Abstract
MRSA infection and colonization have been reported in both companion and food-chain animals, highlighting MRSA as an important veterinary and zoonotic pathogen. Another mec allele, the mecC gene, also confers beta-lactam resistance in Staphylococcus aureus and shows 69% nucleotide identity to mecA. The main aim of this study was to investigate the genotypic and clonal profile of methicillin-resistant S. aureus (MRSA) from cows with mastitis in dairy herds. Thirty-five samples suggestive of bovine subclinical mastitis were evaluated, and S. aureus were detected in all of them using both phenotypic and molecular approaches. According to the multilocus sequence typing (MLST), the S. aureus isolates were assigned in five different STs, 21 (60%) showed ST 742, 6 (17%) ST97, 4 (11%) ST1, 2 (6%) ST30, and 2 (6%) ST126. The presence of mecA was not observed in any of these isolates whereas mecC was detected in nine of them (9/35; 26%). The Panton-Valentine leukocidin (PVL) genes were detected in a total of 4 isolates. Among the 35 isolates analyzed, 26 showed resistance to penicillin. Changes in the S. aureus epidemiology due to the detection of MRSA in milk samples from cows presenting with bovine subclinical mastitis may have consequences for public health in Brazil, challenging the empirical therapy and animal management, with potential medical and social outcomes. To the best of our knowledge, this is the first report describing mecC MRSA in Southeastern Brazil.
Keywords: Staphylococcus aureus, MRSA, mecC, Bovine mastitis
Methicillin-resistant S. aureus (MRSA) are still significant human and animal pathogens, causing serious and lethal infections, with inestimable public health consequences and major economic impacts [1]. MRSA strains normally contain mecA, a gene that encodes for the penicillin-binding protein 2a (PBP2a). This modified enzyme induces resistance to virtually all β-lactam antibiotics, making MRSA a global public health concern [2].
MRSA cases have emerged in production animals in the last years [3]. These bacteria can be transferred to man by direct contact or as food contaminants [4]. A mecA homolog in S. aureus, the mecC, which confers resistance to beta-lactams through a similar mechanisms (production of a PBP2a/2′ with about 63% identity at the amino acid level), was first reported in Denmark and more recently in Czech Republic [3, 5]. The mecC allele has been identified in the so-called livestock-associated-MRSA (LA-MRSA) belonging to different MLST clonal complexes [6, 7]. LA-MRSA has been identified in milk and beef from different livestock animals [3, 8], reinforcing MRSA as a serious threat to public health worldwide. In the present study, we investigated the genotypic and clonal profiles of methicillin-resistant S. aureus (MRSA) from cows presented with mastitis in a dairy herd from the states of Minas Gerais, Rio de Janeiro, and São Paulo.
A total of 35 staphylococci isolates were obtained from milk samples (15 from Minas Gerais, 15 from Rio de Janeiro, and 5 from São Paulo) from cows presented with subclinical mastitis. All milk samples were obtained from April 2015 to December 2015 and kindly provided by Brazilian Agricultural Research Corporation (EMBRAPA) Dairy Cattle, Juiz de Fora, MG. Isolates were confirmed as S. aureus by mass spectrophotometer in a MALDI-TOF (matrix-assisted laser desorption ionization–time off flight—Biotyper-Bruker) and PCR methodology previously described [9].
Antimicrobial susceptibility tests were performed using agar disk-diffusion method on Mueller Hinton agar (Difco), for all S. aureus isolates [10, 11]. The following susceptibility disks were used: cefoxitin (CFO-30 μg), chloramphenicol (CLO-30 μg), ciprofloxacin (CIP-5 μg), clindamycin (CLI-2 μg), erythromycin (ERI-15 μg), gentamicin (GEN-10 μg), rifamycin (RIF-5 μg), tetracycline (TET-30 μg), trimethoprim-sulfamethoxazole (SUT-23,75 μg), nitrofurantoin (NIT-300 μg), and penicillin (PEN-10 U). Also, the MIC for vancomycin was determined by using the Oxoid® M.I.C. Evaluator Strips ™ (M.I.C.E., Thermo Fisher Scientific, Basingstoke, UK) and broth microdiluting using microtiter plates [10].
Detection of mecA, mecC, and lukSF-PVL genes were performed by PCR-based tests as described previously [12–14]. All primer sequences are listed in Table 1. Methicillin-resistant S. aureus (MRSA) strains were also characterized by performing multilocus sequence typing (MLST) [15] and spa typing [16]. To assign the MLST sequence types, the allele sequences were trimmed and analyzed using the S. aureus MLST database (http://www.pubmlst.org). Sequence analysis and phylogeny were performed using the BioEdit Sequence Alignment Editor v7.2.5. Phylogenetic trees were constructed by Maximum Likelihood Tree using MEGA v7.0.21.
Table 1.
Primers used to amplify the mecC and mecA genes in MRSA isolates recovered from bovine mastitis
S. aureus isolates were mostly resistant to penicillin (15 isolates). Also, eight isolates were resistant to tetracycline. In addition, all S. aureus detected were considered susceptible to vancomycin. Results of susceptibility are depicted in Table 2. The presence of mecA could not be observed in any isolate. Notably, mecC was detected in nine S. aureus isolates (26%). The Panton-Valentine leukocidin (PVL) encoding genes were detected in a total of four isolates (11%).
Table 2.
Characterization of Staphylococcus aureus isolates associated with bovine mastitis in the states of Rio de Janeiro, Minas Gerais, and Sao Paulo (Brazil)
| Isolates | Region | Id. | resistance profile | MR | Spa type | ST | CC |
|---|---|---|---|---|---|---|---|
| MB01 | MG | S. aureus | pen | t605 | ST742 | CC97 | |
| MB02 | MG | S. aureus | cli-pen | mecC + | t605 | ST742 | CC97 |
| MB03 | MG | S. aureus | gen-pen-tet | t1298 | ST30 | CC30 | |
| MB04 | RJ | S. aureus | pen | t605 | ST742 | CC97 | |
| MB05 | RJ | S. aureus | – | t591 | ST126 | CC97 | |
| MB06 | RJ | S. aureus | cli-eri-pen | t605 | ST742 | CC97 | |
| MB07 | RJ | S. aureus | pen-tet | t605 | ST742 | CC97 | |
| MB08 | SP | S. aureus | pen | t521 | ST97 | CC97 | |
| MB09 | SP | S. aureus | pen | t605 | ST742 | CC97 | |
| MB10 | RJ | S. aureus | clo-cli-eri-gen-nit-pen-rif-tet | mecC + | t267 | ST97 | CC97 |
| MB11 | MG | S. aureus | pen | t605 | ST742 | CC97 | |
| MB12 | SP | S. aureus | pen | mecC + | t605 | ST742 | CC97 |
| MB13 | SP | S. aureus | pen | t1298 | ST30 | CC30 | |
| MB14 | RJ | S. aureus | pen | mecC + | t605 | ST742 | CC97 |
| MB15 | MG | S. aureus | pen | mecC + | t359 | ST97 | CC97 |
| MB16 | MG | S. aureus | pen | mecC + | t359 | ST97 | CC97 |
| MB17 | MG | S. aureus | – | t605 | ST742 | CC97 | |
| MB18 | MG | S. aureus | – | t605 | ST742 | CC97 | |
| MB19 | MG | S. aureus | pen-tet | t605 | ST742 | CC97 | |
| MB20 | RJ | S. aureus | nit-pen | mecC + | t605 | ST742 | CC97 |
| MB21 | RJ | S. aureus | pen | mecC + | t521 | ST97 | CC97 |
| MB22 | RJ | S. aureus | pen | t605 | ST742 | CC97 | |
| MB23 | MG | S. aureus | – | t127 | ST1 | CC1 | |
| MB24 | RJ | S. aureus | pen-tet | t605 | ST742 | CC97 | |
| MB25 | MG | S. aureus | – | t127 | ST1 | CC1 | |
| MB26 | MG | S. aureus | – | t127 | ST1 | CC1 | |
| MB27 | SP | S. aureus | pen | t605 | ST742 | CC97 | |
| MB28 | SP | S. aureus | oxa-pen | t605 | ST742 | CC97 | |
| MB29 | MG | S. aureus | pen | t605 | ST742 | CC97 | |
| MB30 | RJ | S. aureus | eri-nit-pen-sut | t605 | ST742 | CC97 | |
| MB31 | SP | S. aureus | pen | t605 | ST742 | CC97 | |
| MB32 | MG | S. aureus | – | t127 | ST1 | CC1 | |
| MB33 | MG | S. aureus | cli-eri-pen-tet | Non-typeable | ST126 | CC97 | |
| MB34 | RJ | S. aureus | pen | t605 | ST742 | CC97 | |
| MB35 | RJ | S. aureus | eri-cli-pen-tet | mecC + | t359 | ST97 | CC97 |
MG Minas Gerais. SP São Paulo, RJ Rio de Janeiro, CFO cefoxitin, CLO chloramphenicol, CLI clindamycin, ERI erythromycin, GEN gentamicin, NIT nitrofurantoin, PEN penicillin, RIF rifamycin, SUT trimethoprim-sulfamethoxazole, TET tetracycline, MR methicillin-resistance, ST sequence type, CC clonal complex
The total S. aureus isolates from mastitis were assigned in five different sequence types (STs), whereas 21 isolates (60%) were ST 742, 6 (17%) ST97, four (11%) ST1, two (6%) ST30, and two (6%) ST126 (Table 1). Moreover, the PVL genes were most frequently detected in the ST742 lineage MRSA in a total of 14% (3/21). MRSA isolates harboring the mecC gene were assigned in the ST742.
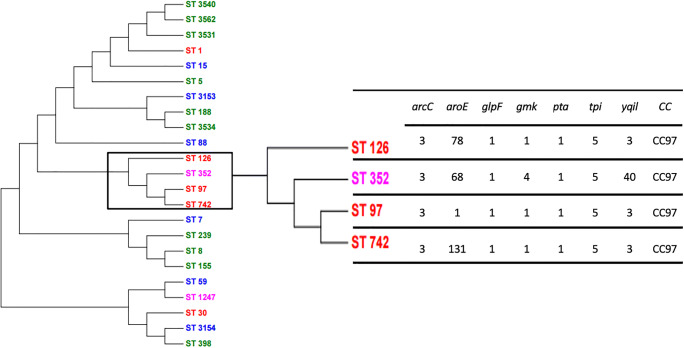
According to phylogenetic analyses, ST 742 and ST97 form a cluster, differing only in the aroE gene. These STs are phylogenetically close to ST126 with differences in aroE gene. Interestingly, the ST352 found in milk samples from Portugal also aligned close to the ST742 and ST97, differing only in the aroE gene (Fig. 1).
Fig. 1.
Maximum likelihood phylogenetic tree of Staphylococcus aureus STs: In red, isolates from this study. In green, other Brazilian isolates. In blue, China isolates. In pink: isolates from Portugal. The STs are accessed at PubMLST.org: Public databases for molecular typing https://pubmlst.org/ Multilocus sequence typing (MLST) databases and software
In Brazil, there are only few studies identifying cases of bovine mastitis caused by MRSA. Recently, Guimarães et al. (2017) reported an outbreak of intramammary infections associated to MRSA in São Paulo [17]. In the present study, we identified a high percentage of MRSA (26%) in bovine subclinical mastitis. This is the first report of mecC-positive LA-MRSA isolates in Southeastern Brazil. Just recently, Silva and co-workers reported the occurrence of a LA-MRSA ST126 harboring the mecC variant in the North of Brazil [18]. MRSA harboring mecC has already been identified in milk samples from animal origin in previous studies elsewhere [7, 12, 19, 20]. Notably, all MRSA isolates harboring the mecC gene in the present study did not express resistance phenotype to oxacillin in the vitro testing (OS-MRSA—oxacillin-susceptible mecC-positive S. aureus). In addition, the mecC isolates were also susceptible to cefoxitin disks. Although we did not test that, cefoxitin-agar screening plates might be a more suitable method for detecting these mecC isolates. In spite of that, from these and other studies, it is clear that a PCR-based method to detect mecC gene is required [21].
The sequence types identified among the mecC isolates studied were also rarely found. MRSA belonging to ST97 had also been identified in milk samples from bovine mastitis in China [22] and Tunisia [23] and also from pigs in Japan [24]. Yet, all MRSA isolates from these studies carried the mecA gene, while in our study, ST97 harboring mecC was for the first time identified. In Brazil, only few studies reported MRSA ST398 [25, 26], ST126, and ST133 [27]. Besides ST97, we identified mecC in a ST746 strain, and to the best of our knowledge, a mecC ST746 has never been reported before. Interestingly, the MRSA ST746 is genetically similar to ST97, varying in the aroE gene. Only recently, Silva and co-workers identified an MRSA ST126 harboring the mecC gene [18]. We identify ST126 among the MSSA isolates, but not among the mecC MRSA detected. Our report, in addition to the study by Silva and collaborators [18], may indicate a more widely spread of mecC among livestock-associated S. aureus in Brazil. Future studies are needed to investigate the extent of this change in S. aureus epidemiology, the animal management, and the potential dissemination of mecC strains in humans, in order to prevent public health and economic impacts. Although we have studied a limited number of milk samples, our data reinforces the entrance of the mecC LA-MRSA in Brazil and the need to include mecC primers when searching for MRSA in clinical specimens from animal and human origins.
Acknowledgments
Maria Aparecida V Paiva E Brito of Brazilian Agricultural Research Corporation (EMBRAPA), Dairy Cattle, Juiz de Fora, MG/Brazil, Laboratory of Prague Studies and Parasites—Faculty of Biology—Federal Fluminense University for yielding the samples.
The authors received financial support of the CNPq (406057/2016-8) and Faperj (Processo E-26/203.293/2017). This study was supported in part by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES)-Finance Code 001.
Compliance with ethical standards
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Bruno Penna, Email: bpenna@id.uff.br.
Evelize Folly, Email: evelizefolly@yahoo.com.br, Email: efolly@id.uff.br.
References
- 1.Hewagama S, Spelman T, Woolley M, McLeod J, Gordon D, Einsiedel L. The epidemiology of Staphylococcus aureus and Panton-Valentine leucocidin (pvl) in Central Australia, 2006-2010. BMC Infect Dis. 2016;16:1–6. doi: 10.1186/s12879-016-1698-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loncaric I, Tichy A, Handler S, Szostak M, Tickert M, Diab-Elschahawi M, Spergser J, Künzel F. Prevalence of methicillin-resistant Staphylococcus sp. (MRS) in different companion animals and determination of risk factors for colonization with MRS. Antibiotics. 2019;8:1–9. doi: 10.3390/antibiotics8020036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petersen A, Stegger M, Heltberg O, Christensen J, Zeuthen A, Knudsen LK, Urth T, Sorum M, Schouls L, Larsen J, Skov R, Larsen AR. Epidemiology of methicillin-resistant Staphylococcus aureus carrying the novel mecC gene in Denmark corroborates a zoonotic reservoir with transmission to humans. Clin Microbiol Infect. 2013;19:E16–E22. doi: 10.1111/1469-0691.12036. [DOI] [PubMed] [Google Scholar]
- 4.Lee HH, Lee GY, Eom HS, Yang SJ. Occurrence and characteristics of methicillin-resistant and -susceptible Staphylococcus aureus isolated from the beef production chain in Korea. Food Sci Anim Resour. 2020;40:401–414. doi: 10.5851/KOSFA.2020.E20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mališová L, Jakubů V, Musílek M, Kekláková J, Žemličková H. Phenotype and genotype characteristics of Staphylococcus aureus resistant to methicillin/oxacillin carrying gene mecC in the Czech Republic from 2002 to 2017. Microb Drug Resist. 2020;26:918–923. doi: 10.1089/mdr.2019.0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fisher EA, Paterson GK. Prevalence and characterisation of methicillin-resistant staphylococci from bovine bulk tank milk in England and Wales. J Glob Antimicrob Resist. 2020;22:139–144. doi: 10.1016/j.jgar.2020.01.013. [DOI] [PubMed] [Google Scholar]
- 7.Giacinti G, Carfora V, Caprioli A, Sagrafoli D, Marri N, Giangolini G, Amoruso R, Iurescia M, Stravino F, Dottarelli S, Feltrin F, Franco A, Amatiste S, Battisti A. Prevalence and characterization of methicillin-resistant Staphylococcus aureus carrying mecA or mecC and methicillin-susceptible Staphylococcus aureus in dairy sheep farms in central Italy. J Dairy Sci. 2017;100:7857–7863. doi: 10.3168/jds.2017-12940. [DOI] [PubMed] [Google Scholar]
- 8.Petinaki E, Spiliopoulou I. Methicillin-resistant Staphylococcus aureus among companion and food-chain animals: impact of human contacts. Clin Microbiol Infect. 2012;18:626–634. doi: 10.1111/j.1469-0691.2012.03881.x. [DOI] [PubMed] [Google Scholar]
- 9.Martineau F, Picard FJ, Ke D, Paradis S, Roy PH, Ouellette M, Bergeron MG. Development of a PCR assay for identification of staphylococci at genus and species levels. J Clin Microbiol. 2001;39(7):2541–2254. doi: 10.1128/JCM.39.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.CLSI (2020) M100 performance standards for antimicrobial susceptibility testing
- 11.CLSI Vet (2018) VET08 Performance Standards for Antimicrobial Disk
- 12.Paterson GK, Morgan FJE, Harrison EM, Peacock SJ, Parkhill J, Zadoks RN, Holmes MA. Prevalence and properties of mecC methicillin-resistant Staphylococcus aureus (mrsa) in bovine bulk tank milk in Great Britain. J Antimicrob Chemother. 2014;69:598–602. doi: 10.1093/jac/dkt417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oliveira DC, de Lencastre H. Multiplex PCR strategy for rapid identification of structural types and variants of the. Antimicrob Agents Chemother. 2002;46:2155–2161. doi: 10.1128/AAC.46.7.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Von Eiff C, Friedrich AW, Peters G, Becker K. Prevalence of genes encoding for members of the staphylococcal leukotoxin family among clinical isolates of Staphylococcus aureus. Diagn Microbiol Infect Dis. 2004;49:157–162. doi: 10.1016/j.diagmicrobio.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 15.Enright MC, Day NPJ, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shopsin B, Gomez M, Montgomery SO, Smith DH, Waddington M, Dodge DE, Bost DA, Riehman M, Naidich S, Kreiswirth BN. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J Clin Microbiol. 1999;37:3556–3563. doi: 10.1128/jcm.37.11.3556-3563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guimarães FF, Manzi MP, Joaquim SF, Richini-Pereira VB, Langoni H. Short communication: outbreak of methicillin-resistant Staphylococcus aureus (MRSA)-associated mastitis in a closed dairy herd. J Dairy Sci. 2017;100:726–730. doi: 10.3168/jds.2016-11700. [DOI] [PubMed] [Google Scholar]
- 18.Silva JG, Araujo WJ, Leite EL, et al (2020) First report of a livestock-associated methicillin-resistant Staphylococcus aureus ST126 harbouring the mecC variant in Brazil Transbound Emerg Dis tbed.13771. 10.1111/tbed.13771 [DOI] [PubMed]
- 19.Schlotter K, Huber-Schlenstedt R, Gangl A, Hotzel H, Monecke S, Müller E, Reißig A, Proft S, Ehricht R. Multiple cases of methicillin-resistant CC130 Staphylococcus aureus harboring mecC in milk and swab samples from a Bavarian dairy herd. J Dairy Sci. 2014;97:2782–2788. doi: 10.3168/jds.2013-7378. [DOI] [PubMed] [Google Scholar]
- 20.Bietrix J, Kolenda C, Sapin A, Haenni M, Madec JY, Bes M, Dupieux C, Tasse J, Laurent F (2019) Persistence and diffusion of mecC-positive CC130 MRSA isolates in dairy farms in Meurthe-et-Moselle county (France). Front Microbiol 10. 10.3389/fmicb.2019.00047 [DOI] [PMC free article] [PubMed]
- 21.Kriegeskorte A, Idelevich EA, Schlattmann A, Layer F, Strommenger B, Denis O, Paterson GK, Holmes MA, Werner G, Becker K (2017) Comparison of different phenotypic approaches to screen and detect mecC-harboring methicillin-resistant Staphylococcus aureus. J Clin Microbiol 56. 10.1128/JCM.00826-17 [DOI] [PMC free article] [PubMed]
- 22.Li T, Lu H, Wang X, Gao Q, Dai Y, Shang J, Li M (2017) Molecular characteristics of Staphylococcus aureus causing bovine mastitis between 2014 and 2015. Front Cell Infect Microbiol 7. 10.3389/fcimb.2017.00127 [DOI] [PMC free article] [PubMed]
- 23.Khemiri M, Abbassi MS, Couto N, Mansouri R, Hammami S, Pomba C. Genetic characterisation of Staphylococcus aureus isolated from milk and nasal samples of healthy cows in Tunisia: first report of ST97-t267-agrI-SCCmecV MRSA of bovine origin in Tunisia. J Glob Antimicrob Resist. 2018;14:161–165. doi: 10.1016/j.jgar.2018.03.013. [DOI] [PubMed] [Google Scholar]
- 24.Sato T, Usui M, Motoya T, Sugiyama T, Tamura Y. Characterisation of meticillin-resistant Staphylococcus aureus ST97 and ST5 isolated from pigs in Japan. J Glob Antimicrob Resist. 2015;3:283–285. doi: 10.1016/j.jgar.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 25.Neto EDA, Pereira RFA, Snyder RE, et al. Emergence of methicillin-resistant Staphylococcus aureus from clonal complex 398 with no livestock association in Brazil. Mem Inst Oswaldo Cruz. 2017;112:647–649. doi: 10.1590/0074-02760170040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silva NCC, Guimarães FF, Manzi MP, Júnior AF, Gómez-Sanz E, Gómez P, Langoni H, Rall VLM, Torres C. Methicillin-resistant Staphylococcus aureus of lineage ST398 as cause of mastitis in cows. Lett Appl Microbiol. 2014;59:665–669. doi: 10.1111/lam.12329. [DOI] [PubMed] [Google Scholar]
- 27.Rossi BF, Bonsaglia ECR, Castilho IG, Dantas STA, Salina A, Langoni H, Pantoja JCF, Budri PE, Fitzgerald-Hughes D, Júnior AF, Rall VLM. Genotyping of long term persistent Staphylococcus aureus in bovine subclinical mastitis. Microb Pathog. 2019;132:45–50. doi: 10.1016/j.micpath.2019.04.031. [DOI] [PubMed] [Google Scholar]