Abstract
Cotton (Gossypium hirsutum L.), a mercantile crop plant, is grown worldwide for fiber and seed oil. As with other economically important crops, cotton is bogged down with many biotic and abiotic stress factors. Towards this, genetic engineering offers numerous protocols to engineer plants for better resilience. However, recalcitrance of cotton to plant tissue culture has been the major constraint for successful in vitro regeneration. Hence, alternate methods that evade tissue culture regeneration have been envisaged. Non tissue culture-based in planta transformation strategies are in vogue due to amenability and ease in the generation of transgenic plants. In the present study, we demonstrate the utility of an in planta transformation protocol and establishment of a stringent selection agent-based screening for the identification of transgenics. The genotype independent nature of the protocol was validated in cotton cv. Pusa 8–6 using GFP. Preliminary transformation efficiency of 28% was achieved with a screening efficiency of 20% in the presence of hygromycin. The proof of T-DNA integration by various molecular and expression analysis in T1 and T2 generations proved that this technique can be employed to generate transgenic cotton.
Keywords: Transgenic cotton, Genotype independent, In planta transformation, Genetic engineering, Agrobacterium tumefaciens
Introduction
Cotton is an important crop globally and planted in an area of 329.49 million hectares (Directorate of Cotton Development Government of India 2017). Despite the use of extra cultivable land, productivity of cotton is severely affected by several biotic and abiotic factors (Mohapatra and Saha 2019). Thus, cotton improvement by introducing genes for superior traits is the need of the hour. Transgenic technology has emerged as a very effective tool for crop improvement, offering the feasibility to selectively introduce one or more genes for mitigation of various stresses (Birch 1997).
In vitro regeneration in cotton has been limited to non-indigenous coker cultivars and its closely related genotypes (Hussain et al. 2009; Zhu et al. 2011) and hence the major obstacle for genetic transformation of cotton. Efforts to improve regenerability in cotton (Aydin et al. 2004; Kumar and Tuli 2004; Wu et al. 2008) were not successful as they required intensive tissue culture skills. This necessitated the need for the development of alternative methods that totally avoided/minimised regeneration to target Agrobacterium into the plant genome. Such protocols were designated as, non-tissue culture-based or in planta transformation approaches. Ability to produce a large number of primary transformants is the major advantage of these tissue culture-independent protocols.
Several research groups have adopted diverse in planta transformation strategies for the generation of transgenic cotton (Rajasekaran et al. 2005; Tian et al. 2010; Jin et al. 2012; Mogali et al. 2013; Pathi and Tuteja. 2013; Vajhala et al. 2013; Kalbande and Patil 2016; Guo et al. 2018). Though apical meristem has been the preferred target in most of these studies, the mode of infection has been different. However, the hypothesis of the standardized protocols have been the introduction of transgenes into differentiating meristematic cells leading to concomitant transgenic expression in the shoots, ultimately resulting in their inheritance (Maher et al. 2020).
The present study also describes an in planta transformation strategy in cotton by targeting the shoot apical meristematic tissues of actively differentiating embryos (Keshamma et al. 2008). The genotype-independent nature of this protocol was demonstrated in different crops like capsicum (Kumar et al. 2009, 2011), pigeonpea (Kumar et al. 2019), and groundnut (Entoori et al. 2008; Keshavareddy et al. 2013). The strategy essentially involves in planta targeting of T-DNA to the differentiating cells of shoot apical meristem and allowing them to grow ex vitro. Since the methodology directs the T-DNA to actively dividing cells of the meristem, the primary transformants (T0 generation plants) produced after transformations are chimeric. This necessitates the identification of transformants to be carried out in T1 generation thus requiring stringent and high throughput screening assays.
We present an effort towards reiterating the genotype independent nature of the previously reported methodology (Keshamma et al. 2008) by examining the feasibility and applicability of GFP as a high throughput screening option for the selection of putative transformants. Additionally, an improved selection agent-based screening for identification of putative transformants has been demonstrated in the present study. This transformation strategy can serve as a credible alternative for the accommodation of genetic modification in cotton for mitigation of varied stresses through transgenesis and also overcome recalcitrance.
Materials and methods
Development of primary transformants
Agrobacterium strain EHA105 containing pCAMBIA1302 harboring GFP (GenBank Accession No: AF234298) gene driven by CaMV35s promoter and hptII as the plant selectable marker was used for transformation. Seeds of cotton cv. P8-6 were surface sterilised and allowed to germinate at 32 °C for 2 days. The germinating seedlings were used as explants for the transformation. The transgenes in the binary vector were directed to the shoot apical meristem, allowed to grow and establish in the net house (Rohini and Rao 2002; Keshamma et al. 2008; Kesiraju and Sreevathsa 2017; Karthik et al. 2020).
In the present study, we investigated two different conditions for the recovery of primary transformants. Initially, seedlings after infection were washed with distilled water, transferred to soilrite and maintained under diffused light. In the second situation, seedlings after infection were placed back into Petri plates with wet filter paper discs, kept in dark and transferred to soilrite on the next day of infection.
Epifluorescence based screening of cotton seedlings for GFP expression
Primary transformants (T0 plants), 36 h after transformation were taken for visualisation of GFP expression under a fluorescence microscope (ZEISS SteREO Discovery V20 microscope) using a 488 nm excitation wavelength, 505–530 band-path filter (which permits visualization of GFP by blue light) to separate GFP and a 560 long-pass filter to determine chlorophyll fluorescence. The seedlings expressing fluorescence were recovered under optimum conditions of 16 h light and 8 h dark photoperiod in growth chambers maintained at 28 °C and transferred into pots filled with soil and maintained in a net house.
In T1 generation, pollen grains from opened flowers of randomly selected plants were collected in 0.5 M phosphate buffer saline (PBS) and observed under the microscope for fluorescence to confirm the inheritance of the transgene.
Screening on hygromycin for the identification of putative transformants
Hygromycin solution of different concentrations (10, 20, 25, 30, 40, 50, 55, 60, 70, 80 and 100 mg/L) was prepared in double distilled water and poured into glass flasks. Overnight-imbibed wild type seeds (in double distilled water) were dropped into them and incubated at 50 rpm for 5 h at 32 °C. The seeds were later transferred to trays filled with autoclaved soilrite and maintained under net house conditions for 8–10 days. Equal number of seeds treated with water were taken as untreated control and planted in a separate tray. The plants were later observed for necrotic symptoms in different hygromycin concentrations. Based on the standardization, the concentration of hygromycin lethal to wild type cotton was used for the identification of putative transformants. Well-established plants were further analysed for transgene integration and expression.
Molecular analysis of transgenic plants for T-DNA integration
PCR analysis
The leaves of transgenic and wild type cotton plants were ground for genomic DNA isolation following a modified cetyltrimethyl ammonium bromide (CTAB) method (Porebski et al. 1997). The PCR reaction mixture (25 µl) containing 1 U Taq DNA polymerase (GeNie, Banglore, Karnataka, India), 1X assay buffer (10 mM pH 9.0 Tris–HCl, 50 mM KCl, 1.5 mM MgCl2, 0.01% gelatin), 150 µM of each dNTP, 0.5 µl of each forward and reverse primer at a final concentration of 0.25 µM and 100 ng of template DNA was used to amplify the transgene. PCR amplification was carried out in a thermal cycler (Eppendorf, Hamburg, Germany) programmed with a hot start of initial denaturation at 95 °C for 5 min followed by 35 cycles of denaturation at 95 °C for 1 min, annealing at 65 °C for GFP and 55 °C for hptII for 1 min, extension at 72 °C for 1 min, final extension at 72 °C for 10 min. Amplified gene products of size 571 bp (GFP) and 700 bp (hptII) were analyzed by electrophoresis (Table 1).
Table 1.
List of primers used in the study
| Primer ID | Primer sequence (5′-3′) |
|---|---|
| GFP forward primer | TGGGCACAAATTTTCTGTCAGTGGA |
| GFP reverse primer | ATGCCATGTGTAATCCCAGCAGCT |
| hptII forward primer | GCTCGATACAAGCCAACCAC |
| hptII reverse primer | CGAAAAGTTCGACAGCGTCTC |
| qPCR GFP forward primer | TCCACACAATCTGCCCTTTC |
| qPCR GFP reverse primer | CTATACAAAGCTAGCCACCACC |
| qPCR Ubiquitin forward primer | ACACGATCGACAACGTTAAGGCGA |
| qPCR Ubiquitin reverse primer | TCAACGCTCCATCTTGTCCTTCGT |
Southern blotting
About 15 μg of genomic DNA from transgenic and wild type plants was digested with HindIII, separated on 0.8% agarose gel and transferred on to a positively charged nylon membrane. The membrane was further hybridised with a DIG-labelled GFP gene fragment as a probe. Hybridization and washing was carried out according to manufacturer’s instructions (Roche Holding AG, Basel, Switzerland). The membrane was exposed to an X-ray film for 2 h in dark and observed for bands to determine the T-DNA copy number in the transgenic plants.
Analysis of transgenic plants for transcript accumulation by RT-PCR
Total RNA was isolated from transgenic and wild type cotton plants using total RNA isolation kit (Spectrum™, Sigma-Aldrich, St. Louis, Missouri, United States) and quantified. cDNA was synthesized from 2.5 µg total RNA according to manufacturer's instructions (SuperScript®VILO™, Invitrogen, Carlsbad, California, United States) and used for transcript quantification. To evaluate transcript accumulation, 1 µl of diluted cDNA mix was used as a template for the amplification of 155 bp ubiquitin and 124 bp GFP fragments (Table 1). Real-time PCR was performed using Aria Mx Real-time PCR system (Agilent Technologies, Santa Clara, California, United States). For this, 1 µl of diluted cDNA template of both transgenics and wild type plants was taken and reaction was set according to the SYBR Green qPCR kit (Agilent). PCR program of 5 min initial denaturation at 95 °C followed by 40 cycles of denaturation at 95 °C for 20 s, annealing at 60 °C for 20 s and extension at 72 °C for 15 s was used. A final melting curve consisting of one cycle of 95 °C for 30 s, 65 °C for 30 s and 95 °C for 30 s was used for ascertaining the variations of GFP expression among transgenic events vis-à-vis wild type. The Ct values of samples were normalized with the Ct value of ubiquitin (internal control) to calculate ΔCt value.
Results and discussion
Transgenic crops are an illustration of successful trait modifications using an array of genes playing crucial roles in crop improvement. Transgenic technology has emerged as an influential tool in plant research globally with diverse techniques and strategies being used in crop improvement programs (Sivasupramaniam et al. 2014; Tian et al 2015; Ma and Zhang 2019).
This study deals with the assessment of a standardized genotype- independent transformation strategy using a non-tissue culture-based Agrobacterium tumefiaciaens-mediated transformation in cotton (Keshamma et al. 2008). Since the use of a previously established protocol does not necessitate success of an experiment, stringent modifications of the protocol in accordance to regional variations are a prerequisite. We present variations in the methodology towards improvement of transformation efficiency, identification and also recovery of transformants using the selectable marker, GFP.
Development of primary transformants
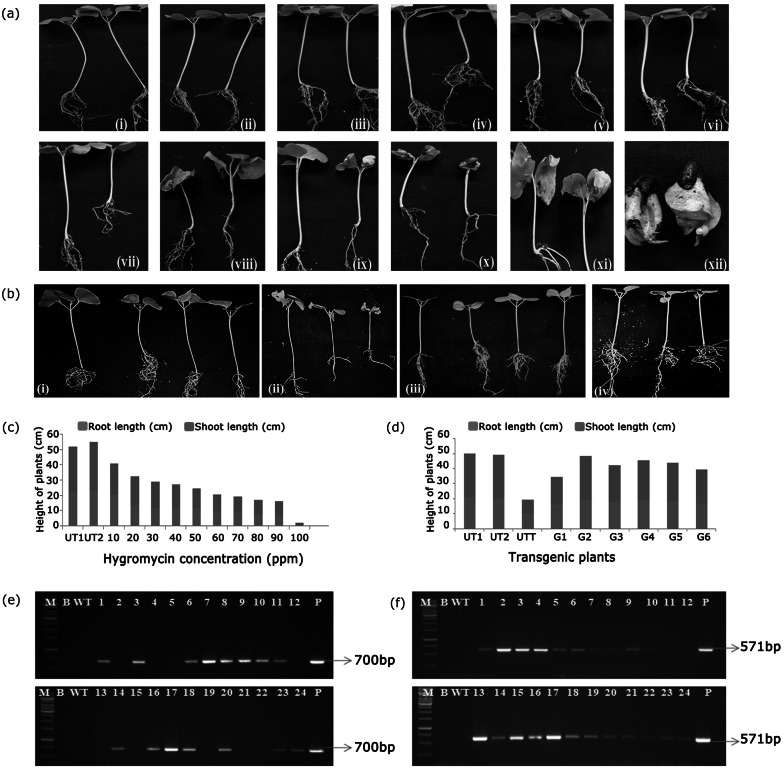
Transgenic cotton plants were generated by apical meristem-targeted Agrobacterium mediated in planta transformation strategy (Fig. 1a i–viii). Two day old germinating seedlings (Fig. 1a i–iii) were pricked at the apical meristem region using an insulin needle (Fig. 1a iv), and dropped into AB minimal media containing tobacco leaf extract (Fig. 1a v, vi) and incubated at 28 °C.
Fig. 1.
a Overview of the apical meristem-targeted in planta transformation strategy for the development of transgenics in cotton. (i) Imbibed seeds allowed to germinate; (ii) Removal of seed coat in the imbibed seeds; (iii) Exposure of the apical meristem in the germinating seedlings; (iv) Pricking of the shoot apical meristem with an insulin needle; (v) and (vi) Agrobacterium infection of seedlings; (vii) Planting of the infected seedlings onto soilrite; (viii) Recovery of primary transformants. b GFP expression-based identification of primary transformants (T0 generation). (i) and (ii) Absence of GFP expression in wild type seedlings, (iii–xi) GFP expression in putative transformants
After transformation, when the infected seedlings were thoroughly washed with sterile water, transferred to autoclaved soilrite and maintained under diffused light, retrieval of seedlings was hampered which resulted in reduced number of recovered primary transformants (batch 1, Table 2). On the other hand, when seedlings were transferred back to the Petri plates (containing filter paper discs soaked with water) after transformation and left overnight in dark, they showed speedy recovery with low embryo mortality (batches 2 and 3 of Table 2). This modification in the protocol resulted in the increased recovery of primary transformants from 10 to 35% (Fig. 1a vii, viii, Table 2).
Table 2.
Transformation efficiency in preliminary cotton transformants of different batches
| Batch no | No. of seeds taken for transformation | No. of plants recovered in 36 h after transformation | No. of plants selected after microscopic observation | No. of selected plants that have recovered and transferred to pots |
|---|---|---|---|---|
| B-1* | 40 | 15 | 4 | 3 |
| B-2** | 50 | 42 | 18 | 13 |
| B-3** | 35 | 26 | 14 | 9 |
| Total | 125 | 83 | 36 | 25 |
*In this batch the transformed seeds were directly transferred to soilrite
**In this batch the transformed seeds were initially transferred to Petri plates and transferred to soilrite on the next day
Primary transformants were initially subjected to recovery for 2 weeks in growth chambers with 16 h light and 8 h dark photoperiod, at 28 °C, supplemented with 1/8th strength of Hoagland solution. Recovered plants were later transferred to pots in the net house.
Preliminary screening of transformants
Microscopic observations aided in the identification of GFP expression in putatively transformed seedlings (Fig. 1b iii–xi) when compared to its absence in wild type seedlings (Fig. 1b i, ii). However, not all the infected seedlings depicted expression of GFP at the infection site which indicated that infection of Agrobacterium was random in nature. The seedlings devoid of fluorescence were discarded while those with the signal were allowed to recover. Out of 125 infected seedlings, 36 showed the presence of GFP and were selected as putative transformants (Table 2), of which 25 plants recovered and were transferred to pots in the net house. Microscopic observations of preliminary transformants (T0 plants) showed that 28% of infected seedlings expressed GFP. The recovered plants were healthy after transferring to soil, developed and set seeds normally. Though the rate of growth was slow in transgenics as compared to wild type plants, the seed pool was uniform and viable. As T0 plants developed using in planta transformation protocol are chimeric, identification of putative transformants and molecular analysis for transgene integration was performed in T1 generation.
Identification of putative transformants by screening under selection pressure
Initial screening studies using hygromycin revealed that wild type cotton seedlings could resist upto 30 mg/L hygromycin and showed normal root and shoot growth (Fig. 2a ii–v). However, 40–60 mg/L hygromycin exhibited variation in root and shoot growth (Fig. 2a vi–ix) with necrotic spots on leaves and stunted growth. Root length retardation was also noticed with increase in the concentration of hygromycin. With further increase in the concentration of the selection agent, plants were unhealthy (Fig. 2a x and xi) and at 100 mg/L hygromycin, there was a stall in the growth (Fig. 2a xii). However, untreated control devoid of hygromycin exhibited normal growth (Fig. 2a i). Thus, 70 mg/L hygromycin was considered for screening and identification of putative transformants (Fig. 2c).
Fig. 2.
Hygromycin screening for the identification of putative transformants. a Standardization of hygromycin concentration in cv. P8–6. Variation in plant response to different hygromycin concentrations (i) Untreated control, (ii) 10 mg/L, (iii) 20 mg/L, (iv) 25 mg/L, (v) 30 mg/L, (vi) 40 mg/L, (vii) 50 mg/L, (viii) 55 mg/L, (ix) 60 mg/L, (x) 70 mg/L, (xi) 80 mg/L, (xii) 100 mg/L. b Response of T1 generation cotton seedlings to 70 mg/L of hygromycin (i) Untreated control, (ii) treated wild type, (iii, iv) Hygromycin-resistant T1 generation transgenic plants. c Graph representing root and shoot lengths of wild type cotton plants in response to different concentrations of hygromycin (UT1, UT2- untreated wild type plants). d Graph representing root and shoot lengths of transgenic T1 plants in response to 70 mg/L hygromycin (UT1, UT2- untreated wild type plants; UTT- untreated wild type, G1–G6 are individual transgenic plants). e and f Molecular analysis of transgenic plants in T1 generation; PCR analysis for the amplification of hptII gene (700 bp amplicon) and GFP gene (571 bp amplicon) fragments, Lane M- 1 Kb marker, Lane B- water blank, Lane WT-wild type DNA (100 µg), Lanes 1–24 DNA (100 µg) of different transgenic plants, Lane P- binary vector DNA (25 ng)
About 20 seeds each of six T0 plants were subjected to hygromycin selection pressure. While the plants exhibiting necrotic symptoms were considered as non-transgenic and discarded. healthy plants were grown in the net house. About 32 plants i.e., 26.6% of seeds were able to resist hygromycin (Fig. 2b iii and iv) which demonstrated that soilrite-based screening strategy was efficient in the identification of putative transformants.
The major variation presented in this study has been the stringent soilrite-based screening for identification of putative transformants, compared to PCR-based screening in the earlier study (Keshamma et al. 2008). Antibiotic-based screening aids in the reduction of escapes with increased stringency towards identification of putative transformants. The plants that survived the hygromycin selection pressure established normally and were at par with the untreated control (Fig. 2 d). Selection of transgenic herbicide tolerant cotton plants developed through in planta transformation strategy using soilrite-based glyphosate screening has also been demonstrated (Karthik et al. 2020) depicting the authenticity and importance of the selection agent in the identification of transformants.
Molecular characterisation of transgenics in T1 generation
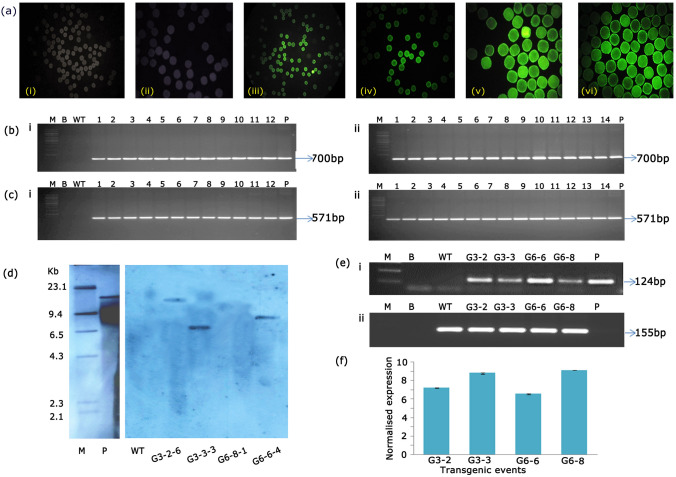
As an initial evidence for T-DNA integration, PCR analysis of 25 T1 generation transgenic plants showed the presence of 700 bp hptII and 571 bp GFP gene fragments (Fig. 2 e, f). Wild type plant DNA did not show any amplification in PCR. Advantage of using screenable markers lies in tracking the inheritance of transgenes (Keshamma et al. 2008; Kumar et al. 2009, 2011), which was ascertained in the present study by the exploitation of GFP expression. Pollen grains were collected randomly from flowers of selected transgenic plants and observed under the microscope. Fluorescence was observed in pollen of transgenic plants (Fig. 3a iii–vi), while those from wild type did not exhibit fluorescence (Fig. 3a i, ii). The presence of transgene in pollen from opened flowers further precisely reconfirmed its inheritance to the next filial generation. Based on PCR analysis and GFP expression in pollen, four T1 generation transgenic plants (G3-2, G3-3, G6-6 and G6-8) were advanced to T2 generation.
Fig. 3.
a Expression of GFP in pollen grains of T1 generation cotton plants. (i, ii) Microscopic images of pollen grains from wild type plants; (iii, iv, v, vi) Microscopic images of pollen grains from transgenic plants (G3-2, G3-3, G6-6 andG6-8). Molecular analysis of transgenic plants in T2 generation b (i, ii) and c (i, ii). PCR analysis for the amplification of hptII (700 bp) and GFP gene (571 bp) fragments. Lanes 1–12 of (i) are progeny plants of G3-2 and G3-3; Lanes 1–14 of (ii) are progeny plants of G6-6 and G6-8, Lane M-1 Kb marker, Lane B- water blank, Lane WT- wild type DNA, Lane P- plasmid (25 ng). d Genomic Southern analysis of transgenic plants, probed with DIG-labelled 571 bp GFP gene; Lane M- Lambda HindIII DNA ladder, Lane P- linearized plasmid pCAMBIA 1302 (10 pg), Lane WT-wild type, Lanes G3-2–5, G3-3–3, G6-8–1, G6-6–4 are T2 generation transgenic plants, e sqRT-PCR amplified products of (i) 124 bp GFP and (ii) 155 bp ubiquitin in transgenic plants. Lane M- 100 bp marker (Thermo scientific), Lane B- water blank, Lane WT-wild type, Lane P- binary vector. f Expression analysis of GFP in transgenic plants by qRT-PCR
Molecular characterisation of transgenics in T2 generation
About 10 seeds from each of the selected plants were sown in pots containing soil under net house conditions. PCR (hptII and GFP) analysis of 30 T2 generation plants confirmed the presence of both the transgenes in all the progeny plants (Fig. 3b i, ii; c i, ii). Further, genomic Southern analysis performed with 4 transgenic plants G3-2–6, G3-3–3, G6-8–1, G6-6–4 demonstrated hybridisation signals in three plants with single copy insertions of the transgene (Fig. 3 d). The absence of the signal in wild type confirmed T-DNA integration in transgenic plants.
Four selected transgenic events G3-2, G3-3, G6-6, G6-8 showed the amplification of a 124 bp GFP fragment confirming its transcript accumulation (Fig. 3e i). The 155 bp ubiquitin (internal control) fragment amplified in both transgenic and wild type plants (Fig. 3e ii) authenticated the PCR. Additionally, qPCR showed varied levels of GFP expression in different events (Fig. 3f) with seven to nine fold increase in GFP transcripts implicating the transgenic integration and inheritance in selected cotton plants.
Establishment and validation of transgenics using genotype-independent transformation protocols have always been pertinent among researchers (Rajasekaran et al. 2005; Tian et al. 2010; Jin et al. 2012; Mogali et al. 2013; Pathi and Tuteja. 2013; Vajhala et al. 2013; Kalbande and Patil 2016; Guo et al. 2018). The present modified in planta transformation strategy is best suited for developing transgenics as evidenced by preliminary transformation efficiency (using screenable marker GFP) of 28.6% which is laudable in a recalcitrant crop like cotton. Further, modification of the earlier protocol increased recovery efficiency by 20% in primary transformants. Adoption of stringent screening procedures in the present study decreased the chance of obtaining false positive plants, as evidenced by 26.6% of T1 generation transgenic plants that were able to withstand the stress of hygromycin and contributed consistently in the rapid screening of the chimeric seed pool. However, ~ 3.33% of the T1 generation plants were advanced to the next generation. Other studies that utilized in planta transformation protocols, demonstrated an efficiency of 0.46–0.93% by pistil drip (Tian et al. 2010), 0.30% by pollen tube pathway (PTP) mediated transformation (Mogali et al. 2013), 6.89% by directing Agrobacterium inoculum to excised cotyledonary leaves at their junction (Kalbande and Patil 2016) and 0.125% by shoot apex mediated transformation (Guo et al. 2018).
Therefore, the present apical meristem targeted in planta strategy has not only proved the genotype independent nature of the protocol but also its utility in producing large number of transgenic plants from which best performing events can be identified using stringent molecular and efficacy analysis. The modifications suggested are of immense importance for increased rate of transformation efficiency in cotton. Incorporation of such high throughput and established strategies in crop improvement programmes can pave way for long standing success of transgenic technology.
Author contributions
KK developed the transgenic plants. KK, PM, AB performed molecular analyses. KK wrote the manuscript. MS critically edited the manuscript. UR provided the fluorescence microscope for GFP expression studies and critically edited the manuscript. RS designed experiments, edited and revised the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that there is no conflict of interest regarding the publication of this article.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Aydin Y, Ipekci Z, Talas-Oğraş T, Zehir A, Bajrovic K, Gozukirmizi N. High frequency somatic embryogenesis in cotton. Biol Plant. 2004;48:491–495. doi: 10.1023/B:BIOP.0000047142.07987.e1. [DOI] [Google Scholar]
- Birch RG. Plant transformation: problems and strategies for practical application. Annu Rev Plant Biol. 1997;48:297–326. doi: 10.1146/annurev.arplant.48.1.297. [DOI] [PubMed] [Google Scholar]
- Directorate of Cotton Development Government of India (2017) Status paper of Indian cotton. Nagpur. Accessed on July 27, 2019 https://www.nfsm.gov.in/StatusPaper/Cotton2016.pdf
- Entoori K, Sreevathsa R, Arthikala MK, Kumar PA, Kumar ARV, Madhusudhan B, Makarla U. A chimeric cry1X gene imparts resistance to Spodoptera litura and Helicoverpa armigera in the transgenic groundnut. Eur Asia J BioSc. 2008;2:53–65. [Google Scholar]
- Guo WF, Wang KY, Wang N, Jun LI, Li GQ, Liu DH. Rapid and convenient transformation of cotton (Gossypium hirsutum L.) using in planta shoot apex via glyphosate selection. J Integr Agric. 2018;17:2196–2203. doi: 10.1016/S2095-3119(17)61865-3. [DOI] [Google Scholar]
- Hussain SS, Rao AQ, Husnain T, Riazuddin S. Cotton somatic embryo morphology affects its conversion to plant. Biol Plant. 2009;53:307–311. doi: 10.1007/s10535-009-0055-6. [DOI] [Google Scholar]
- Jin SX, Liu GZ, Zhu HG, Yang XY, Zhang XL. Transformation of upland cotton (Gossypium hirsutum L.) with gfp gene as a visual marker. J Integr Agric. 2012;11:910–919. doi: 10.1016/S2095-3119(12)60081-1. [DOI] [Google Scholar]
- Kalbande BB, Patil AS. Plant tissue culture independent Agrobacterium tumefaciens mediated In-planta transformation strategy for upland cotton (Gossypium hirsutum) J Genet Eng Biotechnol. 2016;14:9–18. doi: 10.1016/j.jgeb.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthik K, Nandiganti M, Thangaraj A, Singh S, Mishra P, Sharma M, Sreevathsa R. Transgenic cotton (Gossypium hirsutum L.) to combat weed vagaries: utility of an apical meristem-targeted in planta transformation strategy to introgress a modified CP4-EPSPS gene for glyphosate tolerance. Front Plant Sci. 2020 doi: 10.3389/fpls.2020.00768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshamma E, Rohini S, Rao KS, Madhusudhan B, Kumar MU. Tissue culture-independent in planta transformation strategy: an Agrobacterium tumefaciens-mediated gene transfer method to overcome recalcitrance in cotton (Gossypium hirsutumL.) J Cotton Sci. 2008;12:264–272. [Google Scholar]
- Keshavareddy G, Rohini S, Ramu SV, Sundaresha S, Kumar AR, Kumar PA, Udayakumar M. Transgenics in groundnut (Arachis hypogaea L.) expressing cry1AcF gene for resistance to Spodoptera litura (F.) Physiol Mol Biol Plants. 2013;19:343–352. doi: 10.1007/s12298-013-0182-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesiraju K, Sreevathsa R. Apical meristem-targeted In Planta transformation strategy: an overview on its utility in crop improvement. Agri Res and Techol. 2017 doi: 10.19080/ARTOAJ.2017.08.555734. [DOI] [Google Scholar]
- Kumar AM, Reddy KN, Manjulatha M, Arellano ES, Sreevathsa R, Ganeshan G. A rapid, novel and high-throughput identification of putative bell pepper transformants generated through in planta transformation approach. Sci Hortic. 2011;129:898–903. doi: 10.1016/j.scienta.2011.05.001. [DOI] [Google Scholar]
- Kumar AM, Reddy KN, Sreevathsa R, Ganeshan G, Udayakumar M. Towards crop improvement in bell pepper (Capsicum annuum L.): Transgenics (uid A: hpt II) by a tissue-culture-independent Agrobacterium-mediated in planta approach. Sci Hortic. 2009;119:362–370. doi: 10.1016/j.scienta.2008.08.034. [DOI] [Google Scholar]
- Kumar M, Tuli R. Plant regeneration in cotton: A short-term inositol starvation promotes developmental synchrony in somatic embryogenesis. Vitro Cell Dev Biol-Pl. 2004;40:294–298. doi: 10.1079/IVP2004531. [DOI] [Google Scholar]
- Kumar NR, Rathinam M, Singh S, Kesiraju K, Muniyandi V, Singh NK, Dash PK, Sreevathsa R. Assessment of pigeonpea (Cajanus cajan L.) transgenics expressing Bt ICPs, Cry2Aa and Cry1AcF under nethouse containment implicated an effective control against herbivory by Helicoverpa armigera (Hübner) Pest Manag Sci. 2019 doi: 10.1002/ps.5722. [DOI] [PubMed] [Google Scholar]
- Ma W, Zhang T. Next-generation transgenic cotton: pyramiding RNAi with Bt counters insect resistance. In: Zhang B, editor. Transgenic cotton methods in molecular biology. 1. New York: Humana Press; 2019. pp. 245–256. [DOI] [PubMed] [Google Scholar]
- Maher MF, Nasti RA, Vollbrecht M, Starker CG, Clark MD, Voytas DF. Plant gene editing through de novo induction of meristems. Nat Biotechnol. 2020;38:84–89. doi: 10.1038/s41587-019-0337-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogali SC, Khadi BM, Kategeri IS. High efficiency transformation protocol for two Indian cotton (Gossypium hirsutum) varieties via pollen tube pathway. Ind J Agric Sci. 2013;83:949–952. [Google Scholar]
- Mohapatra L, Saha G. Cotton farming in India: alternative perspectives and paradigms. In: Nayak AJRN, editor. Transition strategies for sustainable community systems, The anthropocene: Politik—Economics—Society—Science. 1. Switzerland: Springer; 2019. pp. 195–213. [Google Scholar]
- Pathi KM, Tuteja N. High-frequency regeneration via multiple shoot induction of an elite recalcitrant cotton (Gossypium hirsutum L. cv Narashima) by using embryo apex. Plant Signal Behav. 2013;8:e22763. doi: 10.4161/psb.22763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porebski S, Bailey LG, Baum BR. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep. 1997;15:8–15. doi: 10.1007/BF02772108. [DOI] [Google Scholar]
- Rajasekaran K, Cary JW, Jaynes JM, Cleveland TE. Disease resistance conferred by the expression of a gene encoding a synthetic peptide in transgenic cotton (Gossypium hirsutum L.) plants. Plant Biotechnol J. 2005;3:545–554. doi: 10.1111/j.1467-7652.2005.00145.x. [DOI] [PubMed] [Google Scholar]
- Rohini VK, Rao KS. In planta strategy for gene transfer into plants: embryo transformation. Physiol Mol Biol Plants. 2002;8:161–169. [Google Scholar]
- Sivasupramaniam S, Moar WJ, Ruschke LG, Osborn JA, Jiang C, Sebaugh JL, Greenplate JT. Toxicity and characterization of cotton expressing Bacillus thuringiensis Cry1Ac and Cry2Ab2 proteins for control of lepidopteran pests. J Econ Entomol. 2014;101:546–554. doi: 10.1093/jee/101.2.546. [DOI] [PubMed] [Google Scholar]
- Tian G, Cheng L, Qi X, Ge Z, Niu C, Zhang X, Jin S. Transgenic cotton plants expressing double-stranded RNAs target HMG-CoA reductase (HMGR) gene inhibits the growth, development and survival of cotton bollworms. Int J Biol Sci. 2015;11:1296–1305. doi: 10.7150/ijbs.12463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian ZC, Shen JW, Jun Z, Wang ZG, Tian ZZ. Pistil drip following pollination: a simple in planta Agrobacterium-mediated transformation in cotton. Biotechnol Lett. 2010;32:547–555. doi: 10.1007/s10529-009-0179-y. [DOI] [PubMed] [Google Scholar]
- Vajhala CS, Sadumpati VK, Nunna HR, Puligundla SK, Vudem DR, Khareedu VR. Development of transgenic cotton lines expressing Allium sativum agglutinin (ASAL) for enhanced resistance against major sap-sucking pests. PLoS ONE. 2013;8:e72542. doi: 10.1371/journal.pone.0072542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SJ, Wang HH, Li FF, Chen TZ, Zhang J, Jiang YJ, Zhang TZ. Enhanced Agrobacterium-mediated transformation of embryogenic calli of upland cotton via efficient selection and timely subculture of somatic embryos. Plant Mol Biol Rep. 2008;26:174–185. doi: 10.1007/s11105-008-0032-9. [DOI] [Google Scholar]
- Zhu SJ, Li L, Chen JH, He QL, Fang XX, Ye CY, Mei L. Advance in research and utilization of cotton biotechnology in China. Plant Omics. 2011;4:329–338. [Google Scholar]