using GUS staining Kit (Coolaber, Beijing). Subsequently, the GUS activity was measured as described by Jefferson et al. (1987) and Hou et al. (2016) with minor modification. The leaves were sampled and ground in liquid nitrogen. The 100 mg ground powder was transferred into 1.5 ml tubes, suspended with 400 μL extraction buffer containing 50 mM sodium phosphate (pH 7.2), 0.1% Triton X-100, 0.1% 2-Hydroxy-1-ethanethiol, 10 mM EDTA (pH 8.0) and 1 mg/mL SDS, and centrifuged for 10 min at 12,000 r/min. The supernatant was transferred to 1 mM 4-methylumbelliferyl-β-glucuronide (4-MUG) (Solarbio, Beijing) buffer at 37 °C for 4 h. The fluorescence was measured by using fluorescence spectrophotometer (Fluoroskan Ascent FL, Thermo, USA) at the excitation and emission wavelengths of 365 and 455 nm, respectively. Protein concentration was determined as described by Bradford (1976). The GUS activity was normalized with six MU standards (10 nM, 20 nM, 40 nM, 60 nM, 80 nM, 100 nM) under control conditions and calculated as nmol of 4-MU per mg protein per minute.
Measurement of LUC activity
The LUC activity was measured by using Dual Luciferase Reporter Gene Assay Kit (Beyontime, Shanghai). The 100 mg ground powder was suspended with 200 μL lysis buffer and centrifuged at 15,000 r/min for 5 min. The supernatant was transferred into 100 μL firefly luciferase detection reagent and used for LUC measurement on a multifunctional microplate reader. Subsequently, the 100 μL of REN test working solution was added into the above supernatant, which was used to determine REN activity. The relative activity of LUC was calculated as LUC/REN.
Result
Expression of ZmPP2C26 was inhibited by PEG treatment
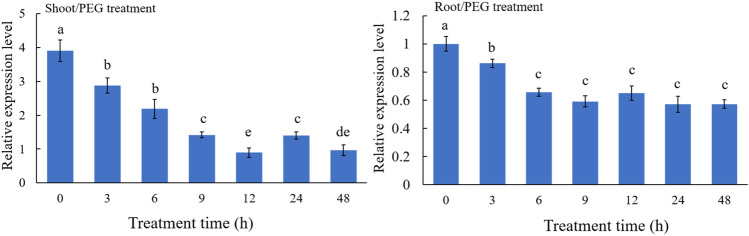
Gene expression patterns can provide important information for gene function. The ZmPP2C26 expression in maize inbred lines 87-1 under PEG treatment was analyzed using qRT-PCR. The results showed that the expression of ZmPP2C26 in maize shoot and root was significantly down-regulated by PEG-6000 treatment, and reached a minimum value at 12 h and 9 h of treatment, respectively (Fig. 1). The finding indicates that ZmPP2C26 genes promoter may be inhibited by some factors under drought stress to down-regulate its expression.
Fig. 1.
Expression pattern of ZmPP2C26 gene. Three-leaf stage of 87-1 seedlings were exposed to water solution supplemented with 16% PEG-6000. Data represent mean ± SD from three biological replicates (n = 3). Different lowercase letters indicate significant differences at P < 0.05
Sequence and cis-elements of ZmPP2C26 promoter
Based on the annotation information of maize genome, the 2175 bp promoter sequence of ZmPP2C26 upstream of the ATG was cloned from maize 87-1 genomic DNA, suggested as a putative full-length promoter, and used to identify cis-acting regulatory elements (Figs. 2 ,3). Apart from the eleven copies of CAAT-box and seven copies of constitutive core elements TATA-box, in the 2175 bp sequence, twelve kinds of putative cis-acting elements including A-Box, ABRE, ABRE4, Box4, CGTCA-motif, DRE1, G-Box, GC-motif, MBS, MYB and Sp1 were identified in the 2175 bp sequence (Table 2).
Fig. 2.
The sequence of ZmPP2C26 promoter. The ‘‘A’’ in ‘‘ATG’’ of ZmPP2C26 gene is designated as ‘‘+ 1”. The predicted cis-acting elements are shown in red color with underline. The core elements CAAT and TATA box are shown in dark grey and white grey background, respectively. The detailed description of elements is listed in Table 2. The black arrow under the sequence indicates the position of primers to amplify different deletion fragments
Fig. 3.
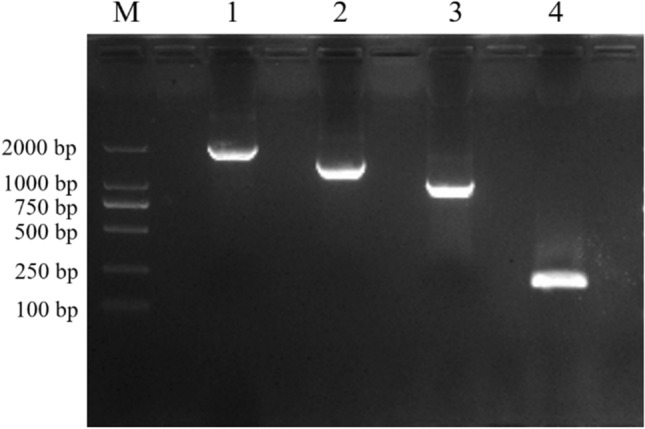
Specific fragments of ZmPP2C26 promoter and three 5′-end deletion fragments amplified by PCR. M: DNA 2000 bp marker. Lanes 1, 2, 3 and 4 represents 2175, 1505, 1084 and 215 bp promoter, respectively and 1.5% agarose gel separation
Table 2.
The cis-acting elements of ZmPP2C26 gene promoter
| Cis element | Number | Description |
|---|---|---|
| AAGAG-motif | 1 | Mediate the regulation of gene expression in signal transduction pathway |
| A-Box | 2 | cis-acting regulatory element |
| ABRE | 1 | cis-acting element involved in abscisic acid responsiveness |
| ABRE4 | 1 | cis-acting element involved in abscisic acid responsiveness |
| Box4 | 4 | Part of a conserved DNA module involved in light responsiveness |
| CAAT-Box | 11 | Common cis-acting element in promoter and enhancer regions |
| CGTCA-motif | 3 | cis-acting regulatory element involved in the MeJA-responsiveness |
| DRE1 | 1 | Dehydration-responsive element |
| G-Box | 1 | cis-acting regulatory element involved in light responsiveness |
| GC-motif | 2 | Enhancer-like element involved in anoxic specific inducibility |
| MBS | 1 | MYB binding site involved in drought response |
| MYB | 1 | MYB cis-acting element |
| Sp1 | 2 | Light responsive element |
| TATA-Box | 7 | Core promoter element around − 30 of transcription start |
Construction of promoter::GUS and promoter::LUC vectors
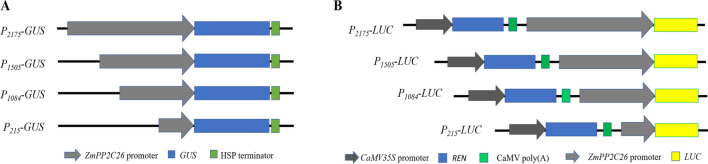
The specific primers (PF0/PR, PF1/PR, PF2/PR, PF3/PR) were used to amplified the 2175 bp promoter and three 5′-end deleted fragment (1505, 1084 and 215 bp) of ZmPP2C26 from maize 87-1 genomic DNA (Fig. 2). As a result, these fragments were specifically amplified and perfectly matched to the reference sequence from maizeGDB, and named as P2175, P1505, P1084 and P215, respectively (Fig. 3). Subsequently, these fragments were cloned into pRI201-GUS and pGreenII 0800-LUC plasmid to generate P2175-GUS, P1505-GUS, P1084-GUS, P215-GUS, P2175-LUC, P1505-LUC, P1084-LUC and P215-LUC plasmid, respectively (Fig. 4).
Fig. 4.
The constructs of promoter deletions of ZmPP2C26. a Diagram of constructs of promoter::GUS. b Diagram of constructs of promoter::LUC. The Renilla luciferase (REN) driven by 35S promoter was used as reference for firefly luciferase (LUC) activity test. The number indicates the length of promoter
The − 1084 to − 215 bp is key region for expression-inhibition
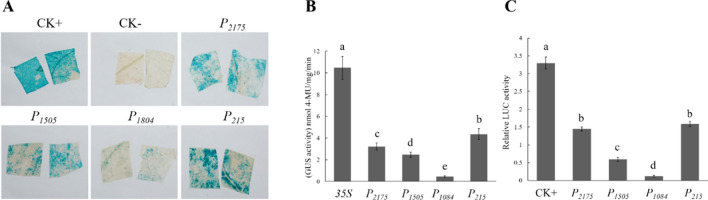
The result of GUS staining showed that the leaf infiltrated with Agrobacterium containing reconstructed constructs exhibited blue color, which was similar to positive control that infiltrated with 35S-GUS (Fig. 5a), indicating that these promoters could drive the gene expression in tobacco leaves. Meanwhile, the GUS and LUC activity assays exhibited that four promoters with different length could drive GUS and LUC expression in different level. However, the activities of GUS and LUC driven by P2175, P1505 and P215 were stronger than that of P1084. Interestingly, P215 showed highest driven-ability among these promoters. The GUS and LUC activities driven by P215 were corresponds to 9.4 and 12.9 folds to P1084, respectively (Fig. 5b, c). These results suggest that the sequence of − 1084 to − 215 bp is key region to inhibit ZmPP2C26 expression. It’s also concluded that the − 215 bp sequence may be the core functional region of ZmPP2C26 promoter, which contains one cope of GC-motif element, ABRE element, DRE1 element and core element TATA-box (Fig. 2).
Fig. 5.
The promoter activity during transient expression in tobacco leaves. P2175, P1505, P1084 and P215 indicate different length promoter. a GUS staining. CK+: infiltrated with 35S-GUS plasmid as positive control; CK−: un-infiltrated wild type. b Fluorometric GUS analysis. c Detection of LUC activity. All values are means (± SE) of three biological replicates. Different lowercase letters indicate significant differences at P < 0.05
The driven-activity of ZmPP2C26 promoter was inhibited by drought stress
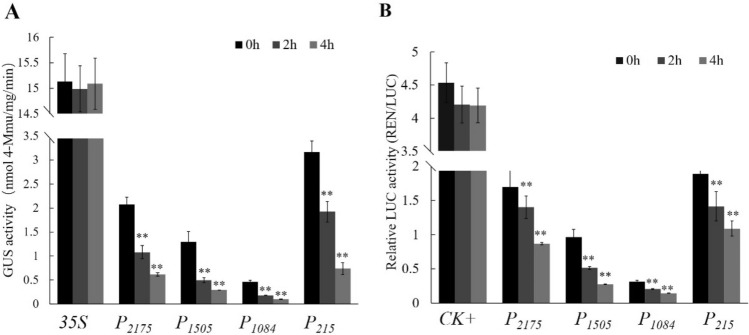
Our previous study showed that ZmPP2C26 negatively regulated drought tolerance in Arabidopsis (Zhang et al. 2018). In order to investigate whether ZmPP2C26 promoter is involved in drought stress, the tobacco leaves infiltrated with Agrobacterium were subjected to 20% PEG-6000. As shown in Fig. 6, the activity of GUS and LUC driven by 35S promoter showed no significant difference after PEG-6000 treatment. However, at 2 and 4 h of PEG-6000 treatment, the activity of GUS and LUC driven by P2175, P1084, P1505 and P215 was significantly reduced compared to control (0 h). Meanwhile, P215 still showed highest driven-ability among these promoters, which was also observed before treatment. Notably, the sequence of − 1084 to − 215 bp contains one MBS elements that is MYB transcription factor binding site involved in drought-response (Fig. 2). Therefore, we speculate that the − 1084 to − 215 bp region of ZmPP2C26 promoter may recruit potential transcription factors to inhibit ZmPP2C26 expression under drought stress and negatively regulate drought tolerance in maize.
Fig. 6.
Quantitative analysis of GUS and LUC activities under PEG-6000 treatment. a GUS activity. b LUC activity. The tobacco leaves were incubated in the liquid 1/2 MS medium supplemented with 20% (w/v) PEG-6000 for 2 and 4 h. The leaves incubated in the liquid 1/2MS medium were used as control. Values represent the mean ± standard deviation among replicates. *P < 0.05, **P < 0.01
Discussion
Protein phosphorylation or dephosphorylation plays pivotal roles in various of signaling cascade and functional modification of transport proteins in plants (He et al. 2019; Lee et al. 2009; Singh et al. 2018). PP2Cs catalyze protein dephosphorylation, which is important for plants growth, development, and stress response. The clade A members of PP2C are key factors in the ABA signaling pathway (Cheng et al. 1999; Yang et al. 2001). The PP2C activity is inhibited by binding ABA and its direct receptor PYL, which negatively regulates ABA signaling. (Lu et al. 2017; Née et al. 2017; Nishimura et al. 2018; Xiang et al. 2017). In our previous study, we found that ZmPP2C26, a member of clade B, negatively regulate drought tolerance in transgenic Arabidopsis (Zhang et al. 2018). Therefore, it is proposed that the upstream promoter of ZmPP2C26 may play a crucial role in drought response in maize. To uncover the hypothesis, the ZmPP2C26 promoter was cloned and evaluated for its driven-ability under drought stress.
Under drought stress, the expression of ZmPP2C26 in root and shoot of maize seedlings was significantly down-regulated (Fig. 1), indicating that its promoter may be inhibited by drought stress. The finding further facilitates us to investigate the function of ZmPP2C26 promoter. As well known, promoter possesses cis-acting elements to drive gene expression (Connors et al. 2002; Yu et al. 2019). In this study, ZmPP2C26 promoter region contains 11 copies of CAAT-box and 7 copies of TATA-box, which are core elements for promoters. Meanwhile, some environmental stimuli responsive elements are found in the ZmPP2C26 promoter, including MBS (related to drought stress), ABRE and ABRE4 (abscisic acid responsive elements), Box4, G-Box and Sp1 (light responsive elements), and CGTCA-motif (MeJA-responsive elements), which may contribute to the resilience of ZmPP2C26 gene to stress response.
The analysis of 5′-end truncated fragments revealed that the sequence of − 215 bp is the key promoter region for ZmPP2C26 expression (Fig. 5 and 6). The TATA-box in this region may be a crucial element for the promoter (Fig. 2). Moreover, the P1084 promoter showed lowest activity and P215 promoter activity was significantly higher than P1505 (Fig. 5 and 6). We found that there was an MBS element (MYB binding site) and no other cis-acting element related to drought stress in the − 1084 to − 215 region (Fig. 2). MYBs, an abundant kinds of transcription factors in plants, are found to regulate the plant tolerance to drought stress (Shan et al. 2012; Wu et al. 2019; Zhao et al. 2019). Therefore, we speculate that the sequence of − 1084 to − 215 bp fragment may be a key part for expression-inhibition under drought stress, and some MYBs may bind to this site to inhibit the ZmPP2C26 expression under drought stress. In addition to MBS, ABRE (ABA-responsive element, − 1505 to − 1084 bp), ABRE4 (− 215 to − 1 bp) and DRE1 (dehydration-responsive element, − 215 to − 1 bp) in the ZmPP2C26 promoter bonded by AREB/ABF or DREB transcription factors may also contribute for drought response through binding (Sharma et al. 2019; Takuya et al. 2010; Yoshihiro et al. 2003). Although they are not confirmed to negatively regulate ZmPP2C26 expression under drought stress, these elements are candidates for further validation.
In summary, the − 215 bp length is enough to drive ZmPP2C26 expression. Besides, the − 1084 to − 215 fragment of ZmPP2C26 promoter may recruit potential transcription factors to inhibit ZmPP2C26 expression under drought stress and negatively regulate drought tolerance in maize. These findings will provide insights into understanding the molecular mechanism of ZmPP2C26 regulating drought tolerance.
Acknowledgements
This study was supported by Sichuan Science and Technology Program (2018JY0470 and 2020YJ0353) and Key Laboratory of Dry-hot Valley Characteristic Bio-Resources Development at university of Sichuan Province (GR-2019-E-01).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Fengling Fu and Haoqiang Yu have contributed equally to this work.
Contributor Information
Fengling Fu, Email: ffl@sicau.edu.cn.
Haoqiang Yu, Email: yhq1801@sicau.edu.cn.
References
- Antoni R, Gonzalezguzman M, Rodriguez L, Rodrigues A, Pizzio GA, Rodrigues P. Selective inhibition of clade a phosphatases type 2c by PYR/PYL/RCAR abscisic acid receptors. Plant Physiol. 2012;158(2):970–980. doi: 10.1104/pp.111.188623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskara GB, Nguyen TT, Verslues PE. Unique drought resistance functions of the highly ABA-induced clade A protein phosphatase 2Cs. Plant Physiol. 2012;160(1):379–395. doi: 10.1104/pp.112.202408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72(1–2):248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Brock AK, Willmann R, Kolb D, Grefen L, Lajunen HM, Bethke G, Lee J, Nurnberger T, Gust AA. The Arabidopsis mitogen-activated protein kinase phosphatase PP2C5 affects seed germination, stomatal aperture, and abscisic acid-inducible gene expression. Plant Physiol. 2010;153(3):1098–1111. doi: 10.1104/pp.110.156109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AY, Ross KE, Kaldis P, Solomon MJ. Dephosphorylation of cyclin-dependent kinases by type 2C protein phosphatases. Genes Dev. 1999;13(22):2946–2957. doi: 10.1101/gad.13.22.2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connors BJ, Miller M, Maynard CA, Powell WA. Cloning and characterization of promoters from American chestnut capable of directing reporter gene expression in transgenicArabidopsis plants. Plant Sci. 2002;163(4):781. [Google Scholar]
- Han L, Li JL, Jin M, Su YH. Functional analysis of a type 2C protein phosphatase gene from Ammopiptanthus mongolicus. Gene. 2018;653:29–42. doi: 10.1016/j.gene.2018.02.015. [DOI] [PubMed] [Google Scholar]
- He ZH, Wu JF, Sun XP, Dai MQ. The maize clade A PP2C phosphatases play critical roles in multiple abiotic stress responses. Int J Mol Sci. 2019;20(14):3573. doi: 10.3390/ijms20143573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou JJ, Jiang PP, Qi SM, Zhang K, He QX, Xu CZ, Ding ZH, Zhang KW, Li KP. Isolation and functional validation of salinity and osmotic stress inducible promoter from the maize type-II H+-pyrophosphatase gene by deletion analysis in transgenic tobacco plants. PLoS ONE. 2016;11(4):e0154041. doi: 10.1371/journal.pone.0154041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Liu L, Xiao B, Li D, Xing X, Kong X, Li D. Enhanced tolerance to low temperature in tobacco by over-expression of a new maize protein phosphatase 2C, ZmPP2C2. J Plant Physiol. 2010;167(15):1307–1315. doi: 10.1016/j.jplph.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, KavanaghTA BMWA. GUS fusion: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6(13):3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julijia U, Alois S, Vaiva K, Zoltan M, Zahra A, Verena U, Chonnanit C, Justyna B, James AHM, Laszlo B, Irute M. MAPK phosphatase AP2C3 induces ectopic proliferation of epidermal cells leading to stomata development in Arabidopsis. PLoS ONE. 2010;5(12):1–18. doi: 10.1371/journal.pone.0015357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu K, Suzuki N, Kuwamura M, Nishikawa Y, Nakatani M, Ohtawa H, Takezawa D, Seki M, Tanaka M, Taji T, Hayashi T, Sakata Y. Group A PP2Cs evolved in land plants as key regulators of intrinsic desiccation tolerance. Nat Commun. 2013;4(2219):375–381. doi: 10.1038/ncomms3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Lan W, Buchanan BB, Luan S. A protein kinase phosphatase pair interacts with an iron channel to regulate ABA signaling in plant guard cells. Proc Natl Acad Sci USA. 2009;106(50):21419–21424. doi: 10.1073/pnas.0910601106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van PY, Rouzé P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30(1):325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan S. Protein phosphatases in plants. Annu Rev Plant Biol. 2003;54(54):63–92. doi: 10.1146/annurev.arplant.54.031902.134743. [DOI] [PubMed] [Google Scholar]
- Lu X, Xiong Q, Cheng T, Li QT, Liu XL, Bi YD, Li W, Zhang WK, Ma B, Lai YC, Du WG, Man WQ, Chen SY, Zhang JS. A PP2C-1 allele underlying a quantitative trait locus enhances soybean 100-seed weight. Mol Plant. 2017;10(5):670–684. doi: 10.1016/j.molp.2017.03.006. [DOI] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324(5930):1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- Meskiene I, Bogre L, Glaser W, Balog J, Brandstotter M, Zwerger K, Ammerer G, Hirt H. MP2C, a plant protein phosphatase 2C, functions as a negative regulator of mitogen-activated protein kinase pathways in yeast and plants. Proc Natl Acad Sci USA. 1998;95(4):1938–1943. doi: 10.1073/pnas.95.4.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Née G, Kramer K, Nakabayashi K, Yuan BJ, Xiang Y, Miatton E, Finkemeier I, Soppe WJJ. DELAY OF GERMINATION1 requires PP2C phosphatases of the ABA signaling pathway to control seed dormancy. Nat Commun. 2017;8(1):72. doi: 10.1038/s41467-017-00113-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Tsuchiya W, Moresco JJ, Hayashi Y, Satoh K, Kaiwa N, Irisa T, Kinoshita T, Schroeder JI, Yates JR, Hirayama T, Yamazaki T. Control of seed dormancy and germination by DOG1-AHG1 PP2C phosphatase complex via binding to heme. Nat Commun. 2018;9(1):2132. doi: 10.1038/s41467-018-04437-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, Alfred SE, Bonetta D, Finkelstein R, Provart NJ, Desveaux D, Rodriguez PL, McCourt P, Zhu JK, Schroeder JI, Volkman BF. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324(5930):1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez. PL. Protein phosphatase 2C (PP2C) function in higher plants. Plant Mol Biol. 1998;38(6):919–927. doi: 10.1023/a:1006054607850. [DOI] [PubMed] [Google Scholar]
- Schweighofer A, Hirt H, Meskiene I. Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci. 2004;9(5):236–243. doi: 10.1016/j.tplants.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Schweighofer A, Kazanaviciute V, Scheikl E, Teige M, Doczi R, Hirt H, Schwanninger M, Merijn KM, Schuurink R, Mauch F, Buchala A, Cardinale F, Meskiene I. The PP2C-type phosphatase AP2C1, which negatively regulates MPK4 and MPK6, modulates innate immunity, jasmonic acid, and ethylene levels in Arabidopsis. Plant Cell. 2007;19(7):2213–2224. doi: 10.1105/tpc.106.049585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan H, Chen SM, Jiang JF, Chen FD, Chen Y, Gu CS, Li PL, Song AP, Zhu XR, Gao HS, Zhou GQ, Li T, Yang X. Heterologous expression of the Chrysanthemum R2R3-MYB transcription factor CmMYB2 enhances drought and salinity tolerance, increases hypersensitivity to ABA and delays flowering in Arabidopsis thaliana. Mol Biotechnol. 2012;51(2):160–173. doi: 10.1007/s12033-011-9451-1. [DOI] [PubMed] [Google Scholar]
- Sharma V, Goel P, Kumar S, Singh AK. An apple transcription factor, MdDREB76, confers salt and drought tolerance in transgenic tobacco by activating the expression of stress-responsive genes. Plant Cell Rep. 2019;38(2):221–241. doi: 10.1007/s00299-018-2364-8. [DOI] [PubMed] [Google Scholar]
- Shubchynskyy V, Boniecka J, Schweighofer A, Simulis J, Kvederaviciute K, Stumpe M, Mauch F, Balazadeh S, Mueller-Roeber B, Boutrot F, Zipfel C, Meskiene I. Protein phosphatase AP2C1 negatively regulates basal resistance and defense responses to Pseudomonas syringae. J Exp Bot. 2017;68(5):1169–1183. doi: 10.1093/jxb/erw485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidonskaya E, Schweighofer A, Shubchynskyy V, Kammerhofer N, Hofmann J, Wieczorek K, Meskiene I. Plant resistance against the parasitic nematode Heterodera schachtii is mediated by MPK3 and MPK6 kinases, which are controlled by the MAPK phosphatase AP2C1 in Arabidopsis. J Exp Bot. 2016;67(1):107–118. doi: 10.1093/jxb/erv440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Yadav AK, Kaur K, Sanyal SK, Jha SK, Fernandes JL, Sharma P, Tokas I, Pandey A, Luan S, Pandey GK. A protein phosphatase 2C, AP2C1, interacts with and negatively regulates the function of CIPK9 under potassium-deficient conditions in Arabidopsis. J Exp Bot. 2018;69(16):4003–4015. doi: 10.1093/jxb/ery182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun FA, Yu HQ, Qu JT, Cao Y, Ding L, Feng WQ, Muhammad HBK, Li WC, Fu FL. Maize ZmBES1/BZR1-5 decreases aba sensitivity and confers tolerance to osmotic stress in transgenic Arabidopsis. Int J Mol Sci. 2020;21(3):996. doi: 10.3390/ijms21030996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takuya Y, Fujita Y, Sayama H, Satoshi K, Kyonoshin M, Junya M, Kazuo S, Kazuko YS. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 2010;61(4):672–685. doi: 10.1111/j.1365-313X.2009.04092.x. [DOI] [PubMed] [Google Scholar]
- Umbrasaite J, Schweighofer A, Kazanaviciute V, Magyar Z, Ayatollahi Z, Unterwurzacher V, Choopayak C, Boniecka J, Murray JA, Bogre L, Meskiene I. MAPK Phosphatase AP2C3 Induces Ectopic Proliferation of Epidermal Cells Leading to Stomata Development in Arabidopsis. PLoS ONE. 2010;5(12):e15357. doi: 10.1371/journal.pone.0015357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YG, Fu FL, Yu HQ, Hu T, Zhang YY, Tao Y, Zhu JK, Zhao Y, Li WC. Interaction network of core ABA signaling components in maize. Plant Mol Biol. 2018;96(3):245–263. doi: 10.1007/s11103-017-0692-7. [DOI] [PubMed] [Google Scholar]
- Wu J, Jiang Y, Liang Y, Chen L, Chen W, Cheng B. Expression of the maize MYB transcription factor ZmMYB3R enhances drought and salt stress tolerance in transgenic plants. Plant Physiol Biochem. 2019;137:179–188. doi: 10.1016/j.plaphy.2019.02.010. [DOI] [PubMed] [Google Scholar]
- Xiang YL, Sun XP, Gao S, Qin F, Dai MQ. Deletion of an endoplasmic reticulum stress response element in a ZmPP2C-A gene facilitates drought tolerance of maize seedlings. Mol Plant. 2017;10(3):456–469. doi: 10.1016/j.molp.2016.10.003. [DOI] [PubMed] [Google Scholar]
- Yang K, Jeonghoe DH, Jang S. Molecular cloning and characterization of a rice PP2C, OsPP2C4. J Plant Biol. 2001;44(1):1–6. [Google Scholar]
- Yoshihiro N, Kazuo N, Zatba KS, Yoh S, Takashi F, Hiroshi A, Mari N, Kazuo S, Kazuko YS. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 2003;34(2):137–148. doi: 10.1046/j.1365-313x.2003.01708.x. [DOI] [PubMed] [Google Scholar]
- Yu HQ, Muhammad HBK, Lu FZ, Sun FA, Qu JT, Liu BL, Li WC, Fu FL. Isolation and identification of a vegetative organ-specific promoter from maize. Physiol Mol Biol Plant. 2019;25(1):277–287. doi: 10.1007/s12298-018-0546-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang PW, Zhang XW, Yu HQ, Lx P, Fu FL, Lx WC. Function of two alternative splice variants of protein phosphatase type 2C gene ZmPP2C26 of maize. J Northwest A&F Univ. 2018;46(7):23–31. [Google Scholar]
- Zhao P, Hou S, Guo X, Jia J, Cheng L. A MYB-related transcription factor from sheep grass, LcMYB2, promotes seed germination and root growth under drought stress. BMC Plant Biol. 2019;19:564. doi: 10.1186/s12870-019-2159-2. [DOI] [PMC free article] [PubMed] [Google Scholar]