Abstract
Bacteriocins are ribosomally synthesized peptides with antibacterial activity against food-borne pathogenic bacteria that cause spoilage, possessing important potential for use as a natural preservative in the food industry. The novel bacteriocin BM1300 produced by Lactobacillus crustorum MN047 was identified after purification in this study. It displayed broad-spectrum antibacterial activity against some selected Gram-positive and Gram-negative bacteria. The minimum inhibitory concentration (MIC) values of BM1300 against Staphylococcus aureus ATCC 25923 and Escherichia coli ATCC 25922 were 13.4 μg/mL and 6.7 μg/mL, respectively. Moreover, BM1300 showed excellent thermal (between 60 and 120 °C), pH (2–11), and chemical (Tween-40, Tween-80, Triton X-100, and EDTA) stabilities. Time-kill curves revealed that BM1300 exhibited bactericidal activity against S. aureus and E. coli. The scanning and transmission electron microscopy indicated that BM1300 acted by disrupting the cell membrane integrity and increasing cell membrane permeabilization of indicator bacteria. The disruption of cell membrane integrity caused by BM1300 was further demonstrated by the uptake of propidium iodide (PI) and the release of intracellular lactate dehydrogenase (LDH) and nucleic acid and proteins. Moreover, BM1300 affected cell cycle distribution to exert antibacterial activity collaboratively. Meanwhile, BM1300 inhibited the growth of S. aureus and E. coli of beef meat and improved the microbiological quality of beef meat. These findings place BM1300 as a potential biopreservative in the food industry.
Electronic supplementary material
The online version of this article (10.1007/s42770-020-00311-3) contains supplementary material, which is available to authorized users.
Keywords: Bacteriocin, Purification, Multiple modes of action, Bactericidal effect, Application
Introduction
Bacteriocins, a huge family of antimicrobial peptides or small proteins, exhibit antibacterial activity against genetically closely bacteria to compete and survive in an ecological system [1]. In recent years, bacteriocins produced by lactic acid bacteria (LAB) have attracted significant interests for their ability to effectively inhibit food-borne pathogenic bacteria and some multidrug-resistant bacteria [2, 3], which can be developed as natural food preservatives and therapeutic antimicrobial agents. To date, wide varieties of bacteriocins have been exploited, such as pediocin PA-1, enterocin AS-48, and lactococcin A [4].
Although a large number of bacteriocins produced by LAB have been studied in recent years, nisin and pediocin PA-1/AcH were approved for use in selected foods only [5, 6]. It should be noted that the narrow-spectrum activity of bacteriocins limits their application in extending shelf life of food products, such as thuricin CD [7], bacteriocin ST91KM [8], and His-tagged colicin 5 [9]. Bacteriocins produced by LAB are generally active against Gram-positive bacteria but exhibit poor activity against Gram-negative bacteria [10]. The reason why Gram-negative bacteria are resistant to these bacteriocins is that their outer membrane is a complex barrier system, preventing the deleterious effect on the inner membrane [11]. Therefore, novel and broad-spectrum bacteriocins which are particularly active against Gram-negative bacteria remain to be investigated.
A clear understanding about modes of action of bacteriocins produced by LAB against pathogens is important to ascertain their effective application as food preservatives in the food industry. Class I bacteriocins, i.e., lantibiotics, usually bind to target molecules (lipid II) acting as the key intermediate in the peptidoglycan biosynthesis machinery of the cell wall, and lastly, peptidoglycan synthesis is hampered. Other lantibiotics get inserted in the cell membrane through targeting lipid II as a docking molecule and lead to pore formation in the cell membrane followed by cell death [4, 12]. Class II bacteriocins generally bind to the transporter mannose phosphotransferase system (Man-PTS) and then insert into the cytoplasmic membrane, causing membrane permeabilization and depolarization by the leakage of ions and intracellular material [13], such as lactococcin A [14] and microcin E492. Class III bacteriocins, i.e., bacteriolysin (e.g., lysostaphin), usually exert the bactericidal activity by target cell lysis for their hydrolytic activity of the cell wall [12]. Lots of bacteriocins produced by LAB have been explored in recent years, but there are some differences among abundant antimicrobial systems. Therefore, specific modes of action of bacteriocins still need to be investigated.
Due to increasing consumer demands toward foods containing less chemical preservatives, the concept of safe and healthy food preservatives has recently intensified [2]. Bacteriocins produced by LAB have promising perspectives to be used as food preservatives. For example, nisin has gained permitted application as a food preservative in over 50 countries [13]. At present, beef meat is regarded as a representation of red meat consumption and preservation of beef is a huge challenge that remains to be solved. However, the majority of Gram-negative bacteria are not sensitive to most of the bacteriocins produced by LAB; few breakthroughs in this regard are reported. Therefore, effective and enhanced bacteriocins produced by LAB against Gram-negative bacteria have excellent potential use in the meat system.
BM1300 was a novel bacteriocin produced by Lactobacillus crustorum MN047 which was isolated from a traditional fermented dairy product koumiss in our previous study [15]. However, its physicochemical properties are unknown; its specific mode of action against target bacteria and the potential use in the food industry are still not investigated. Therefore, in this study, we aimed (i) to characterize bacteriocin BM1300 including its stability and biosafety, (ii) to sufficiently elucidate the modes of action of BM1300 against both Staphylococcus aureus and Escherichia coli, and (iii) to evaluate its inhibitory effects against both S. aureus and E. coli of beef meat during refrigerated storage. The present study provides a foundation for future application of BM1300.
Materials and methods
Microbial strains and preparation of bacteriocin
Bacteriocin BM1300 was heterologously expressed using the E. coli expression system in our previous study [16]. In brief, the purified PCR product was digested with FlyCut™ Nde I and FlyCut™ BamHI and was inserted into pET-28a by ligation with T4 Ligase. This recombinant plasmid containing BM1300 bacteriocin gene was transformed into E. coli BL21 (DE3) competent cells. E. coli BL21 (DE3) containing recombinant vector was incubated in Luria Bertani (LB) broth medium containing kanamycin at 37 °C until an OD600 of 0.4. Cultures were induced by isopropyl β-D-thiogalactoside (IPTG) overnight at 25 °C. Cells were harvested and washed three times with 20 mM Tris-HCl (pH 6.68). At least 5 freeze-thaw cycles were performed to disrupt cells and release bacteriocin BM1300 [16]. The minimum inhibitory concentration (MIC) value of BM1300 was defined as the lowest concentration of BM1300 in which no growth of indicators was observed [17]. It was the parameter for the evaluation of the concentration of BM1300 in the lysate. Before starting the experiments in this study, the MIC was used for the quantification of the bacteriocin unless stated otherwise in this work. All Gram-positive and Gram-negative bacteria in this study were cultivated on LB broth at 37 °C, in which only Vibrio parahaemolyticus was cultured in LB broth containing 3% NaCl. S. aureus ATCC 25923 and E. coli ATCC 25922 were used as indicator bacteria.
Antibacterial spectrum
The antibacterial spectrum of bacteriocin BM1300 was determined, and the selected Gram-positive and Gram-negative bacteria are shown in Table 1. Supernatants of cell disruption (100 μL) were used to test the antibacterial activity, and the zones of growth inhibition surrounding the wells of the test bacteria were measured by the agar well diffusion assay [18]. All measurements were performed in triplicate.
Table 1.
Antimicrobial spectrum of bacteriocin BM1300
| Indicator strains | Indicator diameter of inhibition zone (mm) | Resistant antibiotic |
|---|---|---|
| Gram-positive bacteria | ||
| Bacillus cereus ATCC 14579 | 25.8 ± 0.3 | |
| Staphylococcus aureus ATCC 26111 | 28.0 ± 0.0 | |
| Listeria monocytogenes ATCC 19115 | 21.6 ± 0.2 | |
| Listeria monocytogenes ATCC 15313 | 27 ± 1.0 | |
| Antibiotic-resistant Staphylococcus aureus 1 | 19.1 ± 1.0 | AMP, A/C,T/S,PEN |
| Antibiotic-resistant Staphylococcus aureus 2 | 16.3 ± 0.7 | AMP, PEN, ERY, T/S, CIP |
| Antibiotic-resistant Staphylococcus aureus 3 | 16.8 ± 0.3 | AMP, PEN, T/S, CIP |
| Antibiotic-resistant Staphylococcus aureus 4 | 0 | ERY, FOX, A/C, T/S, PEN |
| Antibiotic-resistant Staphylococcus aureus 5 | 0 | ERY, FOX, AMP, A/C, T/S, PEN |
| Gram-negative bacteria | ||
| Salmonella typhimurium LT2 | 18.5 ± 0.3 | |
| Yersinia enterocolitica 52203 | 17.6 ± 0.2 | |
| Cronobacter sakazakii ATCC BAA-894 | 23.1 ± 0.3 | |
| Salmonella typhimurium ATCC 51005 | 17.6 ± 0.4 | |
| Vibrio parahaemolyticus ATCC 17802 | 13.5 ± 0.5 | |
| Antibiotic-resistant Salmonella 53-1105R | 22.7 ± 0.3 | TCY, FIS, TMP, SMZ |
| Antibiotic-resistant Salmonella 1-1105J | 15.9 ± 0.9 | FIS, NAL |
| Antibiotic-resistant Salmonella 557D | 18.8 ± 0.3 | AMP, A, AMK, CHL, FOX, GAT, GEN, KAN, LVX, NAL, T/S, TCY |
| Antibiotic-resistant C. sakazakii 18-7 (1) | 24.6 ± 0.2 | AMO, A/C, TCY, S, RA, GEN, T/S, FOX |
| Antibiotic-resistant C. sakazakii 18-11 (2) | 15.0 ± 0.0 | AMC, A/C, FOX, TCY, RA, T/S |
| Antibiotic-resistant Escherichia coli SX DMQ004 EC2 | 18.9 ± 0.1 | AMP, KAN, MP, SMZ, CHL, NAL, CIP, TET |
| Antibiotic-resistant Escherichia coli SX WSQ003 EC3 | 18.2 ± 0.3 | AMP, KAN, MP, SMZ, CHL, CFP, TET |
A/C amoxicillin/clavulanic acid, AMC amoxicillin, AMP ampicillin, AMK amikacin, CHL chloramphenicol, CIP ciprofloxacin, CRO ceftriaxone, ERY erythromycin, FIS sulfisoxazole, FOX cefoxitin, GAT gatifloxacin, GEN gentamicin, KAN kanamycin, LVX levofloxacin, MP meropenem, NAL nalidixic acid, PEN penicillin, RA rifampin, S streptomycin, SMZ sulfamethoxazole, T/S trimethoprim/sulfamethoxazole, TCY tetracycline, TMP trimethoprim, TET tetracycline
Purification of BM1300
Crude bacteriocin BM1300 was dialyzed with a dialysis tube (MW 3500) in ultrapure water to remove salt. The active fraction was filter-sterilized (0.22 μm) and loaded onto a HiTrap Q FF anion exchange column at 1 mL/min using an AKTA system (AKTA Purifier 100, GE, Sweden). The column was equilibrated by Tris-HCl (20 mM, pH 6) firstly and then eluted with linear gradient (0-1 M NaCl) in equilibrium buffer (20 mM Tris-HCl). Active fractions from the last step were concentrated in a vacuum freeze concentrator and loaded on an Agilent ZORBAX 300SB-C18 reverse-phase column (250 × 4.6 mm, 5 mm) to purify using an analytical RP-HPLC (Waters 1525, USA). Gradient elution was performed at a flow rate of 1 mL/min with mobile phase A (100% H2O and 0.05% TFA) and mobile phase B (100% acetonitrile) and monitored at 280 nm. The purified BM1300 was applied to LC-MS/MS to identify the BM1300 according to the following steps. The purified BM1300 was subjected to trypsin digestion for 16 h at 37 °C before being desalted by manual Pierce C18 Tips (Thermo Fisher Scientific, USA). The hydrolysis products were analyzed by mass spectrometry using a Q Exactive mass spectrometer (Thermo Fisher Scientific, USA). Mass spectrometry output data were analyzed through the database Mascot (Matrix Science, Boston, MA).
Determination of minimum inhibitory concentration values
The purified BM1300 from RP-HPLC was concentrated and used to measure the MIC values against S. aureus ATCC 25923 and E. coli ATCC 25922 following the protocol as previously described [19]. In brief, aliquots of 50μL purified BM1300 solutions prepared by twofold serial dilutions with phosphate-buffered saline solution (PBS, 0.1 M, pH 7.2) were mixed with 50μL exponential-phase indicator bacteria in a 96-well plate and then incubated for 24 h at 37 °C. Meanwhile, the protein concentrations of each of the above BM1300 diluted solutions (diluted in PBS buffer) in this experiment were determined by BCA Assay Kit (Xi’an Hat, China).
Effects of heat, pH, protease, and chemicals on bacteriocin activity
The residual antibacterial activity of supernatants of cell disruption (bacteriocin BM1300) was determined after the treatments of 60, 80, 100, and 120 °C for 5, 10, and 20 min, respectively. Untreated BM1300 was used as a control. The impact of pH on BM1300 activity was investigated by adjusting pH of bacteriocin solutions (pH 2.0–11.0) with the following buffers (20 mM): glycine-HCl (pH 2.0), citric acid-phosphate (pH 3.0, 4.0, 5.0, 6.0), sodium phosphate (pH 7.0), Tris-HCl (pH 8.0, 9.0), and glycine-NaOH (pH 10.0, 11.0). These solutions were then incubated at 37 °C for 2 h. BM1300 without the treatment was used as a control. BM1300 was coincubated for 4 h at 37 °C with enzyme solutions (10 mg/mL) of pepsin, papain, proteinase K, and α-amylase, respectively, which was aimed at investigating the effects of protease on BM1300 activity. BM1300 without treatment with enzymes was used as a control. BM1300 was exposed to Tween-40, Tween-80, Triton X-100 of 5% (v/v), and EDTA (5 mmol/L) and incubated for 2 h at 37 °C. Untreated bacteriocin solutions and chemicals in the absence of BM1300 were served as controls. The residual antibacterial activity of BM1300 was determined after the above treatments by the agar well diffusion assay using E. coli ATCC 29522 as the indicator strain according to the previously reported methods with some modifications [20].
Hemolytic activity
A hemolysis assay was performed as previously described [21]. The mouse blood cells were washed three times with PBS (0.1 M, pH 7.2) and centrifuged at 1000×g for 10 min. Aliquots of 100μL cells were diluted to 5% (v/v) with PBS (0.1 M, pH 7.2) and added into a 96-well plate. Subsequently, aliquots of 100μL BM1300 solutions prepared by twofold serial dilutions starting at 53.6 μg/mL until 0.11 μg/mL) were also added into the prepared 96-well plate. After incubation for 1 h at 37 °C and centrifugation at 1000×g for 10 min, aliquots of 100 μL of supernatants were added to a new 96-well plate. The absorbance at 540 nm was measured to determine the released hemoglobin. A parallel treatment in the presence of 0.1% (v/v) Triton X-100 was conducted to determine the absorbance of complete hemolysis (100%). Serial twofold dilutions of ampicillin, starting from 53.6 to 0.11 μg/mL, were used as controls.
The hemolysis was calculated as
where ABM1300 is the experimental absorbance of BM1300, Ablack is the absorbance of cell supernatants treated with PBS, and Atriton is the absorbance of 0.1% Triton X-100 lysed cells.
Time-kill curve
The time-kill curve was determined according to the method of Liu et al. [22]. Briefly, the prepared S. aureus and E. coli cell suspensions (106 CFU/mL) were exposed to BM1300 at a final concentration of 2 × MIC. Bacterial growth was monitored at selected times of 0, 0.5, 1, 2, 3, 4, 5, and 6 h. The cultures at these time points were serially diluted 10-fold with sterile water and aliquots of 50μL samples were plated on LB agar plates. Viable colonies were counted after incubation for 24 h at 37 °C. Indicator bacteria grown in the absence of BM1300 were used as controls.
Scanning and transmission electron microscopy
S. aureus cell suspensions (106 CFU/mL) were exposed to BM1300 (1 × MIC and 4 × MIC, respectively) and incubated for 2 h at 37 °C with agitation. Pretreatments of E. coli cells were done following the same method as those for S. aureus. Subsequently, cells were washed three times with PBS (0.1 M, pH 7.2) and then fixed with 2.5% glutaraldehyde overnight. Cell dehydration was performed with gradient ethanol solutions and 100% acetone. The surface of cells was coated with gold (80 nm). A Nova Nano SEM-450 scanning electron microscopy (FEI, USA) was used to visualize the shape and surface characteristic changes of cells treated with BM1300 compared to non-treated cells.
For transmission electron microscope (TEM) analysis, the target bacteria were prepared by following the same method as scanning electron microscope (SEM) experiment before being fixed with 1% osmium tetroxide for 2 h. Thereafter, cells were dehydrated with the gradient ethanol solutions and permeated with white resin. The prepared cells were embedded in an oven at 55 °C for 48 h. Thin sections (70 nm) were prepared using a Leica EM UC7 ultrathin slicer (Leica, Germany) and successively stained with lead citrate and uranyl acetate. Intracellular organization alterations of S. aureus and E. coli were observed using Tecnai G2 Spirit BioTwin transmission electron microscopy (FEI, USA).
Cell membrane integrity analysis
The impact of BM1300 on cell membrane integrity of indicator bacteria was evaluated by measuring the propidium iodide (PI) uptake using a flow cytometer according to the method of Yi et al. [23] with some modifications. S. aureus and E. coli cells (106 CFU/mL) were incubated with BM1300 (final concentration of 2 × MIC) at 37 °C for 2 h. Cells were harvested and washed three times with 0.9% NaCl. Thereafter, propidium iodide solution (100 μg/mL) was added to cell suspensions and incubated for 30 min in the dark. Cells treated with 0.9% NaCl were used as negative controls. PI uptake of cells was analyzed using a FACSCalibur flow cytometer (Becton Dickinson, USA).
The effect of BM1300 on the cell membrane permeability was investigated through the release of lactate dehydrogenase (LDH) and protein and total nucleotide leakage assays. Both S. aureus and E. coli were treated following the method of PI uptake assay. Cell supernatants were collected by centrifugation at 8000×g for 8 min at 0, 1, 2, 3, 4, 5, and 6 h and filtered through 0.22 μm to remove the residual cells. Concentrations of LDH within cell supernatants were measured using a LDH kit (Nanjing Jiancheng, China). The OD values (280 nm and 260 nm) of cell supernatants were also measured by a spectrophotometer (UV-2102PCS, UNICO, USA) to determine the leakage of protein and total nucleotide. Indicator bacteria incubated without BM1300 were used as controls.
Inhibition of biofilm formation
A biofilm formation assay was performed as described previously with some alterations [24]. Overnight cultured indicator bacteria were resuspended in fresh Tryptic Soy Broth (TSB) (OD600 = 0.1). Aliquots of 100μL cell suspensions and BM1300 at the final concentrations of 1/8, 1/4, 1/2, and 1 × MIC were coincubated in a 96-well plate for 24 h at 37 °C. Indicator bacteria treated with PBS (0.1 M, pH 7.2) served as controls. The absorbance at 630 nm was measured by a microplate reader (Bio-Rad, USA). Plates were gently washed to remove the unattached bacteria with PBS (0.1 M, pH 7.2). The biofilm was fixed with 10% methanol for 10 min before wells were stained with 0.1% crystal violet, and then, the absorbance at 570 nm was measured using a microplate reader (Bio-Rad, USA). BFI (biofilm formation index) was calculated as
where SOD570 is the OD570 of the stained attached microorganisms, SCOD570 is the OD570 of the stained blank wells without microorganisms, GOD630 is the OD630 of the cell growth in suspended culture, and GCOD630 is the OD630 of the blank well.
Impact of BM1300 on cell cycle
Impact of BM1300 on S. aureus and E. coli cell cycle was analyzed according to the previously described method [25] with some modifications. In brief, overnight S. aureus and E. coli cells were harvested, washed, and resuspended in fresh LB broth (106 CFU/mL) and incubated with 2 × MIC BM1300 for 2 h at 37 °C. Subsequently, cells were harvested and fixed with 70% ice-cold ethanol overnight. Cells were washed with PBS (0.1 M, pH 7.2) and then incubated with RNase A solution (0.2 mg/mL) for 30 min at 37 °C. Cells were harvested by centrifugation at 6000×g for 5 min and resuspended in PI solution (containing 0.1% Triton X-100), followed by incubation for 30 min in the dark. The contents of DNA were measured by CytoFLEX (Beckman, USA) and analyzed by ModFit LT 5.0 software (Verity Software House, ME, USA). Cells in the absence of BM1300 were used as controls.
Application of BM1300 in beef meat
Fresh beef meat purchased at the local supermarket was cut into even-sized pieces, and 200 g each was placed into several flasks. BM1300 solutions (120 mL, final concentrations of 1 × MIC and 4 × MIC against S. aureus and E. coli, respectively) were added to the prepared flasks and incubated for 15 min. Beef meat treated with 20 mM Tris-HCl served as a control. After being dried at room temperature, aliquots of exponential-phase S. aureus and E. coli (5 mL, 106 CFU/mL) were sprayed on the beef meat and stored at 4 °C, respectively. Beef meat (5 g) of each group was weighed at 0, 1, 2, 3, 4, 6, 8, and 10 day and ground in 10-mL sterile PBS (0.1 M, pH 7.2). Aliquots of 50μL supernatants were tenfold diluted with PBS and plated on S. aureus–selective Baird Parker agar medium, E. coli–selective MacConkey agar medium, and LB agar plates, respectively. Viable colonies of S. aureus and E. coli and total microbial counts were counted after being cultivated for 24 h at 37 °C.
Statistical analysis
All experiments were performed for three independent replicates. Data are expressed as mean ± standard deviation. Student’s t test was used for all comparisons between two groups. One-way ANOVA was used to determine significant differences between three or more groups (*p < 0.05, **p < 0.01, ***p < 0.001).
Results
Antibacterial spectrum
The antibacterial spectrum of BM1300 against selected Gram-positive and Gram-negative bacteria is presented in Table 1. BM1300 significantly inhibited not only Gram-positive bacteria including Bacillus cereus, Listeria monocytogenes, S. aureus but also Gram-negative bacteria such as Cronobacter sakazakii, Salmonella typhimurium, and Yersinia enterocolitica, with the diameter of inhibition zone being more than 13 mm. Moreover, BM1300 exhibited desirable antibacterial activity against a wide range of antibiotic-resistant bacteria, including antibiotic-resistant C. sakazakii, E. coli, and Salmonella. The broad-spectrum antibacterial activity of BM1300 indicated that it had potential as a natural food preservative in the food industry.
Purification of BM1300 and MIC
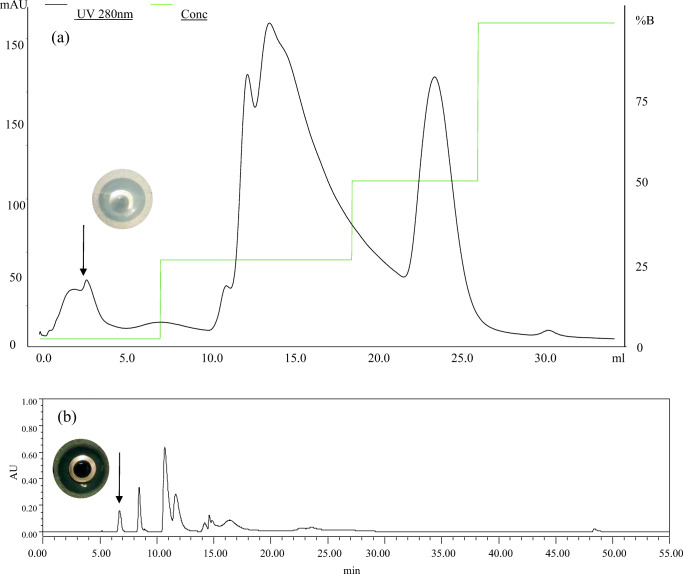
Crude BM1300 which retained the N-terminal His tag and the thrombin cleavage sequence was firstly dialyzed to desalt and remove low molecular protein (< 3.5 KDa). As shown in Fig. 1, the bacteriocin sample was purified by an anion-exchange column, at which five peaks were obtained and massive heteroproteins were moved. The active peak (arrow in Fig. 1a) was collected and then applied to the final purification of RP-HPLC. Subsequently, the chromatogram showed that nine clear peaks were isolated (Fig. 1b), after which the active peak (arrow in Fig. 1b) was taken manually and concentrated. Eight target fragments of purified BM1300 were identified and analyzed by LC-MS/MS (Fig. 2) with the coverage of 76.39% (Fig. S1). Amino acid sequences of both the beginning and middle parts were covered, suggesting that BM1300 was the core bacteriocin for mostly full coverage and correct expression (Fig. 2). The unidentified fragments of IEFW and FVLRSFKNLVIKR might be caused by the loss in desalting using manual C18 Tips. The MIC values of BM1300 against S. aureus ATCC 25923 and E. coli ATCC 25922 were 13.4 μg/mL and 6.7 μg/mL, respectively.
Fig. 1.
Purification of bacteriocin BM1300 by chromatographic columns. a Anion exchange column and inhibition zone of the arrowed peak. Black line is 280 nm, and green line is conc. b RP-HPLC and inhibition zone of the arrowed peak. The y-axis in b is the absorbance value (arbitrary units (AU)) of bacteriocin under 280 nm
Fig. 2.
The BM1300 residue fragments identified by LC-MS/MS were aligned with the full amino acid sequence. The identified amino acid fragments were highlighted in yellow color
Effect of heat, pH, protease, and chemicals on BM1300
The impact of heat, pH, protease, and chemicals on BM1300 stability are shown in Table 2. The residual activities were 94.56%, 91.06%, 90.07%, and 89.46% after being heated for 20 min at 60, 80, 100, and 120 °C, suggesting its remarkable thermal stability. As shown in Table 2, pH variation had little effect on BM1300 activity. BM1300 had higher activity in the pH range of 8–11 than at 2–4. Pepsin, papain, and proteinase K decreased the BM1300 activity, suggesting that BM1300 was slightly sensitive to protease (Table 2). The antibacterial activity of BM1300 after being treated with catalase and α-amylase was still stable, suggesting that the activity was not derived from hydrogen peroxide and carbohydrate moieties. Moreover, BM1300 was relatively stable (> 90% of residual inhibitory activity) to chemicals (EDTA, Tween-40, Tween-80, and Triton X-100) investigated in this experiment.
Table 2.
Stability of the bacteriocin BM1300 to heat, pH, protease, and chemicals
| Treatment | Residual inhibitory activity (%) |
|---|---|
| Temperature | |
| 60 °C, 5 min | 97.4% ± 1.3 |
| 60 °C, 10 min | 95.2% ± 1.3 |
| 60 °C, 20 min | 94.6% ± 0.4 |
| 80 °C, 5 min | 93.0% ± 0.9 |
| 80 °C, 10 min | 92.3% ± 0.4 |
| 80 °C, 20 min | 91.1% ± 1.0 |
| 100 °C, 5 min | 92.3% ± 1.0 |
| 100 °C, 10 min | 90.9% ± 1.3 |
| 100 °C, 20 min | 90.1% ± 0.9 |
| 120 °C, 5 min | 90.6% ± 0.3 |
| 120 °C, 10 min | 90.6% ± 0.4 |
| 120 °C, 20 min | 89.5% ± 0.8 |
| pH | |
| 2 | 89.5% ± 4.0 |
| 3 | 93.6% ± 0.5 |
| 4 | 94.0% ± 0.5 |
| 5 | 97.7% ± 2.5 |
| 6 | 96.6% ± 1.5 |
| 7 | 96.6% ± 2.5 |
| 8 | 97.4% ± 2.0 |
| 9 | 101.9% ± 0.5 |
| 10 | 100.8% ± 1.5 |
| 11 | 101.1% ± 1.0 |
| Protease | |
| Pepsin | 73.61% ± 2.5 |
| Papain | 78.87% ± 1.5 |
| Proteinase K | 65.86% ± 2.5 |
| Catalase | 96.77% ± 2.0 |
| α-Amylase | 97.74% ± 1.0 |
| Chemicals | |
| EDTA (5 mmol/L) | 97.2% ± 0.8 |
| Tween-40 (5% v/v) | 94.7% ± 1.2 |
| Tween-80 (5% v/v) | 98.3% ± 1.5 |
| Triton-100 (5% v/v) | 98.3% ± 1.8 |
Hemolytic activity of BM1300
In this work, the hemolytic activity assay was carried out to observe the hemolysis caused by increased concentrations of BM1300 to confirm it is biologically safe. As shown in Fig. S2, BM1300 showed less than 1.5% hemolytic activity among tested concentrations. According to the data obtained, hemolysis caused by the highest concentrations used (53.6 μg/mL) was very low (less than 1.2%). On the contrary, ampicillin had 2% hemolysis in a low concentration range, which was more than BM1300. It was remarkable that lysis caused by BM1300 on mouse erythrocytes required concentrations much higher than those required for antibacterial activity.
Time-kill curves
The time-kill curve assay is aimed at investigating the mode of action of BM1300. As shown in Fig. 3, exposure of 2 × MIC BM1300 for 1 h resulted in a decrease of 3.44 and 5.40 log10 CFU/mL (greater than 3 log10 CFU/mL) for S. aureus and E. coli, respectively. It suggested that BM1300 damaged S. aureus and E. coli through the quick-acting mode. In particular, we found that BM11300 had a better bactericidal effect (99.9% [3log10] reduction of bacterial inoculum) [22] against E. coli compared to S. aureus. The viable counts of S. aureus and E. coli continually decreased in a time-dependent manner. After 6 h, a significant reduction of viable counts relative to the initial density was observed for S. aureus and E. coli (reduced by 5.28 and 7.18 log10 CFU/mL, respectively). These results suggested that BM1300 had bactericidal modes of action against S. aureus and E. coli in a time-dependent manner.
Fig. 3.
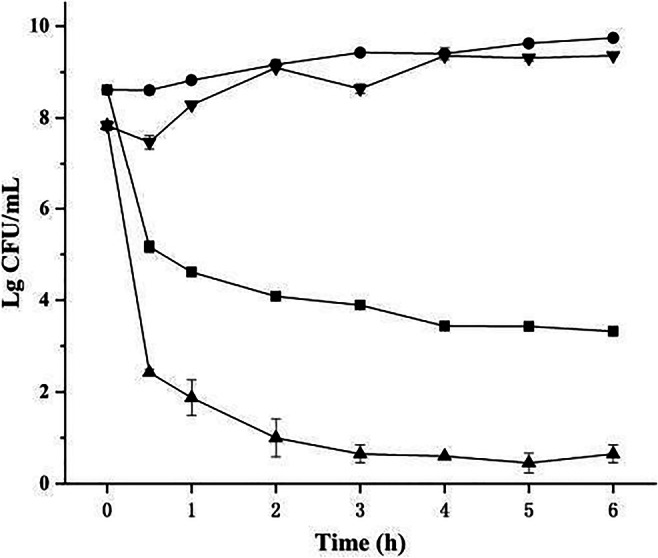
Time-kill curve against S. aureus (circle is the control, and square is treated by BM1300 at 2 × MIC) and E. coli (inverted triangle is the control, and triangle is treated by BM1300 at 2 × MIC). Data are expressed as mean ± SD. Each individual treatment is run in triplicate
SEM and TEM
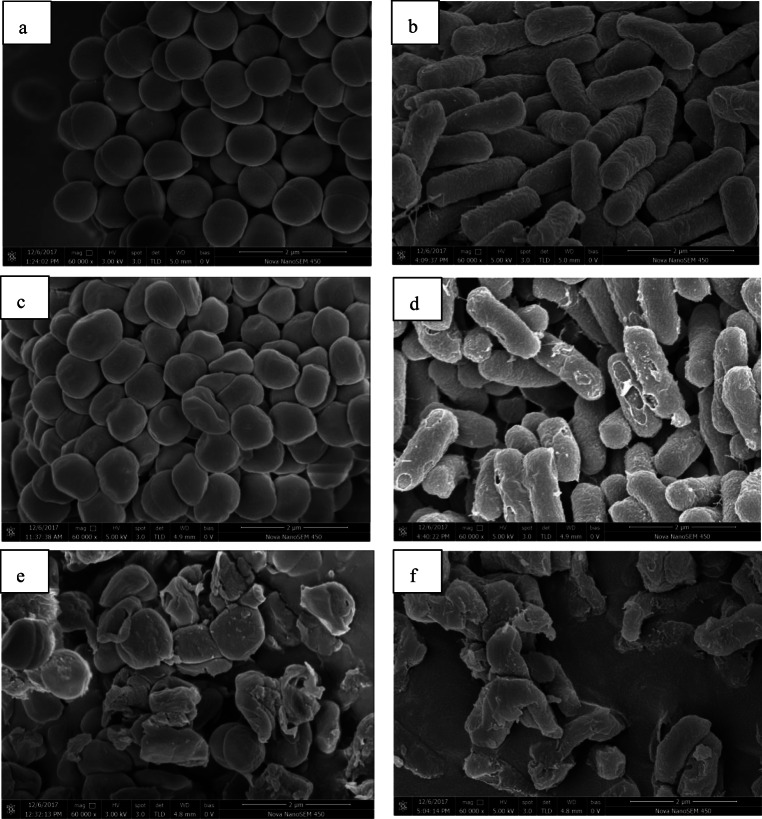
SEM analysis was investigated to observe the morphology changes of target bacteria caused by BM1300. As shown in Fig. 4a, untreated S. aureus cells showed typical plump cell and smooth cell membranes. In contrast, S. aureus cells treated with 1 × MIC BM1300 exhibited noticeable sunken deformations of shape with some cavities (Fig. 4c). When S. aureus cells were exposed to 4 × MIC BM1300, the cell walls were distorted completely and the majority of cells could not maintain their normal shape, indicating there was abnormal cell breakage and cell lysis (Fig. 4e). Moreover, compared with untreated E. coli which had rod-shaped and pump profile, membrane damage was observed on the surface of E. coli cells after treatment with 1 × MIC BM1300 (Fig. 4d). Massive damage caused by 4 × MIC BM1300 was more evident with the evidence of abnormal cell breakage and cell lysis (Fig. 4f). There were apparent pores, incomplete and rough features on the surface of E. coli cells; there were even almost no intact rod cells due to severe cell lysis. Obviously, these results indicated that BM1300 induced alterations in the morphology of S. aureus and E. coli in a concentration-dependent manner.
Fig. 4.
SEM images of S. aureus (control (a); treated by BM1300 at 1 × MIC (c); treated by BM1300 at 4 × MIC (e)) and E. coli (control (b); treated by BM1300 at 1 × MIC (d); treated by BM1300 at 4 × MIC (f))
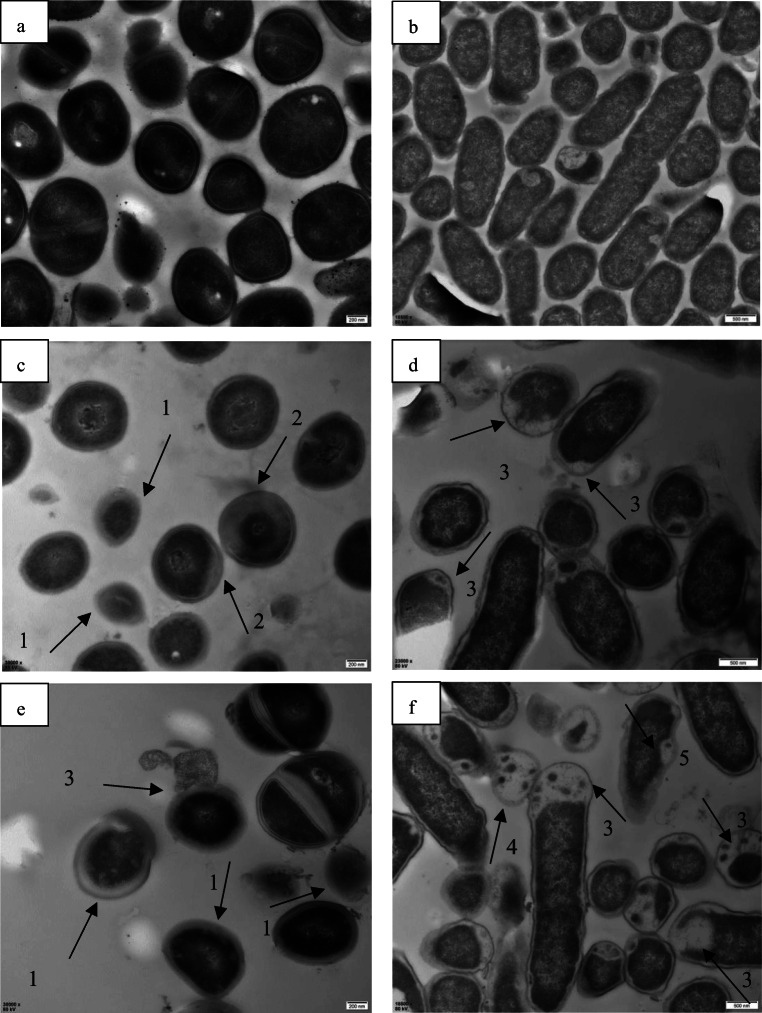
TEM was performed to observe the intracellular organization changes of target cells caused by BM1300. As shown in Fig. 5c, there was no clear delineation between the S. aureus cell wall and the cytoplasm membrane after treatment with 1 × MIC BM1300. In contrast, following the treatment with 4 × MIC BM1300, severe separation between the cytoplasm membrane and the cell wall, as well as heterogeneous electron density, could be observed (Fig. 5e). TEM images of E. coli treated by 1 × MIC BM1300 demonstrated that uneven cytoplasm and cytoplasmic vacuolization that resulted from the leakage of the intracellular cytoplasmic contents could be found (Fig. 5d). There were a large number of cells with vacuolization due to loss of cytoplasmic substance after treatment with 4 × MIC BM1300 (Fig. 5f). TEM images clearly demonstrated that BM1300 induced the leakage of cytoplasmic contents of S. aureus and E. coli due to the significantly damaged permeability and integrity of the cell membranes.
Fig. 5.
TEM images of S. aureus (control (a); treated by BM1300 at 1 × MIC (c); treated by BM1300 at 4 × MIC (e)) and E. coli (control (b); treated by BM1300 at 1 × MIC (d); treated by BM1300 at 4 × MIC (f)). No clear delineation between cell wall and cytoplasm membrane and plasmolysis appearance of cell (arrows 1#), heterogeneous electron density (arrows 2#), loss of cytoplasm (arrows 3#), vacuolization (arrows 4#), and formation of pores (arrows #5) are observed
Effect of BM1300 on cell membrane integrity
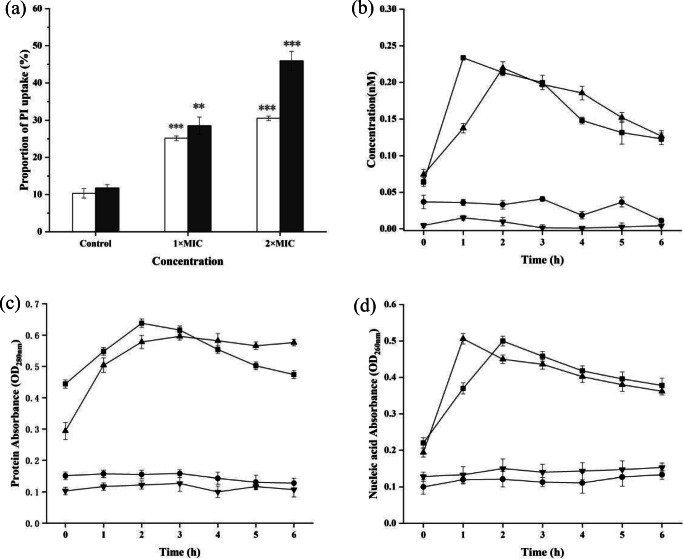
The cell membrane integrity was investigated by a flow cytometer combined with the fluorescent dye PI, which could distinguish membrane-damaged cells from intact cells [26]. The results showed that there were 25.2% positive S. aureus cells of PI uptake caused by 1 × MIC BM1300 (Fig. 6a), and the 30.5% PI fluorescent signal of cells treated by 4 × MIC BM1300 indicated the increase of cytoplasmic membrane permeability. More massive damage was observed for E. coli and PI uptake of cells increased from 11.8 (control) to 28.5% (1 × MIC) and 45.9% (4 × MIC), respectively. These results indicated that the significantly damaged cell membrane was induced by BM1300 in a concentration-dependent manner, which was in accordance with the TEM analysis. Meanwhile, it was consistent with results that BM1300 exhibited stronger antibacterial activity against E. coli.
Fig. 6.
Cell membrane integrity of S. aureus and E. coli. a PI uptake by flow cytometry analysis (white bar is S. aureus, and black bar is E. coli). b LDH release of S. aureus (circle is the control, and square is treated by BM1300 at 2 × MIC) and E. coli (inverted triangle is the control, and triangle is treated by BM1300 at 2 × MIC). c Protein release of S. aureus (circle is the control, and square is treated by BM1300 at 2 × MIC) and E. coli (inverted triangle is the control, and triangle is treated by BM1300 at 2 × MIC). d Total nucleic acid leakage of S. aureus (circle is the control, and square is treated by BM1300 at 2 × MIC) and E. coli (inverted triangle is the control, and triangle is treated by BM1300 at 2 × MIC). Data are expressed as mean ± SD. Each individual treatment is run in triplicate. *p < 0.05, **p < 0.001, and ***p < 0.001 were considered significantly different compared with the controls
We also determined whether BM1300 could permeabilize target cells to release intracellular substance in a time-dependent manner. As shown in Fig. 6b, BM1300 induced a rapid leakage of LDH of S. aureus and reached 0.22 nM at 2 h, after which there was a slight decrease. The concentration of LDH of E. coli reached a maximum value of 0.23 nM at 1 h, suggesting BM1300 was quick-acted against E. coli than S. aureus. The decrease of LDH concentrations may be a consequence of digestion by enzymes after the rapid death of the target cells [27]. These results demonstrated that BM1300 damaged the cell membrane integrity with the selective pores, causing the leakage of LDH of target cells in a time-dependent manner.
BM1300 resulted in the leakage of protein and nucleotide in a time-dependent manner (Fig. 6c, d). Treatment with BM1300 led the absorbance values for external protein of S. aureus to significantly increase from 0.44 to 0.638 at 2 h, followed by a slight decreasing trend between 3 and 6 h. The efflux of protein from E. coli increased from 0.29 to 0.596 at 2 h, which was greater than that released from S. aureus. On the contrary, both S. aureus and E. coli without BM1300 treatment maintained low levels for external protein and nucleotide. After treatment with 2 × MIC BM1300 for 2 h, the absorbance values for S. aureus nucleic acids increased from 0.22 to 0.50 (Fig. 6d) and then also showed a slight decreasing trend along the intervals evaluated. In contrast, external nucleic acids levels of E. coli cells increased from 0.19 to 0.50 within 1 h (Fig. 6d). These results demonstrated that the disruption of cell membrane integrity caused by BM1300 led to cell death.
Inhibition of biofilm formation
To evaluate the effect of the BM1300 on biofilm formation, the biofilm formation index (BFI) of S. aureus and E. coli treated by BM1300 was analyzed (Fig. 7). BM1300 exhibited bactericidal activity against both S. aureus and E. coli at 2 × MIC (Fig. 3). Therefore, we investigated the effect on biofilm formation at lower concentrations. Results revealed that BM1300 significantly reduced the biofilm formation in a concentration-dependent manner. The 1/8 × MIC BM1300 led to a significant 73.8% and 46.4% decrease of BFI for S. aureus and E. coli, respectively. Moreover, this inhibitory effect reached a maximum value of 96% (S. aureus) and 69.2% (E. coli) at 1 × MIC, indicating its antibiofilm activity against S. aureus was better than that against E. coli.
Fig. 7.
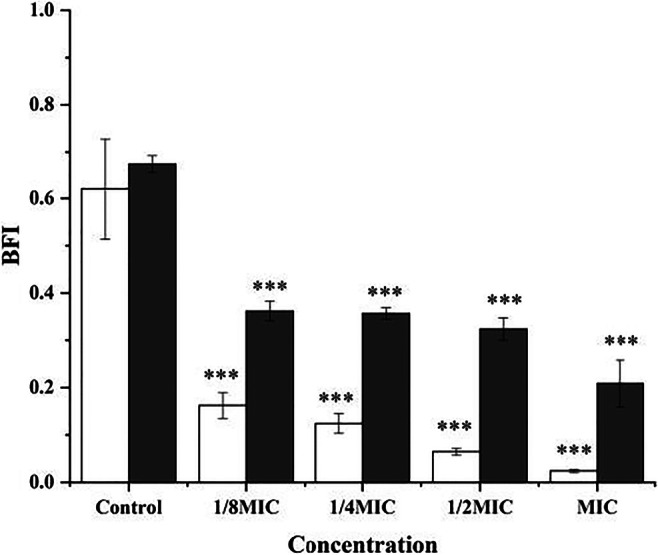
Inhibition of biofilm formation against S. aureus (white bar) and E. coli (black bar) at 1/8, 1/4, 1/2, and 1 × MIC BM1300. Data are expressed as mean ± SD. Each individual treatment is run in triplicate. *p < 0.05, **p < 0.001, and ***p < 0.001 were considered significantly different compared with the controls
Effect of BM1300 on cell cycle
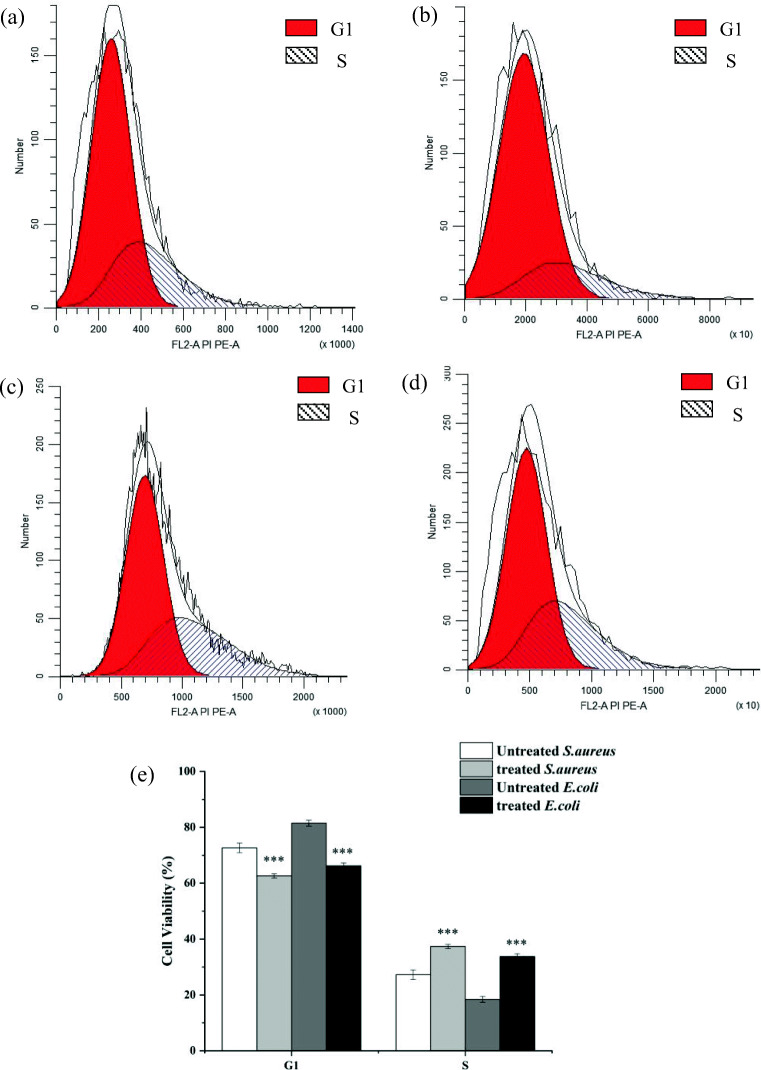
Membrane permeabilization caused by BM1300 would impact cell, and it may subsequently impact cell cycle. To determine if BM1300 had impact on cell cycle of S. aureus and E. coli, we investigated the distribution alterations of cell cycle treated by BM1300. For S. aureus, the proportion of cells in phase R (or in phase S) significantly increased from 27.32 to 37.37% after treatment with BM1300, while cells in phase I (or in phase G1) decreased from 72.68 to 62.62% (Fig. 8e). Moreover, there was a similar tendency for E. coli treated by BM1300. The proportion of cells in G1 phase decreased from 81.53 to 66.27% and cells in S phase increased from 18.46 to 33.73% (Fig. 8e). These results suggested that the effect of BM1300 on the cell cycle of E. coli was more obvious than S. aureus, which was a reason why BM1300 exhibited stronger antibacterial activity toward E. coli.
Fig. 8.
The cell cycle distribution (G1 and S phases) of S. aureus (control (a) and treated with 2 × MIC BM1300 (c)) and E. coli (control (b) and treated with 2 × MIC BM1300 (d)). Corresponding quantitative results reflecting the relationship between DNA content and cell numbers (e). Data are expressed as mean ± SD. Each individual treatment is run in triplicate. *p < 0.05, **p < 0.001, and ***p < 0.001 were considered significantly different compared with the controls
Control of S. aureus and E. coli in beef meat
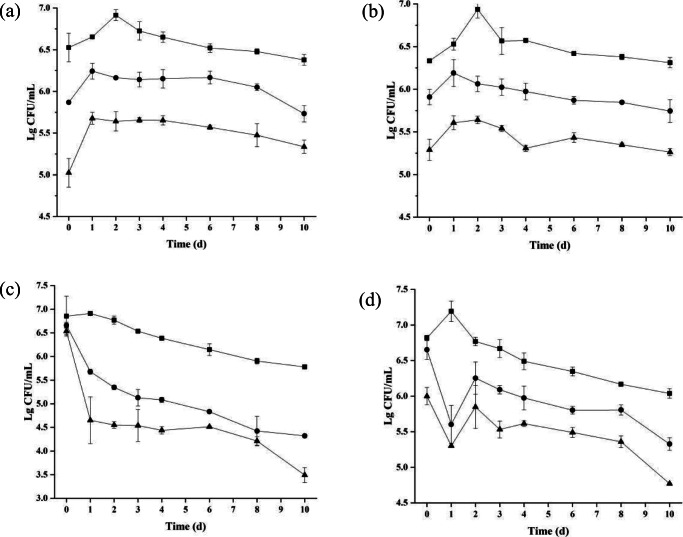
Survival S. aureus, E. coli, and total microbial counts in beef meat after treatment with BM1300 were monitored during a 10-day storage period (Fig. 9). As shown in Fig. 9a, counts of S. aureus in the absence of BM1300 continuously increased and then remained stable for the control. In contrast, the counts of S. aureus treated with 1 × MIC and 4 × MIC BM1300 decreased to 5.87 and 5.02 log10 CFU/mL following initial application, respectively. After maintaining a steady growth trend of colonies, the two treatment groups remained stable. The viable counts of S. aureus were lower than that of the control in a concentration-dependent manner (Fig. 9a). On the other hand, the effect of total bacterial counts caused by BM1300 was similar to the changes of the viable counts of S. aureus (Fig. 9b), indicating BM1300 had a bacteriostatic effect against total bacterial counts in beef meat. Viable counts of E. coli treated by 1 × MIC and 4 × MIC BM1300 reduced by 0.99 and 1.88 log10 CFU/mL, respectively, at the first day compared to the initial day (Fig. 9c). And a stable decrease of E. coli was observed from the second day to the end of the experiment. Meanwhile, the addition of 1 × MIC and 4 × MIC BM1300 also reduced total microbial counts at the first day (reduced to 5.60 and 5.30 log10 CFU/mL, respectively) (Fig. 9d). Subsequently, total bacterial counts had a slight increase because there were some other bacteria that cannot be inhibited. Therefore, these data demonstrated that BM1300 could effectively inhibit the growth of E. coli and corresponding total bacterial counts during refrigerated storage, and the antibacterial effect against E. coli was better than that of S. aureus.
Fig. 9.
The viable counts of S. aureus (a), corresponding total bacterial counts (b), and E. coli (c), corresponding total bacterial counts (d), of beef. Square, circle, and triangle are the controls, treated by BM1300 at 1 × MIC and treated by BM1300 at 4 × MIC, respectively. Data are expressed as mean ± SD. Each individual treatment is run in triplicate
Discussion
Pathogenic S. aureus, L. monocytogenes, B. cereus, S. typhimurium, C. sakazakii, Y. enterocolitica, and E. coli are considered the leading cause of food poisoning via contaminating food. Bacteriocin BM1300 was active against these food-borne pathogenic bacteria, which showed promising potential use in the prevention or treatment of these pathogens. In general, bacteriocins produced by LAB could significantly inhibit closely related strains but possess weak antibacterial activity to Gram-negative bacteria. The broad-spectrum antibacterial activity of BM1300 was similar to pentocin 31-1 [28] and enterocin LR/6 [29]. But BM1300 could inhibit Salmonella, B. cereus, and S. aureus, which was different from the above bacteriocins. This characteristic feature of broad-spectrum antibacterial activity of BM1300 was in agreement with many antimicrobial peptides. MIC values against bacteria were 5–100 μg/mL for the majority of bacteriocins [23]. Lower MICs against both S. aureus and E. coli were obtained for BM1300 than that previously reported for plantaricin JLA-9 [17], bifidocin A [26], and enterocin-B [30]. Obviously, BM1300 with low MICs possessed the potential use as a food preservative in the food industry.
Good stability of bacteriocins is essential for their application under different complex physicochemical conditions. It was reported that some bacteriocins were similar to BM1300 and were stable after heat treatment for 10–20 min at 121 °C, such as plantarincin JY22 [31] and pentocin MQ1 [2]. BM1300 was stable at a wide pH range of 2–11, particularly at alkaline conditions. However, nisin displayed limited activity at neutral and alkaline conditions [32], thus becoming a disadvantage of application in the food industry. In contrast, BM1300 possessed this advantage in the food processing. Moreover, BM1300 shared similar pH stability with plantaricin 163 [33], and the stability of BM1300 after treatment with chemicals was similar to pentocin MQ1 [2]. The desirable combined characteristics of thermal, pH, and chemical stabilities of BM1300 favored its future application as a food preservative subjected to complicated processing conditions.
Time-kill curve experiment showed that BM1300 exhibited bactericidal modes of action against both S. aureus and E. coli at 2 × MIC. Moreover, BM1300 exerted a more efficient bactericidal effect toward E. coli than S. aureus. Both the SEM and TEM images distinctly showed that BM1300 caused S. aureus cells inward depression and even lysis, which led to cell death. On the contrary, BM1300 led to the increase of cell membrane permeability, pore forming, and leakage of cytoplasmic contents of E. coli. As previously reported, chitosan also could disrupt the organization of the OM and further increase its membrane permeability [34]. Thereafter, BM1300 may interact with the outer membrane protein receptor and cross the outer membrane barrier leading to cell death [4]. Moreover, the leakage of intracellular macromolecules might appear which resulted from the pore formation of target cells [22]. The PI uptake and release of LDH and protein and nucleotide experiments indicated that the cell membrane integrity was damaged. There were some non-selective pores formed by low concentration BM1300, and more severe disruption caused by high concentration BM1300 occurred, in which the cell walls exhibited noticeable hollowness and even dissolved completely, causing the leakage of intracellular contents. With bacteriocin aggregation, pore formation and ion leakage (protein and nucleic acid) even cell lysis caused by BM1300 occurred. Therefore, the changes of cell membrane integrity and cell membrane permeability resulted in complete cell disintegration. These results were similar to the antibacterial mechanism of plantaricin LPL-1 [35] and bacteriocin CHQS [36]. This mode of action was different from the non-pore-forming bacteriocin lactococcin 972 [37] and fermencin SA715 [38]. Therefore, the primary mode of action of BM1300 on S. aureus and E. coli cells could be the disruption of cell membrane integrity.
After disrupting the cell membrane, BM1300 would act on inside targets. We found that cell arrests occurred at S phase after the treatment with BM1300. Cell cycle arrests caused by BM1300 contributed to its antibacterial activity against S. aureus and E. coli. Based on the above results, BM1300 might firstly disrupt the cell membrane through increasing cell membrane permeability or pore formation, and then BM1300 was allowed to enter cells to disrupt the cell cycle to exert its antibacterial activity. Moreover, BM1300 probably inhibit or hamper gene expression, which is an effective way to block the synthesis of normal enzyme and receptor or mRNA. Finally, the metabolism disruption of the life cycle of bacteria, including DNA replication, results in cell death [25]. This, however, is subject to further investigation.
We also investigated whether antibiofilm activity of BM1300 and destruction of cell membrane caused by BM1300 played a synergistic role in inhibiting the growth of target cells. We found that BM1300 significantly reduced the biofilm formation of S. aureus and E. coli. The reason why antibiofilm activity of BM1300 against S. aureus was better than that against E. coli was different cell membrane structure of target cells [39]. The biofilm architecture formed by E. coli was much thicker and heterogeneous, acting as a more effective diffusion barrier against BM1300 [40]. BM1300 caused a reduction (> 80%) of the biofilm formation in S. aureus, which was similar with human cathelicidin peptide LL37 [41]. The reduction of biofilm formation in E. coli after treatment with 1 × MIC BM1300 was close to the effect of L-K6 (6.25 μM) on Streptococcus mutans [42]. It is well known that the biofilms of pathogenic microorganisms on equipment surfaces can lead to potential hygienic risks [43]. Since the effective methods of removing and controlling biofilm are limited at present, therefore, BM1300 has the potential use as a natural biopreservative to inhibit the biofilm formation.
BM1300 showed low hemolytic activity against mouse blood cells (Fig. S2), indicating that it was biologically safe. At present, microbial deterioration is one of the most common factors of beef meat spoilage [44]. BM1300 could substantially and effectively inhibit the growth of S. aureus and E. coli in beef meat under storage. It was reported that some bacteriocins showed similar results; for example, bacteriocin DY4-2 produced by Lactobacillus plantarum exhibited an inhibitory effect on microbial growth in fillets [34]. Although the biopreservative capability of several bacteriocins has been reported [2, 3, 45], few reports indicated that they could control the growth of S. aureus, E. coli, and total bacterial counts of beef meat. BM1300 improved the microbiological quality of beef meat, and its potential capability for preserving beef meat was demonstrated in this study.
Conclusion
In conclusion, the novel bacteriocin BM1300 was purified and exhibited broad-spectrum antibacterial activity against selected Gram-positive bacteria and Gram-negative bacteria. BM1300 possessed excellent thermal, pH, and chemical stabilities. BM1300 had bactericidal modes of action against S. aureus and E. coli. BM1300 disrupted and made pores on the cytoplasmic membrane in S. aureus and E. coli cells. Moreover, BM1300 led to the plasmolysis of S. aureus and leakage of intracellular materials of E. coli. Meanwhile, BM1300 exhibited antibacterial activity through the inhibition of biofilm formation and the disruption of cell cycle distribution. BM1300 could inhibit the growth of S. aureus, E. coli, and total bacterial counts in beef meat, suggesting its potential application for food preservation.
Electronic supplementary material
(DOCX 1.42 mb)
Funding information
This research was supported by the Special Fund for Agroscientific Research in the Public Interest (Grant No. 201503135) of China.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Messaoudi S, Manai M, Kergourlay G, Prevost H, Connil N, Chobert JM, Dousset X. Lactobacillus salivarius: bacteriocin and probiotic activity. Food Microbiol. 2013;36(2):296–304. doi: 10.1016/j.fm.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 2.Wayah SB, Philip K (2018) Pentocin MQ1: a novel, broad-spectrum, pore-forming bacteriocin from Lactobacillus pentosus CS2 with quorum sensing regulatory mechanism and biopreservative potential. Front Microbiol 9. 10.3389/fmicb.2018.00564 [DOI] [PMC free article] [PubMed]
- 3.Perez RH, Zendo T, Sonomoto K. Novel bacteriocins from lactic acid bacteria (LAB): various structures and applications. Microb Cell Factories. 2014;13:S3. doi: 10.1186/1475-2859-13-s1-s3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cotter PD, Ross RP, Hill C. Bacteriocins - a viable alternative to antibiotics? Nat Rev Microbiol. 2013;11(2):95–105. doi: 10.1038/nrmicro2937. [DOI] [PubMed] [Google Scholar]
- 5.Barbosa AAT, Mantovani HC, Jain S. Bacteriocins from lactic acid bacteria and their potential in the preservation of fruit products. Crit Rev Biotechnol. 2017;37(7):852–864. doi: 10.1080/07388551.2016.1262323. [DOI] [PubMed] [Google Scholar]
- 6.Saraniya A, Jeevaratnam K. Purfication and moe of action of antilisterial bacteriocins produced by lactobacillus pentosus SJ65 isolated from uttappam batter. J Food Biochem. 2014;38(6):612–619. doi: 10.1111/jfbc.12098. [DOI] [Google Scholar]
- 7.Rea MC, Sit CS, Clayton E, O’Connor PM, Whittal RM, Zheng J, Vederas JC, Ross RP, Hill C. Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile. Proc Natl Acad Sci USA. 2010;107(20):9352–9357. doi: 10.1073/pnas.0913554107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pieterse R, Todorov SD, Dicks LMT. Bacteriocin ST91KM, produced by Streptococcus gallolyticus subsp macedonicus ST91KM, is a narrow-spectrum peptide active against bacteria associated with mastitis in dairy cattle. Can J Microbiol. 2008;54(7):525–531. doi: 10.1139/w08-040. [DOI] [PubMed] [Google Scholar]
- 9.Yang H, Wan L, Li XW, Cai HW, Chen LH, Li SF, Li YP, Cheng JQ, Liu XF. High level expression of His-tagged colicin 5 in E. coli and characterization of its narrow-spectrum bactericidal activity and pore-forming action. Protein Expr Purif. 2007;54(2):309–317. doi: 10.1016/j.pep.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 10.Gao YR, Li DP, Sheng Y, Liu XY. Mode of action of sakacin C2 against Escherichia coli. Food Control. 2011;22(5):657–661. doi: 10.1016/j.foodcont.2010.07.010. [DOI] [Google Scholar]
- 11.Cao-Hoang L, Marechal PA, Le-Thanh M, Gervais P. Synergistic action of rapid chilling and nisin on the inactivation of Escherichia coli. Appl Microbiol Biotechnol. 2008;79(1):105–109. doi: 10.1007/s00253-008-1402-9. [DOI] [PubMed] [Google Scholar]
- 12.Bali V, Panesar PS, Bera MB, Kennedy JF. Bacteriocins: recent trends and potential applications. Crit Rev Food Sci Nutr. 2016;56(5):817–834. doi: 10.1080/10408398.2012.729231. [DOI] [PubMed] [Google Scholar]
- 13.Alvarez-Sieiro P, Montalban-Lopez M, Mu DD, Kuipers OP. Bacteriocins of lactic acid bacteria: extending the family. Appl Microbiol Biotechnol. 2016;100(7):2939–2951. doi: 10.1007/s00253-016-7343-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diep DB, Skaugen M, Salehian Z, Holo H, Nes IF. Common mechanisms of target cell recognition and immunity for class II bacteriocins. Proc Natl Acad Sci USA. 2007;104(7):2384–2389. doi: 10.1073/pnas.0608775104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yi LH, Dang Y, Wu JL, Zhang LH, Liu XJ, Liu BF, Zhou Y, Lu X. Purification and characterization of a novel bacteriocin produced by Lactobacillus crustorum MN047 isolated from koumiss from Xinjiang, China. J Dairy Sci. 2016;99(9):7002–7015. doi: 10.3168/jds.2016-11166. [DOI] [PubMed] [Google Scholar]
- 16.Yi LH, Luo LL, Lu X. Heterologous expression of two novel bacteriocins produced by Lactobacillus crustorum MN047 and application of BM1157 in control of Listeria monocytogenes. Food Control. 2018;86:374–382. doi: 10.1016/j.foodcont.2017.11.042. [DOI] [Google Scholar]
- 17.Zhao SM, Han JZ, Bie XM, Lu ZX, Zhang C, Lv FX. Purification and characterization of plantaricin JLA-9: a novel bacteriocin against Bacillus spp. produced by Lactobacillus plantarum JLA-9 from Suan-Tsai, a traditional Chinese fermented cabbage. J Agric Food Chem. 2016;64(13):2754–2764. doi: 10.1021/acs.jafc.5b05717. [DOI] [PubMed] [Google Scholar]
- 18.Yu A, Ying W, Liang X, Yi H, Zuo Z, Xu X, Zhang D, Yu C, Xue H. Purification and partial characterization of M1-UVs300, a novel bacteriocin produced by Lactobacillus plantarum isolated from fermented sausage. Food Control. 2017;81:S0956713517302724. [Google Scholar]
- 19.Yi LH, Luo LL, Lu X (2018) Efficient exploitation of multiple novel bacteriocins by combination of complete genome and peptidome. Front Microbiol 9. 10.3389/fmicb.2018.01567 [DOI] [PMC free article] [PubMed]
- 20.Lu X, Hu P, Dang Y, Liu BF. Purification and partial characterization of a novel bacteriocin produced by Lactobacillus casei TN-2 isolated from fermented camel milk (Shubat) of Xinjiang Uygur Autonomous Region, China. Food Control. 2014;43:276–283. doi: 10.1016/j.foodcont.2014.03.020. [DOI] [Google Scholar]
- 21.Chen YQ, Min C, Sang M, Han YY, Ma XA, Xue XQ, Zhang SQ. A cationic amphiphilic peptide ABP-CM4 exhibits selective cytotoxicity against leukemia cells. Peptides. 2010;31(8):1504–1510. doi: 10.1016/j.peptides.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 22.Liu GR, Song ZQ, Yang XL, Gao YK, Wang CT, Sun BG. Antibacterial mechanism of bifidocin A, a novel broad-spectrum bacteriocin produced by Bifidobacterium animalis BB04. Food Control. 2016;62:309–316. doi: 10.1016/j.foodcont.2015.10.033. [DOI] [Google Scholar]
- 23.Yi LH, Li X, Luo LL, Lu YY, Yan H, Qiao Z, Lu X. A novel bacteriocin BMP11 and its antibacterial mechanism on cell envelope of Listeria monocytogenes and Cronobacter sakazakii. Food Control. 2018;91:160–169. doi: 10.1016/j.foodcont.2018.03.038. [DOI] [Google Scholar]
- 24.Segev-Zarko L, Saar-Dover R, Brumfeld V, Mangoni ML, Shai Y. Mechanisms of biofilm inhibition and degradation by antimicrobial peptides. Biochem J. 2015;468:259–270. doi: 10.1042/bj20141251. [DOI] [PubMed] [Google Scholar]
- 25.Li LR, Shi YH, Cheserek MJ, Su GF, Le GW. Antibacterial activity and dual mechanisms of peptide analog derived from cell-penetrating peptide against Salmonella typhimurium and Streptococcus pyogenes. Appl Microbiol Biotechnol. 2013;97(4):1711–1723. doi: 10.1007/s00253-012-4352-1. [DOI] [PubMed] [Google Scholar]
- 26.Liu GR, Ren GM, Zhao L, Cheng L, Wang CT, Sun BG. Antibacterial activity and mechanism of bifidocin A against Listeria monocytogenes. Food Control. 2017;73:854–861. doi: 10.1016/j.foodcont.2016.09.036. [DOI] [Google Scholar]
- 27.Debeer D, Srinivasan R, Stewart PS. Direct measurement of chlorine penetration into biofilms during disinfection. Appl Environ Microbiol. 1994;60(12):4339–4344. doi: 10.1586/erm.11.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu GR, Lv Y, Li PL, Zhou K, Zhang JL. Pentocin 31-1, an anti-Listeria bacteriocin produced by Lactobacillus pentosus 31-1 isolated from Xuan-Wei Ham, a traditional China fermented meat product. Food Control. 2008;19(4):353–359. doi: 10.1016/j.foodcont.2007.04.010. [DOI] [Google Scholar]
- 29.Kumar M, Srivastava S. Antilisterial activity of a broad-spectrum bacteriocin, enterocin LR/6 from Enterococcus faecium LR/6. Appl Biochem Biotechnol. 2010;162(3):698–706. doi: 10.1007/s12010-009-8851-1. [DOI] [PubMed] [Google Scholar]
- 30.Ankaiah D, Palanichamy E, Antonyraj CB, Ayyanna R, Perumal V, Ahamed SIB, Arul V. Cloning, overexpression, purification of bacteriocin enterocin-B and structural analysis, interaction determination of enterocin-A, B against pathogenic bacteria and human cancer cells. Int J Biol Macromol. 2018;116:502–512. doi: 10.1016/j.ijbiomac.2018.05.002. [DOI] [PubMed] [Google Scholar]
- 31.Lv XR, Miao LH, Ma HH, Bai FL, Lin Y, Sun MT, Li JR. Purification, characterization and action mechanism of plantaricin JY22, a novel bacteriocin against Bacillus cereus produced by Lactobacillus plantarum JY22 from golden carp intestine. Food Sci Biotechnol. 2018;27(3):695–703. doi: 10.1007/s10068-017-0280-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DelvesBroughton J, Blackburn P, Evans RJ, Hugenholtz J. Applications of the bacteriocin, nisin. Anton Leeuw Int J Gen Mol Microbiol. 1996;69(2):193–202. doi: 10.1007/bf00399424. [DOI] [PubMed] [Google Scholar]
- 33.Hu MZ, Zhao HZ, Zhang C, Yu JS, Lu ZX. Purification and characterization of plantaricin 163, a novel bacteriocin produced by Lactobacillus plantarum 163 isolated from traditional Chinese fermented vegetables. J Agric Food Chem. 2013;61(47):11676–11682. doi: 10.1021/jf403370y. [DOI] [PubMed] [Google Scholar]
- 34.Lou MM, Zhu B, Muhammad I, Li B, Xie GL, Wang YL, Li HY, Sun GC. Antibacterial activity and mechanism of action of chitosan solutions against apricot fruit rot pathogen Burkholderia seminalis. Carbohydr Res. 2011;346(11):1294–1301. doi: 10.1016/j.carres.2011.04.042. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y, Qin Y, Zhang Y, Wu R, Li P. Antibacterial mechanism of plantaricin LPL-1, a novel class IIa bacteriocin against Listeria monocytogenes. Food Control. 2019;97:87–93. doi: 10.1016/j.foodcont.2018.10.025. [DOI] [Google Scholar]
- 36.Cao S, Du R, Zhao F, Xiao H, Han Y, Zhou Z. The mode of action of bacteriocin CHQS, a high antibacterial activity bacteriocin produced by Enterococcus faecalis TG2. Food Control. 2019;96:470–478. doi: 10.1016/j.foodcont.2018.09.028. [DOI] [Google Scholar]
- 37.Martinez B, Zomer AL, Rodriguez A, Kok J, Kuipers OP. Cell envelope stress induced by the bacteriocin Lcn972 is sensed by the lactococcal two-component system CesSR. Mol Microbiol. 2007;64(2):473–486. doi: 10.1111/j.1365-2958.2007.05668.x. [DOI] [PubMed] [Google Scholar]
- 38.Wayah SB, Philip K. Characterization, yield optimization, scale up and biopreservative potential of fermencin SA715, a novel bacteriocin from Lactobacillus fermentum GA715 of goat milk origin. Microb Cell Factories. 2018;17:125. doi: 10.1186/S12934-018-0972-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mataraci E, Dosler S. In vitro activities of antibiotics and antimicrobial cationic peptides alone and in combination against methicillin-resistant Staphylococcus aureus biofilms. Antimicrob Agents Chemother. 2012;56(12):6366–6371. doi: 10.1128/aac.01180-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chung PY, Khanum R. Antimicrobial peptides as potential anti-biofilm agents against multidrugresistant bacteria. J Microbiol Immunol Infect. 2017;50(4):405–410. doi: 10.1016/j.jmii.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 41.Hell E, Giske CG, Nelson A, Romling U, Marchini G. Human cathelicidin peptide LL37 inhibits both attachment capability and biofilm formation of Staphylococcus epidermidis. Lett Appl Microbiol. 2010;50(2):211–215. doi: 10.1111/j.1472-765X.2009.02778.x. [DOI] [PubMed] [Google Scholar]
- 42.Shang DJ, Liang H, Wei S, Yan X, Yang QZ, Sun Y. Effects of antimicrobial peptide L-K6, a temporin-1CEb analog on oral pathogen growth, Streptococcus mutans biofilm formation, and anti-inflammatory activity. Appl Microbiol Biotechnol. 2014;98(20):8685–8695. doi: 10.1007/s00253-014-5927-9. [DOI] [PubMed] [Google Scholar]
- 43.Shi XM, Zhu XN. Biofilm formation and food safety in food industries. Trends Food Sci Technol. 2009;20(9):407–413. doi: 10.1016/j.tifs.2009.01.054. [DOI] [Google Scholar]
- 44.Ellis DI, Broadhurst D, Goodacre R. Rapid and quantitative detection of the microbial spoilage of beef by Fourier transform infrared spectroscopy and machine learning. Anal Chim Acta. 2004;514(2):193–201. doi: 10.1016/j.aca.2004.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aspri M, O’Connor PM, Field D, Cotter PD, Ross P, Hill C, Papademas P. Application of bacteriocin-producing Enterococcus faecium isolated from donkey milk, in the bio-control of Listeria monocytogenes in fresh whey cheese. Int Dairy J. 2017;73:1–9. doi: 10.1016/j.idairyj.2017.04.008. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 1.42 mb)